Biochemical Atypia in Russian Neisseria gonorrhoeae Clinical Isolates Belonging to the G807 NG-MAST Genogroup/ST1594 MLST
Abstract
1. Introduction
2. Materials and Methods
2.1. Specimens (Clinical Isolates) and N. gonorrhoeae Genome Information
2.2. Genome Sequencing
2.3. Genome Assembly
2.4. Bioinformatics Analysis
3. Results
3.1. General Characterization of Genomes of Russian Clinical Isolates of G807 NG-MAST GENOGROUP/ST1594 MLST
3.2. Identification of Genetic Aspects of Biochemical Atypia
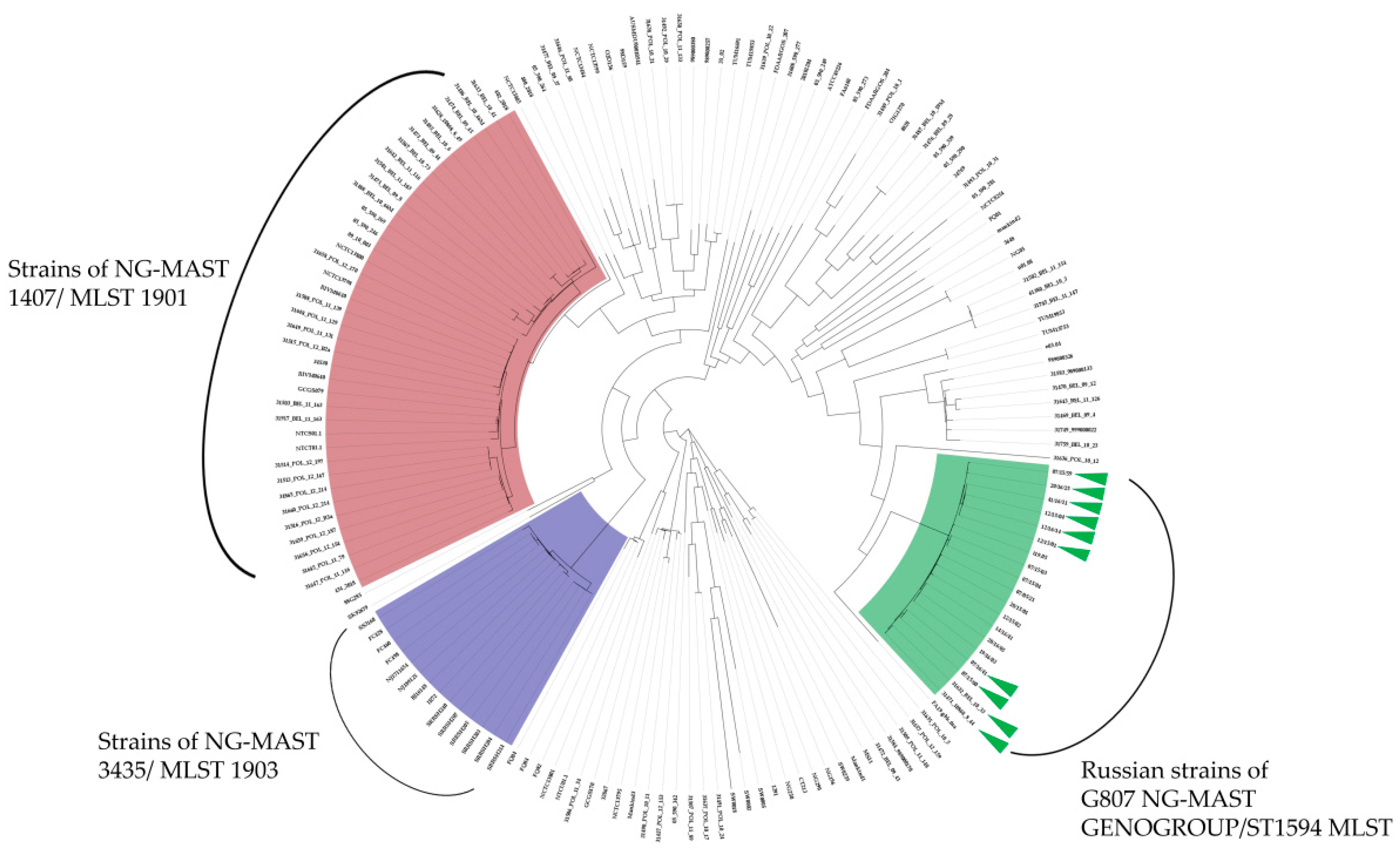
3.3. Phylogenetic Analysis of the G807 NG-MAST GENOGROUP/ST1594 MLST in the Global Population of N. gonorrhoeae
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Unemo, M.; Shafer, W.M. Antimicrobial resistance in Neisseria gonorrhoeae in the 21st century: Past, evolution, and future. Clin. Microbiol. Rev. 2014, 27, 587–613. [Google Scholar] [CrossRef] [PubMed]
- WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. Available online: https://www.who.int/ru/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 4 November 2022).
- Plakhova, X.I.; Petrova, N.P.; Nikonorov, A.A.; Kubanov, A.A. Biochemical atypia in the modern russian strains of Neisseria gonorrhoeae. Klin. Lab. Diagn. (Russ. Clin. Lab. Diagn.) 2020, 65, 507–511. [Google Scholar] [CrossRef] [PubMed]
- Shaskolskiy, B.; Kravtsov, D.; Kandinov, I.; Gorshkova, S.; Kubanov, A.; Solomka, V.; Deryabin, D.; Dementieva, E.; Gryadunov, D. Comparative Whole-Genome Analysis of Neisseria gonorrhoeae Isolates Revealed Changes in the Gonococcal Genetic Island and Specific Genes as a Link to Antimicrobial Resistance. Front. Cell. Infect. Microbiol. 2022, 12, 831336. [Google Scholar] [CrossRef] [PubMed]
- Mlynarczyk-Bonikowska, B.; Kowalewski, C.; Krolak-Ulinska, A.; Marusza, W. Molecular Mechanisms of Drug Resistance and Epidemiology of Multidrug-Resistant Variants of Neisseria gonorrhoeae. Int. J. Mol. Sci. 2022, 23, 10499. [Google Scholar] [CrossRef] [PubMed]
- Martin, I.M.C.; Ison, C.A.; Aanensen, D.M.; Fenton, K.A.; Sprat, B.G. Rapid sequence-based identification of gonococcal transmission clusters in a large metropolitan area. J. Infect. Dis. 2004, 189, 1497–1505. [Google Scholar] [CrossRef] [PubMed]
- Jolley, K.A. Multi-locus sequence typing. In Meningococcal Diseases: Methods and Protocols; Humana Press: Totowa, NJ, USA, 2001; pp. 173–186. [Google Scholar]
- Ilina, E.N.; Malakhova, M.V.; Bodoev, I.N.; Oparina, N.Y.; Filimonova, A.V.; Govorun, V.M. Mutation in ribosomal protein S5 leads to spectinomycin resistance in Neisseria gonorrhoeae. Front. Microbiol. 2013, 4, 186. [Google Scholar] [CrossRef] [PubMed]
- Unemo, M.; Golparian, D.; Skogen, V.; Olsen, A.O.; Moi, H.; Syversen, G.; Hjelmevoll, S.O. Neisseria gonorrhoeae strain with high-level resistance to spectinomycin due to a novel resistance mechanism (mutated ribosomal protein S5) verified in Norway. Antimicrob. Agents Chemother. 2013, 57, 1057–1061. [Google Scholar] [CrossRef]
- Morse, S.; Stein, S.; Hines, J. Glucose metabolism in Neisseria gonorrhoeae. J. Bacteriol. 1974, 120, 702–714. [Google Scholar] [CrossRef]
- Eyre, D.W.; Town, K.; Street, T.; Barker, L.; Sanderson, N.; Cole, M.J.; Mohammed, H.; Pitt, R.; Gobin, M.; Irish, C.; et al. Detection in the United Kingdom of the Neisseria gonorrhoeae FC428 clone, with ceftriaxone resistance and intermediate resistance to azithromycin, October to December 2018. Eurosurveillance 2019, 24, 1900147. [Google Scholar] [CrossRef]
- Fedarovich, A.; Cook, E.; Tomberg, J.; Nicholas, R.A.; Davies, C. Structural effect of the Asp345a insertion in penicillin-binding protein 2 from penicillin-resistant strains of Neisseria gonorrhoeae. Biochemistry 2014, 53, 7596–7603. [Google Scholar] [CrossRef]
- Town, K.; Harris, S.; Sánchez-Busó, L.; Cole, M.J.; Pitt, R.; Fifer, H.; Mohammed, H.; Field, N.; Hughes, G. Genomic and Phenotypic Variability in Neisseria gonorrhoeae Antimicrobial Susceptibility, England. Emerg. Infect. Dis. 2020, 26, 505–515. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.T.; Seifert, H.S. Phase variation leads to the misidentification of a Neisseria gonorrhoeae virulence gene. PLoS ONE 2013, 8, e72183. [Google Scholar] [CrossRef] [PubMed]
- Juni, E.; Heym, G.A. Studies of some naturally occurring auxotrophs of Neisseria gonorrhoeae. J. Gen. Microbiol. 1980, 121, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Blackmore, T.; Hererra, G.; Shi, S.; Bridgewater, P.; Wheeler, L.; Byrne, J. Characterization of prolyl iminopeptidase-deficient Neisseria gonorrhoeae. J. Clin. Microbiol. 2005, 43, 4189–4190. [Google Scholar] [CrossRef] [PubMed]
- Harrison, O.B.; Cehovin, A.; Skett, J.; Jolley, K.A.; Massari, P.; Genco, C.A.; Tang, C.M.; Maiden, M.C.J. Neisseria gonorrhoeae Population Genomics: Use of the Gonococcal Core Genome to Improve Surveillance of Antimicrobial Resistance. J. Infect. Dis. 2020, 222, 1816–1825. [Google Scholar] [CrossRef] [PubMed]
- Johansson, E.; Fredlund, H.; Unemo, M. Prevalence, phenotypic and genetic characteristics of prolyliminopeptidase-negative Neisseria gonorrhoeae isolates in Sweden during 2000–2007. APMIS 2009, 117, 900–904. [Google Scholar] [CrossRef]
- Unemo, M.; Palmer, H.M.; Blackmore, T.; Herrera, G.; Fredlund, H.; Limnios, A.; Nguyen, N.; Tapsall, J. Global transmission of prolyliminopeptidase-negative Neisseria gonorrhoeae strains: Implications for changes in diagnostic strategies. Sex. Transm. Infect. 2007, 83, 47–51. [Google Scholar] [CrossRef]
- Murray, G.G.R.; Charlesworth, J.; Miller, E.L.; Casey, M.J.; Lloyd, C.T.; Gottschalk, M.; Tucker, A.W.D.; Welch, J.J.; Weinert, L.A. Genome Reduction Is Associated with Bacterial Pathogenicity across Different Scales of Temporal and Ecological Divergence. Mol. Biol. Evol. 2021, 38, 1570–1579. [Google Scholar] [CrossRef]
- Buchanan, R.; Ball, D.; Dolphin, H.; Dave, J. Matrix-assisted laser desorption-ionization time-of-flight mass spectrometry for the identification of Neisseria gonorrhoeae. Clin. Microbiol. Infect. 2016, 22, 815.e5–815.e7. [Google Scholar] [CrossRef]
- Carannante, A.; De Carolis, E.; Vacca, P.; Vella, A.; Vocale, C.; De Francesco, M.A.; Cusini, M.; Del Re, S.; Dal Conte, I.; Cristaudo, A.; et al. Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) for identification and clustering of Neisseria gonorrhoeae. BMC Microbiol. 2015, 15, 142. [Google Scholar] [CrossRef]
- Pleininger, S.; Indra, A.; Golparian, D.; Heger, F.; Schindler, S.; Jacobsson, S.; Heidler, S.; Unemo, M. Extensively drug-resistant (XDR) Neisseria gonorrhoeae causing possible gonorrhoea treatment failure with ceftriaxone plus azithromycin in Austria, April 2022. Eurosurveillance 2022, 27, 2200455. [Google Scholar] [CrossRef] [PubMed]
- Anselmo, A.; Ciammaruconi, A.; Carannante, A.; Neri, A.; Fazio, C.; Fortunato, A.; Palozzi, A.M.; Vacca, P.; Fillo, S.; Lista, F.; et al. Draft Genome Sequence of Neisseria gonorrhoeae Sequence Type 1407, a Multidrug-Resistant Clinical Isolate. Genome Announc. 2015, 3, e00903-15. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Nakayama, S.I.; Osawa, K.; Yoshida, H.; Arakawa, S.; Furubayashi, K.I.; Kameoka, H.; Shimuta, K.; Kawahata, T.; Unemo, M.; et al. Clonal expansion and spread of the ceftriaxone-resistant Neisseria gonorrhoeae strain FC428, identified in Japan in 2015, and closely related isolates. J. Antimicrob. Chemother. 2019, 74, 1812–1819. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.; Li, Y.; Xiu, L.; Zhang, C.; Fu, Y.; Jiang, C.; Tang, L.; Peng, J. Identification of multidrug-resistant Neisseria gonorrhoeae isolates with combined resistance to both ceftriaxone and azithromycin, China, 2017–2018. Emerg. Microbes Infect. 2019, 8, 1546–1549. [Google Scholar] [CrossRef]
- Kenyon, C.; Laumen, J.; Van Dijck, C.; De Baetselier, I.; Abdelatti, S.; Manoharan-Basil, S.S.; Unemo, M. Gonorrhoea treatment combined with population-level general cephalosporin and quinolone consumption may select for Neisseria gonorrhoeae antimicrobial resistance at the levels of NG-MAST genogroup: An ecological study in Europe. J. Glob. Antimicrob. Resist. 2020, 23, 377–384. [Google Scholar] [CrossRef]
- Golparian, D.; Sánchez-Busó, L.; Cole, M.; Unemo, M. Neisseria gonorrhoeae Sequence Typing for Antimicrobial Resistance (NG-STAR) clonal complexes are consistent with genomic phylogeny and provide simple nomenclature, rapid visualization and antimicrobial resistance (AMR) lineage predictions. J. Antimicrob. Chemother 2021, 76, 940–944. [Google Scholar] [CrossRef]


| Number in the SSCDC * Collection | Source of Obtaining (Isolation Site, Sex, Age, Region) | Biochemical Activity Test Results | NG-MAST and MLST Type |
|---|---|---|---|
| 07/15/03 | ECS **; W, 25, Arkhangelsk region | TBA ***; 99% Neisseria gonorrhoeae | 807/1594 |
| 07/15/04 | US; M, 18, Arkhangelsk region | TBA, 99% Neisseria gonorrhoeae | 12,529 (tbpB C → T in the position 381) |
| 07/15/59 | US; M, 34, Arkhangelsk region | ABA (defective amino acids and glucose metabolism); 94% Neisseria cinerea **** | 1531 (porB G → A in the position 127) |
| 07/15/60 | US; M, 26, Arkhangelsk region | ABA (defective amino acids and glucose metabolism); 95% Moraxella catharrhalis **** | 807 |
| 07/16/41 | US; M, 22, Arkhangelsk region | ABA (defective amino acids and glucose metabolism); 97% Moraxella catharrhalis **** | 807 |
| 12/15/01 | US; M, 29, Chuvash Republic | ABA (defective amino acids metabolism); Low discrimination Moraxella catharrhalis/Neisseria gonorrhoeae **** | 5941 (porB A → G in the position 341) |
| 12/15/02 | US; M, 26, Chuvash Republic | TBA, 99% Neisseria gonorrhoeae | 807 |
| 12/15/04 | US; M, 43, Chuvash Republic | ABA (defective amino acids and glucose metabolism); Low discrimination Moraxella catharrhalis/Neisseria cinerea **** | 807 |
| 12/16/14 | US; M, 25, Chuvash Republic | ABA (defective glucose metabolism); 96% Neisseria gonorrhoeae | 807 |
| 14/16/41 | US; M, 41, Arkhangelsk region | TBA, 99% Neisseria gonorrhoeae | 807 |
| 19/16/03 | ECS; W, 23, Omsk region | TBA, 99% Neisseria gonorrhoeae | 807 |
| 20/16/05 | US; W, 31, Kaluga Region | TBA, 99% Neisseria gonorrhoeae | 807 |
| 20/16/25 | US; M, 52, Kaluga Region | ABA (defective amino acids metabolism); 92% Neisseria gonorrhoeae | 9576 (porB T → G in the position 349) |
| 28/15/01 | US; M, 25, Penza region | TBA, 99% Neisseria gonorrhoeae | 9570 (porB G → A in the position 340) |
| 41/16/11 | US; M, 30, Tomsk region | ABA (defective amino acids metabolism); Low discrimination Moraxella catharrhalis/Neisseria gonorrhoeae//Neisseria meningitidis **** | 807 |
| 07/05/21 | Arkhangelsk region | TBA, 99% Neisseria gonorrhoeae | 807 |
| Sample | Contigs | Length of Contigs. mb | N50 of Contigs | GC of Contigs (%) | Genes | CDS | ncRNA | rRNA | tRNA | tmRNA | Accession |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 07/15/03 | 135 | 2.3 | 43.5 | 52.5 | 2165 | 2095 | 16 | 3 | 50 | 1 | JANQDD000000000 |
| 07/15/04 | 99 | 2.3 | 53.1 | 52.5 | 2210 | 2140 | 16 | 3 | 50 | 1 | JANUFX000000000 |
| 07/15/59 | 93 | 2.3 | 67.6 | 52.5 | 2208 | 2141 | 16 | 3 | 47 | 1 | JANUFY000000000 |
| 07/15/60 | 110 | 2.3 | 48.5 | 52.5 | 2211 | 2140 | 17 | 3 | 50 | 1 | JANUFZ000000000 |
| 07/16/41 | 124 | 2.3 | 48.3 | 52.5 | 2210 | 2139 | 17 | 3 | 50 | 1 | JANUGA000000000 |
| 12/15/01 | 98 | 2.2 | 53.3 | 52.5 | 2159 | 2090 | 15 | 3 | 50 | 1 | JANUGB000000000 |
| 12/15/02 | 96 | 2.2 | 55.3 | 52.5 | 2164 | 2095 | 15 | 3 | 50 | 1 | JANUGC000000000 |
| 12/15/04 | 77 | 2.2 | 67.2 | 52.5 | 2163 | 2094 | 16 | 3 | 49 | 1 | JANUGD000000000 |
| 12/16/14 | 122 | 2.3 | 52.4 | 52.4 | 2162 | 2093 | 16 | 3 | 49 | 1 | JANUGE000000000 |
| 14/16/41 | 113 | 2.2 | 53.6 | 52.4 | 2160 | 2091 | 15 | 3 | 50 | 1 | JANUGF000000000 |
| 19/16/03 | 99 | 2.2 | 59.0 | 52.5 | 2164 | 2095 | 15 | 3 | 50 | 1 | JANUGG000000000 |
| 20/16/05 | 116 | 2.2 | 53.2 | 52.5 | 2169 | 2100 | 15 | 3 | 50 | 1 | JANUGH000000000 |
| 20/16/25 | 123 | 2.3 | 50.1 | 52.4 | 2167 | 2097 | 16 | 3 | 50 | 1 | JANUGI000000000 |
| 28/15/01 | 111 | 2.2 | 53.4 | 52.4 | 2165 | 2096 | 15 | 3 | 50 | 1 | JANUGJ000000000 |
| 41/16/11 | 97 | 2.2 | 48.7 | 52.5 | 2157 | 2088 | 15 | 3 | 50 | 1 | JANUGK000000000 |
| 07/05/21 | 116 | 2.3 | 48.3 | 52.4 | 2166 | 2097 | 15 | 3 | 50 | 1 | JANUGL000000000 |
| FA19 | 1 | 2.2 | - | 52.4 | 2332 | 2261 | 4 | 3 | 55 | N/A | NZ_CP012026 |
| Gene | Product | SNP Position in the FA19 Genome | Type of Substitution | Synonymity of Substitution |
|---|---|---|---|---|
| DapC | succinyldiaminopimelate transaminase | 193,614 | G → A | Nonsyn (Glu→Lys) |
| VT05_RS07040 | galactose mutarotase—pseudogen (frameshifted) | 1,331,807 | C → T | - |
| rpsE | 30S ribosomal protein S5 | 1,607,962 | G → A | Nonsyn (Ala→Ser) |
| VT05_RS09720 | Na+/H+ antiporter family protein | 1,800,365 | C → T | Syn |
| nth | endonuclease III | 2,119,191 | C → T | Nonsyn/STOP codon |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nosov, N.; Kubanov, A.; Solomka, V.; Deryabin, D. Biochemical Atypia in Russian Neisseria gonorrhoeae Clinical Isolates Belonging to the G807 NG-MAST Genogroup/ST1594 MLST. Microorganisms 2022, 10, 2271. https://doi.org/10.3390/microorganisms10112271
Nosov N, Kubanov A, Solomka V, Deryabin D. Biochemical Atypia in Russian Neisseria gonorrhoeae Clinical Isolates Belonging to the G807 NG-MAST Genogroup/ST1594 MLST. Microorganisms. 2022; 10(11):2271. https://doi.org/10.3390/microorganisms10112271
Chicago/Turabian StyleNosov, Nikita, Alexey Kubanov, Viktoria Solomka, and Dmitry Deryabin. 2022. "Biochemical Atypia in Russian Neisseria gonorrhoeae Clinical Isolates Belonging to the G807 NG-MAST Genogroup/ST1594 MLST" Microorganisms 10, no. 11: 2271. https://doi.org/10.3390/microorganisms10112271
APA StyleNosov, N., Kubanov, A., Solomka, V., & Deryabin, D. (2022). Biochemical Atypia in Russian Neisseria gonorrhoeae Clinical Isolates Belonging to the G807 NG-MAST Genogroup/ST1594 MLST. Microorganisms, 10(11), 2271. https://doi.org/10.3390/microorganisms10112271






