Metabolic Characteristics of Porcine LA-MRSA CC398 and CC9 Isolates from Germany and China via Biolog Phenotype MicroArrayTM
Abstract
1. Introduction
2. Materials and Methods
2.1. Selection of Representative Bacterial Isolates
2.2. Whole-Genome Sequencing, Assembly and Annotation
2.3. Molecular and Phylogenetic Analyses
2.4. Biolog PM Assay Performance and Data Evaluation
3. Results and Discussion
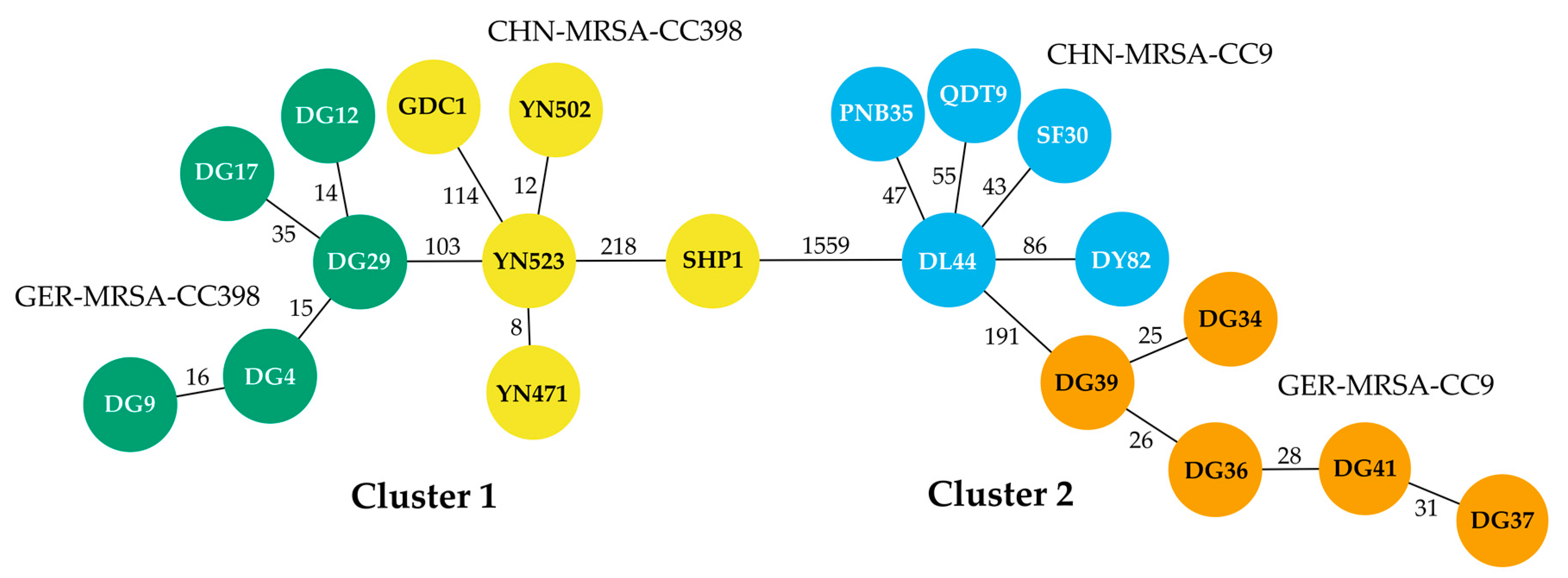
3.1. Molecular and Phylogenetic Analyses
3.2. Biolog PM Assay Analysis
3.2.1. General Metabolic Properties of Porcine LA-MRSA from Germany and China
3.2.2. Statistical Evaluation of the Biolog PM Assay Data
3.2.2.1. AUC sPLS-DA
3.2.2.2. Combined LT, MH, PT, and Slope sPLS-DA Variable Selection
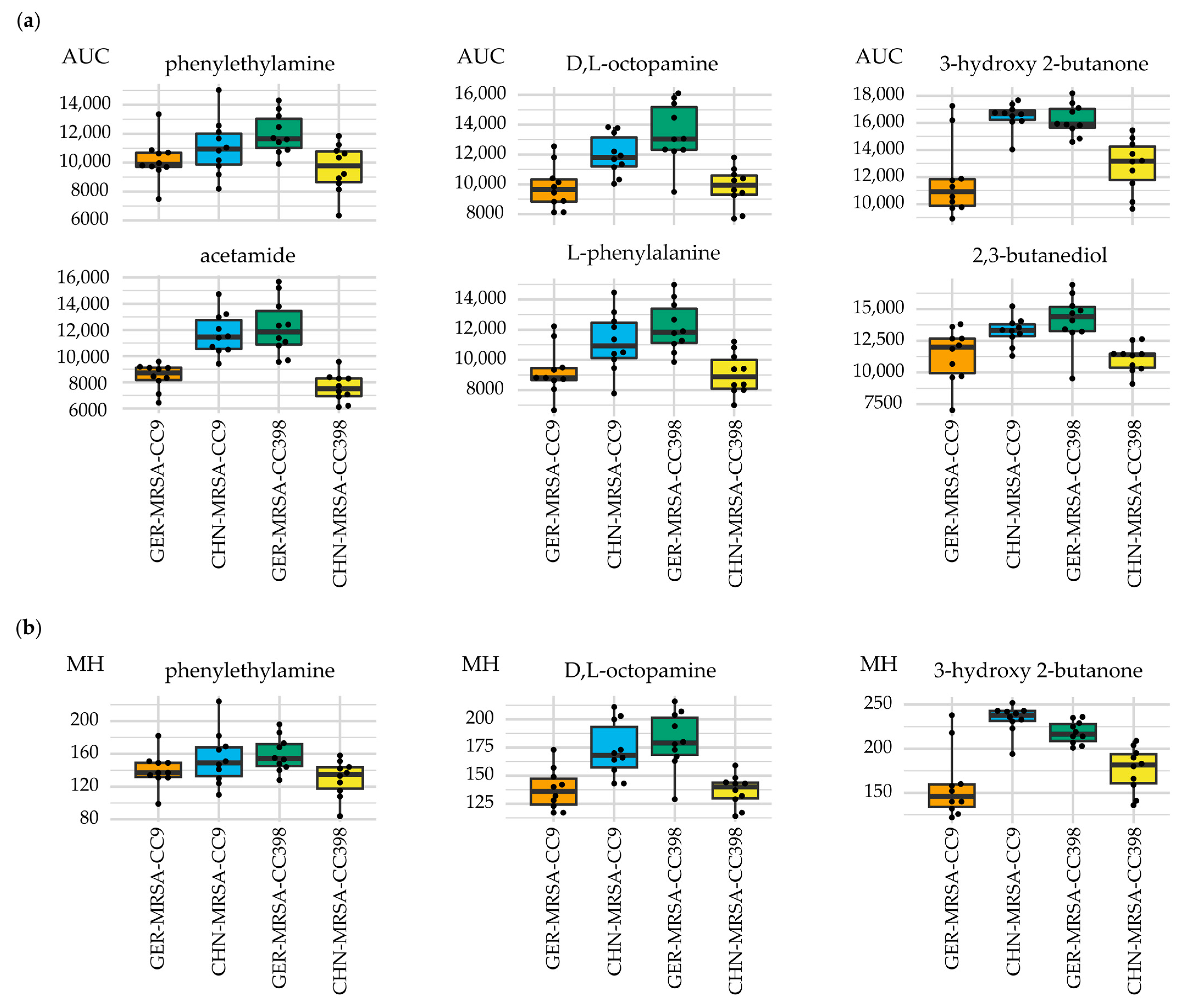
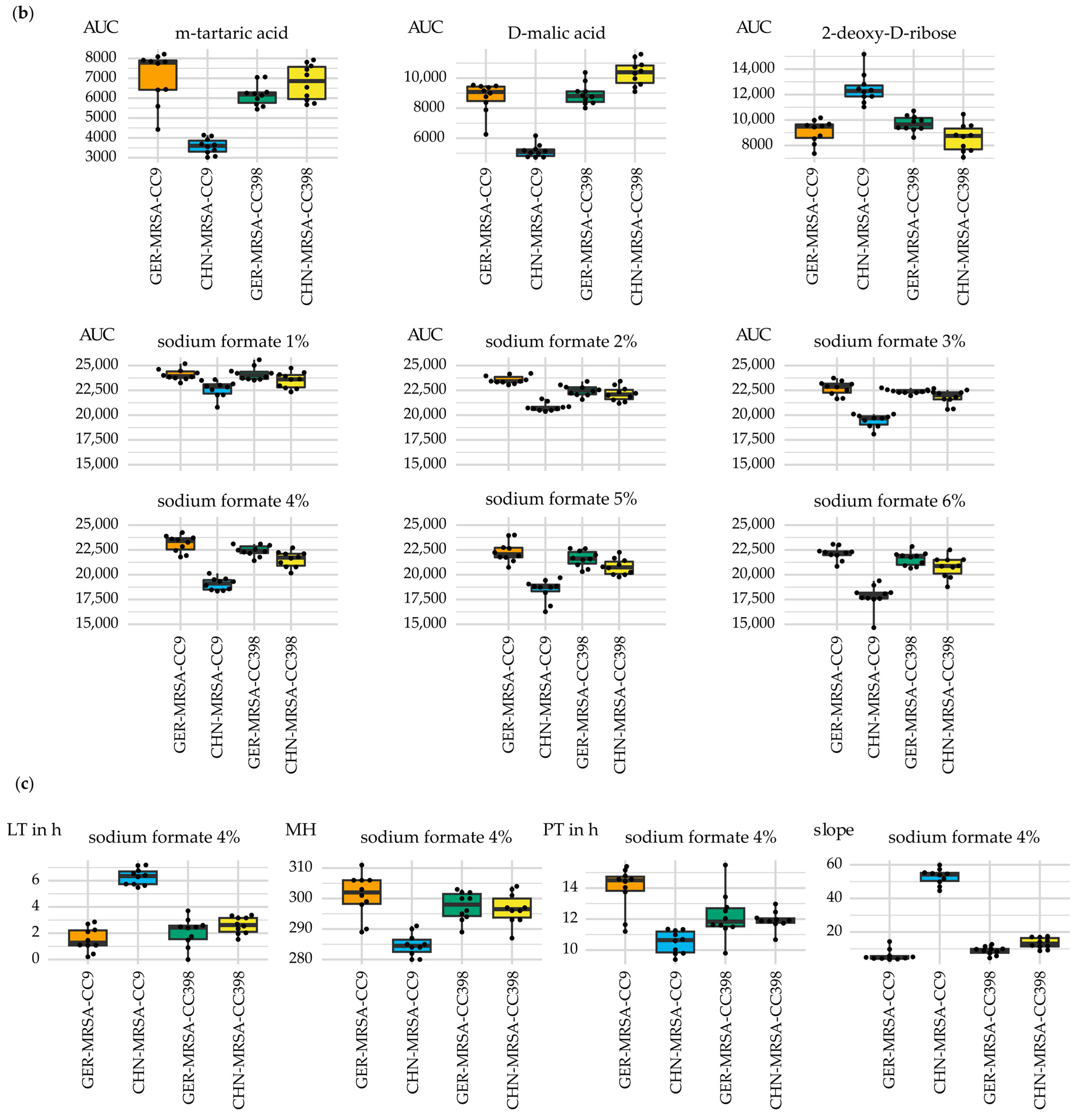
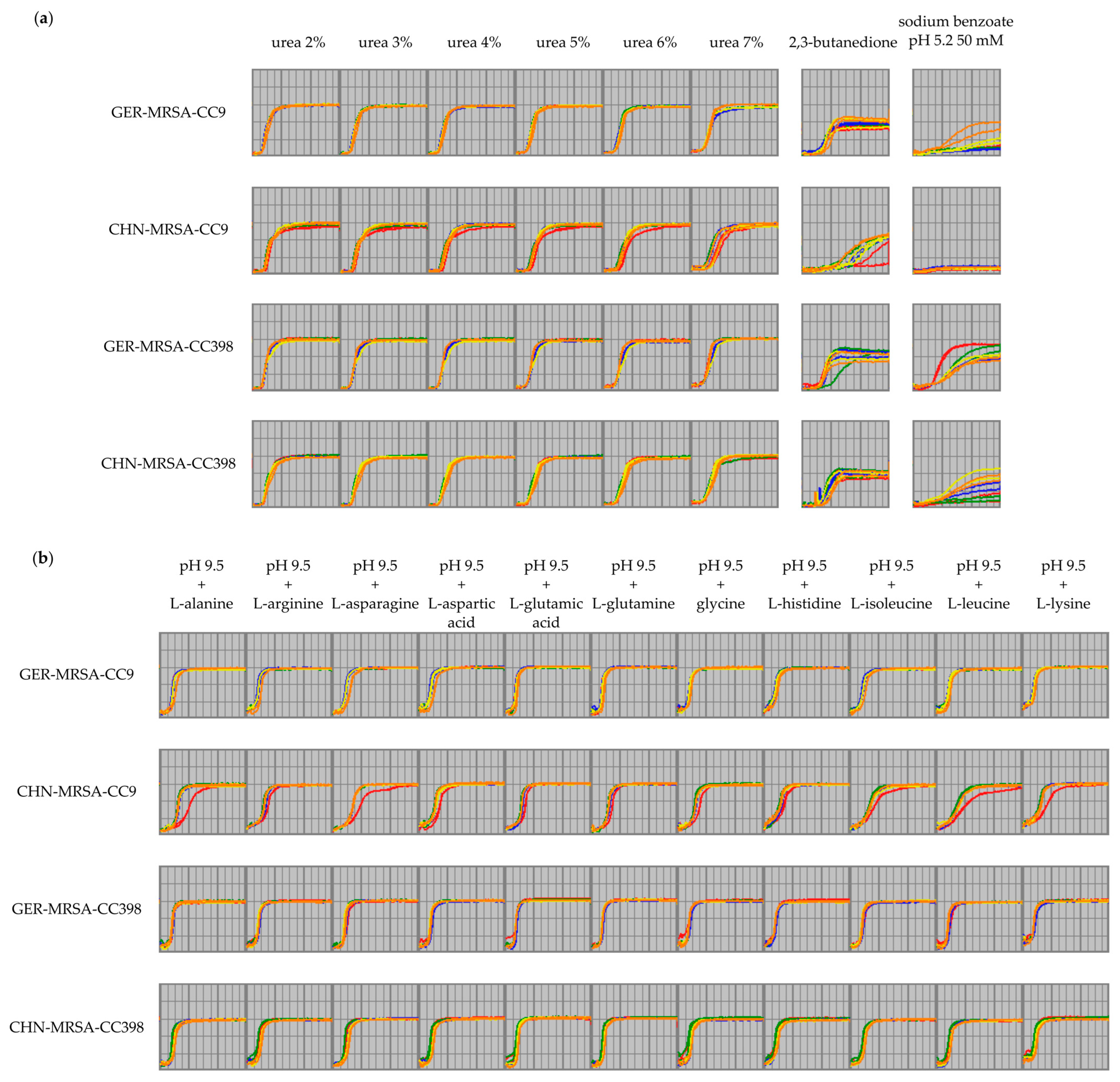
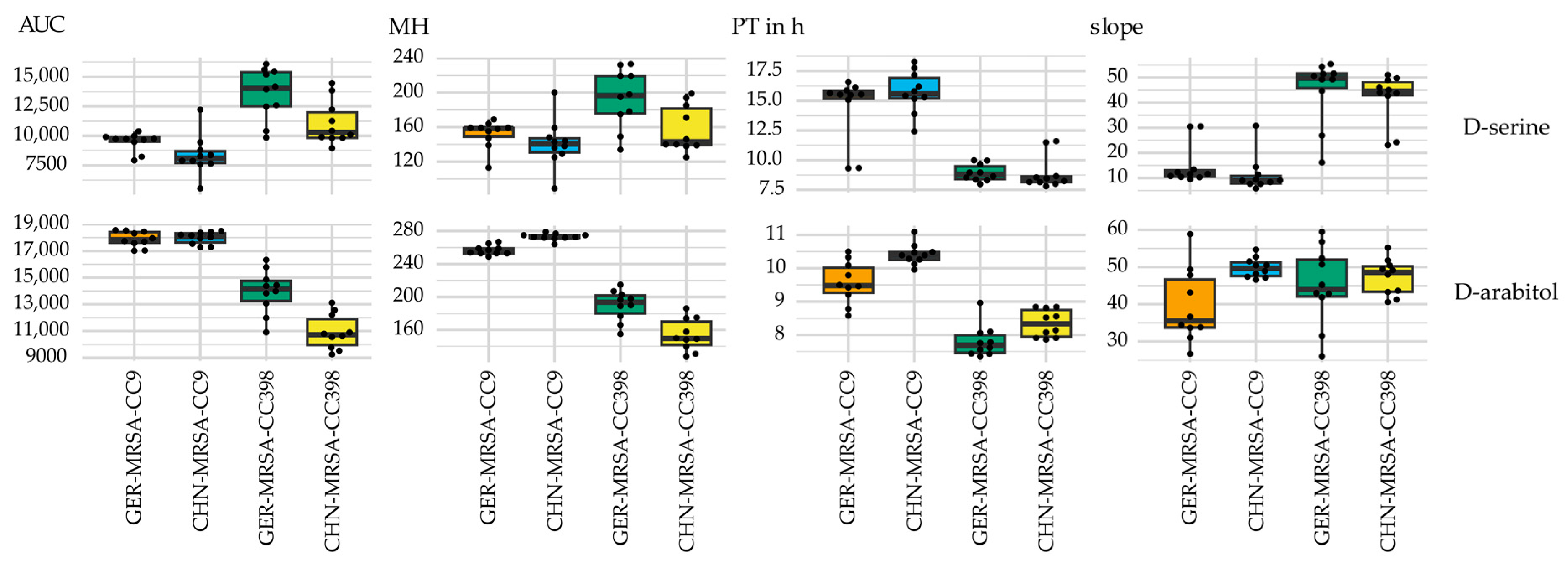
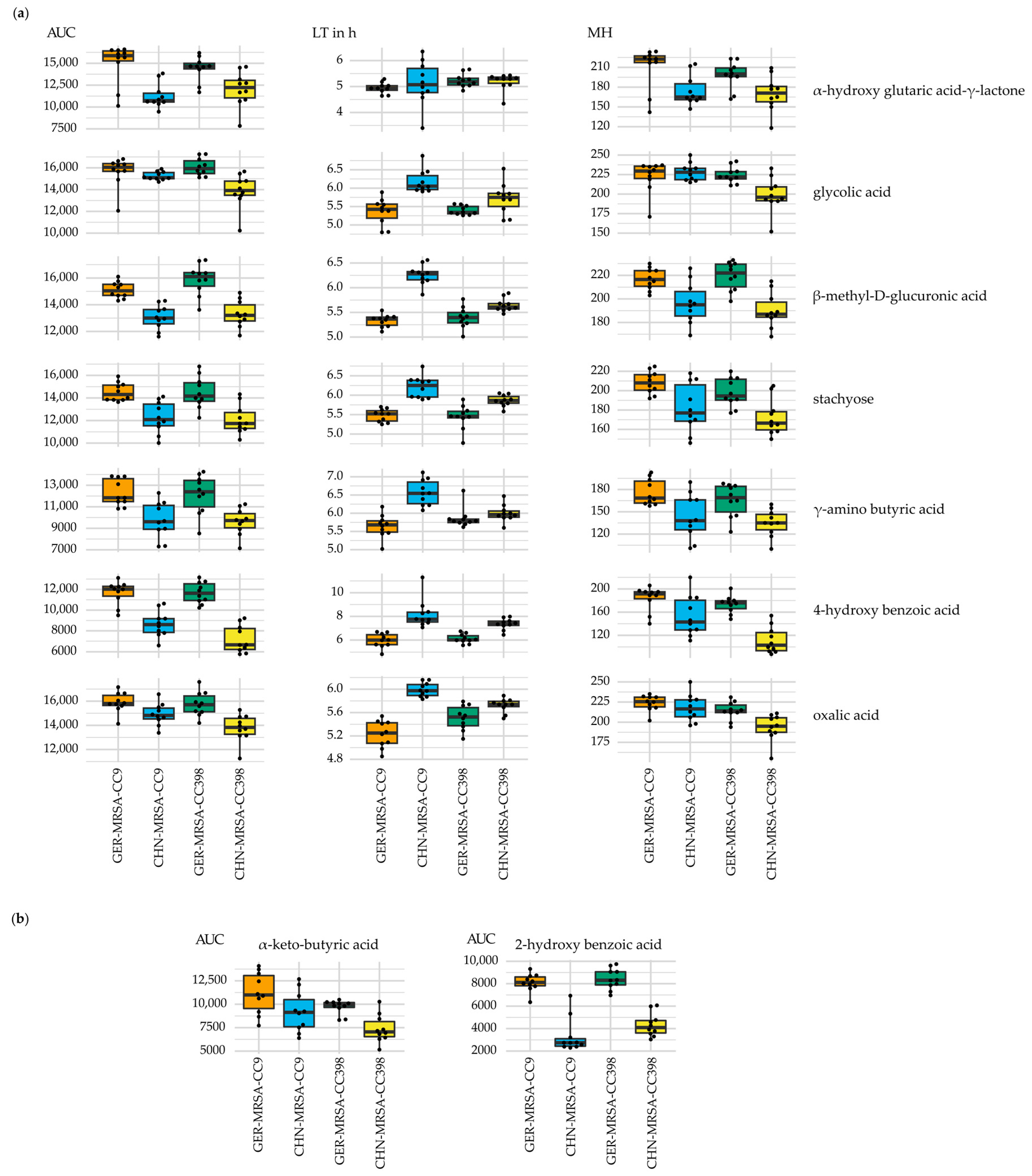
3.2.3. Relevant Metabolic Varieties between the Four MRSA Groups
3.2.3.1. Metabolic Differences between the Dominant and Rare Lineage within a CC
3.2.3.2. Distinctive Metabolic Features of CHN-MRSA-CC9
3.2.3.3. Metabolic Diversity Based on the Assignment to a CC
3.2.3.4. Metabolic Diversity Attributed to the Country of Origin
3.2.3.5. Other Metabolic Differences between the Groups
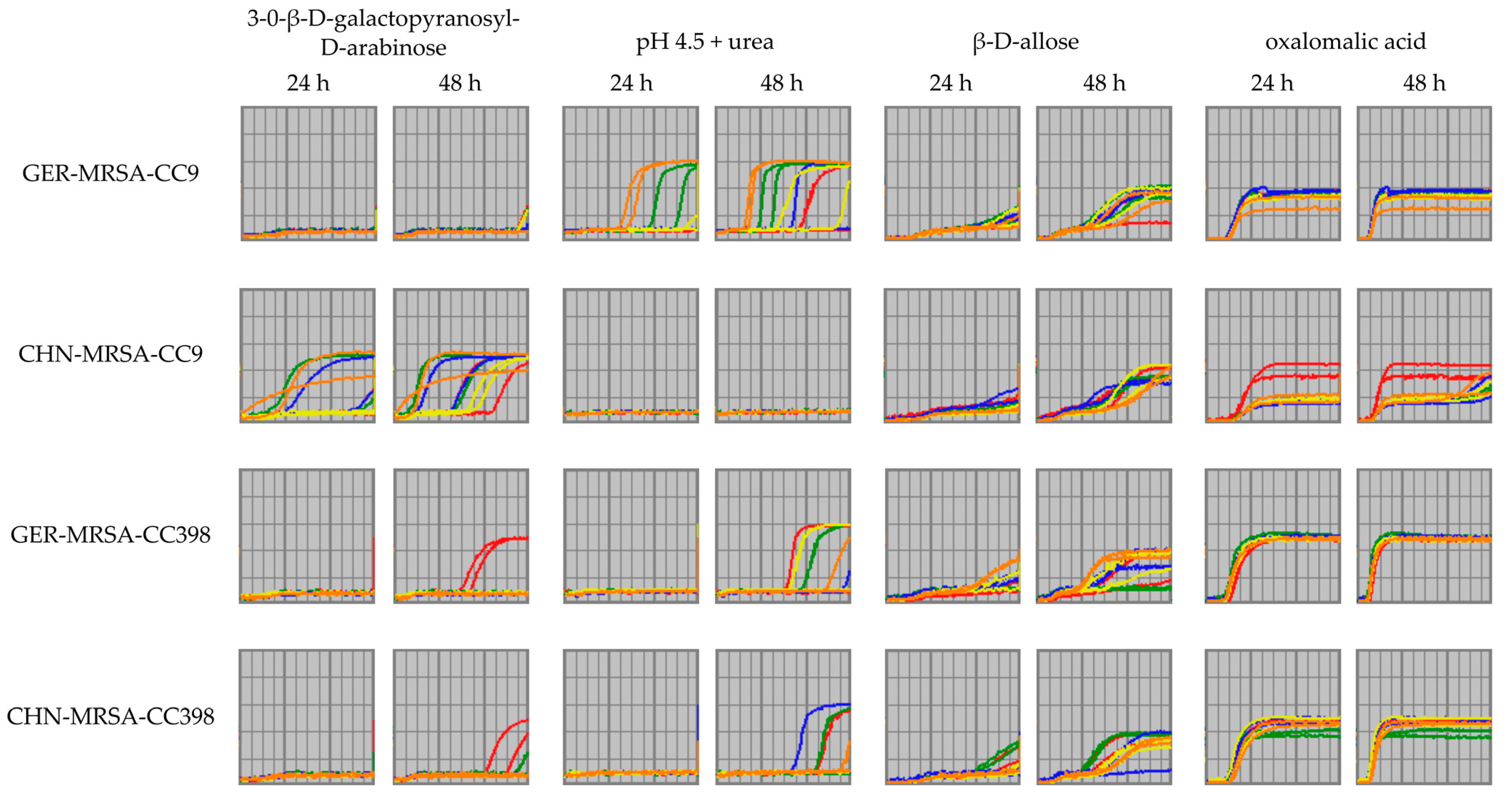
3.2.3.6. Metabolic Variations Detectable Only after a 48 h Incubation Period
3.3. Potential Limitations of the Study
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Graveland, H.; Duim, B.; van Duijkeren, E.; Heederik, D.; Wagenaar, J.A. Livestock-associated methicillin-resistant Staphylococcus aureus in animals and humans. Int. J. Med Microbiol. 2011, 301, 630–634. [Google Scholar] [CrossRef] [PubMed]
- Voss, A.; Loeffen, F.; Bakker, J.; Klaassen, C.; Wulf, M. Methicillin-resistant Staphylococcus aureus in pig farming. Emerg. Infect. Dis. 2005, 11, 1965–1966. [Google Scholar] [CrossRef]
- Armand-Lefevre, L.; Ruimy, R.; Andremont, A. Clonal comparison of Staphylococcus aureus isolates from healthy pig farmers, human controls, and pigs. Emerg. Infect. Dis. 2005, 11, 711–714. [Google Scholar] [CrossRef] [PubMed]
- Vanderhaeghen, W.; Hermans, K.; Haesebrouck, F.; Butaye, P. Methicillin-resistant Staphylococcus aureus (MRSA) in food production animals. Epidemiol. Infect. 2010, 138, 606–625. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.C.; Pearson, N. The Emergence of Staphylococcus aureus ST398. Vector Borne Zoonotic Dis. 2011, 11, 327–339. [Google Scholar] [CrossRef] [PubMed]
- Cuny, C.; Köck, R.; Witte, W. Livestock associated MRSA (LA-MRSA) and its relevance for humans in Germany. Int. J. Med Microbiol. 2013, 303, 331–337. [Google Scholar] [CrossRef]
- Dong, Q.; Liu, Y.; Li, W.; Liu, Y.; Ye, X. Cross-species transmission risk of livestock-associated MRSA: A systematic review and Bayesian meta-analysis of global data. Prev. Vet. Med. 2021, 194, 105429. [Google Scholar] [CrossRef]
- Kadlec, K.; Ehricht, R.; Monecke, S.; Steinacker, U.; Kaspar, H.; Mankertz, J.; Schwarz, S. Diversity of antimicrobial resistance pheno- and genotypes of methicillin-resistant Staphylococcus aureus ST398 from diseased swine. J. Antimicrob. Chemother. 2009, 64, 1156–1164. [Google Scholar] [CrossRef]
- Monecke, S.; Coombs, G.; Shore, A.C.; Coleman, D.C.; Akpaka, P.; Borg, M.; Chow, H.; Ip, M.; Jatzwauk, L.; Jonas, D.; et al. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS ONE 2011, 6, e17936. [Google Scholar] [CrossRef]
- Price, L.B.; Stegger, M.; Hasman, H.; Aziz, M.; Larsen, J.; Andersen, P.S.; Pearson, T.; Waters, A.E.; Foster, J.T.; Schupp, J.; et al. Staphylococcus aureus CC398: Host adaptation and emergence of methicillin resistance in livestock. mBio 2012, 3, e00305–e00311. [Google Scholar] [CrossRef]
- Guardabassi, L.; O’Donoghue, M.; Moodley, A.; Ho, J.; Boost, M. Novel lineage of methicillin-resistant Staphylococcus aureus, Hong Kong. Emerg. Infect. Dis. 2009, 15, 1998–2000. [Google Scholar] [CrossRef] [PubMed]
- Wagenaar, J.A.; Yue, H.; Pritchard, J.; Broekhuizen-Stins, M.; Huijsdens, X.; Mevius, D.J.; Bosch, T.; Van Duijkeren, E. Unexpected sequence types in livestock associated methicillin-resistant Staphylococcus aureus (MRSA): MRSA ST9 and a single locus variant of ST9 in pig farming in China. Vet. Microbiol. 2009, 139, 405–409. [Google Scholar] [CrossRef] [PubMed]
- Chuang, Y.-Y.; Huang, Y.-C. Livestock-associated meticillin-resistant Staphylococcus aureus in Asia: An emerging issue? Int. J. Antimicrob. Agents 2015, 45, 334–340. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Liu, J.-H.; Zhang, X.-F.; Wang, J.; Ma, Z.-B.; Chen, L.; Zeng, Z.-L. Emergence of methicillin-resistant Staphylococcus aureus ST398 in pigs in China. Int. J. Antimicrob. Agents 2018, 51, 275–276. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.; Li, J.; Ali, T.; Kalim, K.; Wang, H.; Song, L.; Li, Z.; Ren, X.; Ma, F.; Zou, M.; et al. Emergence of livestock-associated MRSA ST398 from bulk tank milk, China. J. Antimicrob. Chemother. 2020, 75, 3471–3474. [Google Scholar] [CrossRef]
- Cui, M.; Ali, T.; Li, J.; Song, L.; Shen, S.; Li, T.; Zhang, C.; Cheng, M.; Zhao, Q.; Wang, H. New clues about the global MRSA ST398: Emergence of MRSA ST398 from pigs in Qinghai, China. Int. J. Food Microbiol. 2022, 378, 109820. [Google Scholar] [CrossRef]
- Li, X.; Xie, L.; Huang, H.; Li, Z.; Li, G.; Liu, P.; Xiao, D.; Zhang, X.; Xiong, W.; Zeng, Z. Prevalence of Livestock-Associated MRSA ST398 in a Swine Slaughterhouse in Guangzhou, China. Front. Microbiol. 2022, 13, 914764. [Google Scholar] [CrossRef]
- Ballhausen, B.; Kriegeskorte, A.; van Alen, S.; Jung, P.; Kock, R.; Peters, G.; Bischoff, M.; Becker, K. The pathogenicity and host adaptation of livestock-associated MRSA CC398. Vet. Microbiol. 2017, 200, 39–45. [Google Scholar] [CrossRef]
- Li, J.; Jiang, N.; Ke, Y.; Feßler, A.T.; Wang, Y.; Schwarz, S.; Wu, C. Characterization of pig-associated methicillin-resistant Staphylococcus aureus. Vet. Microbiol. 2017, 201, 183–187. [Google Scholar] [CrossRef]
- Monecke, S.; Slickers, P.; Gawlik, D.; Muller, E.; Reissig, A.; Ruppelt-Lorz, A.; de Jackel, S.C.; Feßler, A.T.; Frank, M.; Hotzel, H.; et al. Variability of SCCmec elements in livestock-associated CC398 MRSA. Vet. Microbiol. 2018, 217, 36–46. [Google Scholar] [CrossRef]
- Butaye, P.; Argudín, M.A.; Smith, T.C. Livestock-Associated MRSA and Its Current Evolution. Curr. Clin. Microbiol. Rep. 2016, 3, 19–31. [Google Scholar] [CrossRef]
- Yu, F.; Cienfuegos-Gallet Astrid, V.; Cunningham Marcus, H.; Jin, Y.; Wang, B.; Kreiswirth Barry, N.; Chen, L. Molecular Evolution and Adaptation of Livestock-Associated Methicillin-Resistant Staphylococcus aureus (LA-MRSA) Sequence Type 9. mSystems 2021, 6, e00492-21. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Krüger, H.; Tao, J.; Wang, Y.; Feßler, A.T.; Bai, R.; Wang, S.; Dong, Y.; Shen, J.; Wang, Y.; et al. Comparative analysis of genomic characteristics, fitness and virulence of MRSA ST398 and ST9 isolated from China and Germany. Emerg. Microbes Infect. 2021, 10, 1481–1494. [Google Scholar] [CrossRef] [PubMed]
- Biolog Inc., Hayward, CA, USA. Phenotype MicroArrays for Microbial Cells. Available online: https://www.biolog.com/products-portfolio-overview/phenotype-microarrays-for-microbial-cells/ (accessed on 4 August 2022).
- Mackie, A.M.; Hassan, K.A.; Paulsen, I.T.; Tetu, S.G. Biolog Phenotype Microarrays for phenotypic characterization of microbial cells. Methods Mol. Biol. 2014, 1096, 123–130. [Google Scholar] [CrossRef]
- Biolog Inc., Hayward, CA, USA. Bibliography. Phenotype MicroArrays for Microbial Cells. Available online: https://www.biolog.com/support/bibliography/ (accessed on 4 August 2022).
- von Eiff, C.; McNamara, P.; Becker, K.; Bates, D.; Lei, X.H.; Ziman, M.; Bochner, B.R.; Peters, G.; Proctor, R.A. Phenotype microarray profiling of Staphylococcus aureus menD and hemB mutants with the small-colony-variant phenotype. J. Bacteriol. 2006, 188, 687–693. [Google Scholar] [CrossRef]
- Lucas, A.L.; Manna, A.C. Phenotypic characterization of sarR mutant in Staphylococcus aureus. Microb. Pathog. 2013, 57, 52–61. [Google Scholar] [CrossRef]
- Marchi, E.; Furi, L.; Arioli, S.; Morrissey, I.; Di Lorenzo, V.; Mora, D.; Giovannetti, L.; Oggioni, M.R.; Viti, C. Novel insight into antimicrobial resistance and sensitivity phenotypes associated to qac and norA genotypes in Staphylococcus aureus. Microbiol. Res. 2015, 170, 184–194. [Google Scholar] [CrossRef]
- Vaillant, J.J.; Cunningham, S.A.; Patel, R. Antibiotic susceptibility testing of Staphylococcus aureus using the Biolog OmniLog® system, a metabolic phenotyping assay. Diagn. Microbiol. Infect. Dis. 2022, 104, 115759. [Google Scholar] [CrossRef]
- Truong-Bolduc, Q.C.; Dunman, P.M.; Strahilevitz, J.; Projan, S.J.; Hooper, D.C. MgrA is a multiple regulator of two new efflux pumps in Staphylococcus aureus. J. Bacteriol. 2005, 187, 2395–2405. [Google Scholar] [CrossRef]
- Sreedharan, S.; Oram, M.; Jensen, B.; Peterson, L.R.; Fisher, L.M. DNA gyrase gyrA mutations in ciprofloxacin-resistant strains of Staphylococcus aureus: Close similarity with quinolone resistance mutations in Escherichia coli. J. Bacteriol. 1990, 172, 7260–7262. [Google Scholar] [CrossRef]
- Noguchi, N.; Okihara, T.; Namiki, Y.; Kumaki, Y.; Yamanaka, Y.; Koyama, M.; Wakasugi, K.; Sasatsu, M. Susceptibility and resistance genes to fluoroquinolones in methicillin-resistant Staphylococcus aureus isolated in 2002. Int. J. Antimicrob. Agents 2005, 25, 374–379. [Google Scholar] [CrossRef] [PubMed]
- Hauschild, T.; Feßler, A.T.; Billerbeck, C.; Wendlandt, S.; Kaspar, H.; Mankertz, J.; Schwarz, S.; Kadlec, K. Target gene mutations among methicillin-resistant Staphylococcus aureus and methicillin-susceptible S. aureus with elevated MICs of enrofloxacin obtained from diseased food-producing animals or food of animal origin. J. Antimicrob. Chemother. 2012, 67, 1791–1793. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nurk, S.; Bankevich, A.; Antipov, D.; Gurevich, A.; Korobeynikov, A.; Lapidus, A.; Prjibelsky, A.; Pyshkin, A.; Sirotkin, A.; Sirotkin, Y.; et al. Assembling Genomes and Mini-metagenomes from Highly Chimeric Reads. In Proceedings of the 17th Annual International Conference on Research in Computational Molecular Biology, Beijing, China, 7–10 April 2013; pp. 158–170. [Google Scholar]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genomics 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Inouye, M.; Dashnow, H.; Raven, L.A.; Schultz, M.B.; Pope, B.J.; Tomita, T.; Zobel, J.; Holt, K.E. SRST2: Rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 2014, 6, 90. [Google Scholar] [CrossRef]
- Feldgarden, M.; Brover, V.; Haft, D.H.; Prasad, A.B.; Slotta, D.J.; Tolstoy, I.; Tyson, G.H.; Zhao, S.; Hsu, C.-H.; McDermott, P.F.; et al. Validating the AMRFinder Tool and Resistance Gene Database by Using Antimicrobial Resistance Genotype-Phenotype Correlations in a Collection of Isolates. Antimicrob. Agents Chemother. 2019, 63, e00483-19. [Google Scholar] [CrossRef]
- Zankari, E.; Allesøe, R.; Joensen, K.G.; Cavaco, L.M.; Lund, O.; Aarestrup, F.M. PointFinder: A novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 2017, 72, 2764–2768. [Google Scholar] [CrossRef]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontén, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef]
- Bartels, M.D.; Petersen, A.; Worning, P.; Nielsen, J.B.; Larner-Svensson, H.; Johansen, H.K.; Andersen, L.P.; Jarløv, J.O.; Boye, K.; Larsen, A.R.; et al. Comparing whole-genome sequencing with Sanger sequencing for spa typing of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 2014, 52, 4305–4308. [Google Scholar] [CrossRef]
- SCCmecFinder 1.2. Available online: https://cge.food.dtu.dk/services/SCCmecFinder/ (accessed on 4 August 2022).
- Goering, R.V.; Morrison, D.; Al-Doori, Z.; Edwards, G.F.S.; Gemmell, C.G. Usefulness of mec-associated direct repeat unit (dru) typing in the epidemiological analysis of highly clonal methicillin-resistant Staphylococcus aureus in Scotland. Clin. Microbiol. Infect. 2008, 14, 964–969. [Google Scholar] [CrossRef]
- Jünemann, S.; Sedlazeck, F.J.; Prior, K.; Albersmeier, A.; John, U.; Kalinowski, J.; Mellmann, A.; Goesmann, A.; von Haeseler, A.; Stoye, J.; et al. Updating benchtop sequencing performance comparison. Nat. Biotechnol. 2013, 31, 294–296. [Google Scholar] [CrossRef] [PubMed]
- Leopold, S.R.; Goering, R.V.; Witten, A.; Harmsen, D.; Mellmann, A.; Tang, Y.W. Bacterial Whole-Genome Sequencing Revisited: Portable, Scalable, and Standardized Analysis for Typing and Detection of Virulence and Antibiotic Resistance Genes. J. Clin. Microbiol. 2014, 52, 2365–2370. [Google Scholar] [CrossRef] [PubMed]
- Lê Cao, K.-A.; Boitard, S.; Besse, P. Sparse PLS discriminant analysis: Biologically relevant feature selection and graphical displays for multiclass problems. BMC Bioinform. 2011, 12, 253. [Google Scholar] [CrossRef] [PubMed]
- Rohart, F.; Gautier, B.; Singh, A.; Lê Cao, K.-A. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef]
- Federal Office for Consumer Protection and Food Safety (BVL). BVL-Report 11.2, Zoonosen-Monitoring 2015; BVL: Berlin, Germany, 2016; pp. 28–29. [Google Scholar]
- van Belkum, A.; Melles, D.C.; Peeters, J.K.; van Leeuwen, W.B.; van Duijkeren, E.; Huijsdens, X.W.; Spalburg, E.; de Neeling, A.J.; Verbrugh, H.A.; Dutch Working Party on Surveillance and Research of MRSA-SOM. Methicillin-resistant and -susceptible Staphylococcus aureus sequence type 398 in pigs and humans. Emerg. Infect. Dis. 2008, 14, 479–483. [Google Scholar] [CrossRef]
- Cuny, C.; Layer, F.; Köck, R.; Werner, G.; Witte, W. Methicillin Susceptible Staphylococcus aureus (MSSA) of Clonal Complex CC398, t571 from Infections in Humans Are Still Rare in Germany. PLoS ONE 2013, 8, e83165. [Google Scholar] [CrossRef]
- Vandendriessche, S.; Kadlec, K.; Schwarz, S.; Denis, O. Methicillin-susceptible Staphylococcus aureus ST398-t571 harbouring the macrolide-lincosamide-streptogramin B resistance gene erm(T) in Belgian hospitals. J. Antimicrob. Chemother. 2011, 66, 2455–2459. [Google Scholar] [CrossRef]
- Chen, H.; Liu, Y.; Jiang, X.; Chen, M.; Wang, H. Rapid change of methicillin-resistant Staphylococcus aureus clones in a Chinese tertiary care hospital over a 15-year period. Antimicrob. Agents Chemother. 2010, 54, 1842–1847. [Google Scholar] [CrossRef]
- Jimenez, J.N.; Velez, L.A.; Mediavilla, J.R.; Ocampo, A.M.; Vanegas, J.M.; Rodriguez, E.A.; Kreiswirth, B.N.; Correa, M.M. Livestock-associated methicillin-susceptible Staphylococcus aureus ST398 infection in woman, Colombia. Emerg. Infect. Dis. 2011, 17, 1970–1971. [Google Scholar] [CrossRef]
- van der Mee-Marquet, N.; François, P.; Domelier-Valentin, A.-S.; Coulomb, F.; Decreux, C.; Hombrock-Allet, C.; Lehiani, O.; Neveu, C.; Ratovohery, D.; Schrenzel, J.; et al. Emergence of Unusual Bloodstream Infections Associated with Pig-Borne—Like Staphylococcus aureus ST398 in France. Clin. Infect. Dis. 2011, 52, 152–153. [Google Scholar] [CrossRef][Green Version]
- Bonnet, I.; Millon, B.; Meugnier, H.; Vandenesch, F.; Maurin, M.; Pavese, P.; Boisset, S. High prevalence of spa type t571 among methicillin-susceptible Staphylococcus aureus from bacteremic patients in a French University Hospital. PLoS ONE 2018, 13, e0204977. [Google Scholar] [CrossRef] [PubMed]
- Wardyn, S.E.; Stegger, M.; Price, L.B.; Smith, T.C. Whole-Genome Analysis of Recurrent Staphylococcus aureus t571/ST398 Infection in Farmer, Iowa, USA. Emerg. Infect. Dis. 2018, 24, 153–154. [Google Scholar] [CrossRef] [PubMed]
- Krishnasamy, V.; Otte, J.; Silbergeld, E. Antimicrobial use in Chinese swine and broiler poultry production. Antimicrob. Resist. Infect. Control 2015, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Larsen, J.; Clasen, J.; Hansen, J.E.; Paulander, W.; Petersen, A.; Larsen, A.R.; Frees, D. Copresence of tet (K) and tet (M) in Livestock-Associated Methicillin-Resistant Staphylococcus aureus Clonal Complex 398 Is Associated with Increased Fitness during Exposure to Sublethal Concentrations of Tetracycline. Antimicrob. Agents Chemother. 2016, 60, 4401–4403. [Google Scholar] [CrossRef]
- Cunningham, S.A.; Chia, N.; Jeraldo, P.R.; Quest, D.J.; Johnson, J.A.; Boxrud, D.J.; Taylor, A.J.; Chen, J.; Jenkins, G.D.; Drucker, T.M.; et al. Comparison of Whole-Genome Sequencing Methods for Analysis of Three Methicillin-Resistant Staphylococcus aureus Outbreaks. J. Clin. Microbiol. 2017, 55, 1946–1953. [Google Scholar] [CrossRef] [PubMed]
- Somerville, G.A. Staphylococcus aureus Metabolism and Physiology. In Staphylococcus Genetics and Physiology; Somerville, G.A., Ed.; Caister Academic Press: Norfolk, UK, 2016; pp. 107–117. [Google Scholar]
- Frees, D.; Ingmer, H. Stress Responses in Staphylococcus aureus. In Staphylococcus Genetics and Physiology; Somerville, G.A., Ed.; Caister Academic Press: Norfolk, UK, 2016; pp. 221–248. [Google Scholar]
- Lanciotti, R.; Sinigaglia, M.; Gardini, F.; Vannini, L.; Guerzoni, M.E. Growth/no growth interfaces of Bacillus cereus, Staphylococcus aureus and Salmonella enteritidis in model systems based on water activity, pH, temperature and ethanol concentration. Food Microbiol. 2001, 18, 659–668. [Google Scholar] [CrossRef]
- Iyer, V.; Raut, J.; Dasgupta, A. Impact of pH on growth of Staphylococcus epidermidis and Staphylococcus aureus in vitro. J. Med Microbiol. 2021, 70, 001421. [Google Scholar] [CrossRef]
- Richardson, A.R. Virulence and Metabolism. Microbiol. Spectr. 2019, 7, GPP3-0011-2018. [Google Scholar] [CrossRef]
- Halsey Cortney, R.; Lei, S.; Wax Jacqueline, K.; Lehman Mckenzie, K.; Nuxoll Austin, S.; Steinke, L.; Sadykov, M.; Powers, R.; Fey Paul, D. Amino Acid Catabolism in Staphylococcus aureus and the Function of Carbon Catabolite Repression. mBio 2017, 8, e01434-16. [Google Scholar] [CrossRef]
- Vogwill, T.; MacLean, R.C. The genetic basis of the fitness costs of antimicrobial resistance: A meta-analysis approach. Evol. Appl. 2015, 8, 284–295. [Google Scholar] [CrossRef]
- Jin, Y.; Yu, X.; Chen, Y.; Chen, W.; Shen, P.; Luo, Q.; Zhang, S.; Kong, X.; Zheng, B.; Xiao, Y. Characterization of highly virulent community-associated methicillin-resistant Staphylococcus aureus ST9-SCCmec XII causing bloodstream infection in China. Emerg. Microbes Infect. 2020, 9, 2526–2535. [Google Scholar] [CrossRef] [PubMed]
- Gladstone, G.P. The Nutrition of Staphylococcus aureus; Nitrogen Requirements. Br. J. Exp. Pathol. 1937, 18, 322–333. [Google Scholar]
- Downs, D.M. Understanding microbial metabolism. Annu. Rev. Microbiol. 2006, 60, 533–559. [Google Scholar] [CrossRef] [PubMed]


| ID | Year | MLST | spa | dru | SCCmec | Resistance Phenotype 1 | Resistance Genotype 2 |
|---|---|---|---|---|---|---|---|
| CHN-MRSA-CC9 | |||||||
| DL44 | 2009 | ST9 | t899 | dt12w | XII(9C2) | bla, tet, ery, cli, gen, cip, ffn, tia, tmp | mecA, blaZ (2x), tet(L), erm(C), lnu(B), lsa(E), aacA-aphD, aadD, aadE, spw, cat(pC221), fexA, dfrG 3,4 |
| QDT9 | 2014 | ST9 | t899 | dt12w | XII(9C2) | bla, tet, ery, cli, gen, cip, ffn, tia, tmp | mecA, blaZ (2x), tet(L), erm(C), lnu(B), lsa(E), aacA-aphD, aadD, aadE, spw, fexA, dfrG 3,4 |
| PNB35 | 2009 | ST9 | t899 | dt12w | XII(9C2) | bla, tet, ery, cli, gen, cip, ffn, tia, tmp | mecA, blaZ, tet(L), lnu(B), lsa(E), aadD, aadE, spw, fexA, dfrG 3,4 |
| SF30 | 2013 | ST9 | t899 | dt8av | XII(9C2) | bla, tet, ery, cli, gen, cip, ffn, tia, tmp | mecA, blaZ (2x), tet(L), lnu(B), lsa(E), aadD, aadE, spw, fexA, dfrG 3,4 |
| DY82 | 2014 | ST9 | t1939 | dt10du | XII(9C2) | bla, tet, ery, cli, gen, cip, ffn, tia, tmp | mecA, blaZ, tet(K), tet(L), erm(C), lnu(B), lsa(E), aacA-aphD, aadD, aadE, spw, str, fexA, dfrG 4 |
| CHN-MRSA-CC398 | |||||||
| GDC1 5 | 2016 | ST398 | t034 | dt11ax | Vc(5C2&5) | bla, tet, ery, cli, ffn, tia, tmp | mecA, blaZ, tet(K), tet(M), erm(C), lnu(B), lsa(E), spc, fexA, dfrG, cfr |
| SHP1 5 | 2016 | ST398 | t571 | dt9bw | V(5C2) | bla, tet, ery, cli, gen, cip, tia, tmp | mecA, blaZ (2x), tet(L), tet(M), erm(T), lnu(B), lsa(E), aacA-aphD, aadD, aadE, spw, fexA, dfrG 3 |
| YN471 | 2017 | ST398 | t034 | dt6j | Vc(5C2&5) | bla, tet, ery, cli, tia, tmp | mecA, blaZ, tet(K), tet(M), erm(A), vga(E), spc, dfrG |
| YN523 | 2017 | ST398 | t034 | dt6j | Vc(5C2&5) | bla, tet, ery, cli, tia, tmp | mecA, blaZ, tet(K), tet(M), erm(A), vga(E), spc, dfrG |
| YN502 | 2017 | ST398 | t034 | dt6j | Vc(5C2&5) | bla, tet, ery, cli, tia, tmp | mecA, blaZ, tet(K), tet(M), erm(A), erm(C), vga(E), spc, dfrG |
| GER-MRSA-CC9 | |||||||
| DG34 | 2015 | ST9 | t15199 | dt10a | IV(2B) | bla, cip | mecA, blaZ, str 3,4 |
| DG36 | 2015 | ST9 | t1430 | dt10a | IV(2B) | bla, cip | mecA, blaZ, str 3,4 |
| DG37 | 2015 | ST9 | t1430 | dt10a | IV(2B) | bla, cli, cip | mecA, blaZ, str3,4 |
| DG39 | 2015 | ST9 | t1430 | dt10a | IV(2B) | bla, tet, ery, cli, cip, tmp | mecA, blaZ, tet(L), erm(B), aadD, dfrK3,4 |
| DG41 | 2017 | ST9 | t1430 | dt10a | IV(2B) | bla, cli, cip | mecA, blaZ, str 3,4 |
| GER-MRSA-CC398 | |||||||
| DG4 | 2008 | ST398 | t011 | dt11a | Vc(5C2&5) | bla, tet | mecA, blaZ, tet(K), tet(M), str |
| DG9 | 2008 | ST398 | t011 | dt11a | Vc(5C2&5) | bla, tet, tmp | mecA, blaZ, tet(K), tet(L), tet(M), str, dfrK |
| DG12 | 2008 | ST398 | t011 | dt11a | Vc(5C2&5) | bla, tet, ery, cli, gen, tmp | mecA, blaZ, tet(K), tet(M), erm(C), str, dfrG |
| DG17 | 2008 | ST398 | t011 | dt11a | Vc(5C2&5) | bla, tet, ery, cli, cip | mecA, blaZ, tet(K), tet(M), erm(C), str 3 |
| DG29 | 2004 | ST398 | t011 | dt11a | Vc(5C2&5) | bla, tet, cli, gen | mecA, blaZ, tet(K), tet(M), aacA-aphD, str |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krüger-Haker, H.; Ji, X.; Bartel, A.; Feßler, A.T.; Hanke, D.; Jiang, N.; Tedin, K.; Maurischat, S.; Wang, Y.; Wu, C.; et al. Metabolic Characteristics of Porcine LA-MRSA CC398 and CC9 Isolates from Germany and China via Biolog Phenotype MicroArrayTM. Microorganisms 2022, 10, 2116. https://doi.org/10.3390/microorganisms10112116
Krüger-Haker H, Ji X, Bartel A, Feßler AT, Hanke D, Jiang N, Tedin K, Maurischat S, Wang Y, Wu C, et al. Metabolic Characteristics of Porcine LA-MRSA CC398 and CC9 Isolates from Germany and China via Biolog Phenotype MicroArrayTM. Microorganisms. 2022; 10(11):2116. https://doi.org/10.3390/microorganisms10112116
Chicago/Turabian StyleKrüger-Haker, Henrike, Xing Ji, Alexander Bartel, Andrea T. Feßler, Dennis Hanke, Nansong Jiang, Karsten Tedin, Sven Maurischat, Yang Wang, Congming Wu, and et al. 2022. "Metabolic Characteristics of Porcine LA-MRSA CC398 and CC9 Isolates from Germany and China via Biolog Phenotype MicroArrayTM" Microorganisms 10, no. 11: 2116. https://doi.org/10.3390/microorganisms10112116
APA StyleKrüger-Haker, H., Ji, X., Bartel, A., Feßler, A. T., Hanke, D., Jiang, N., Tedin, K., Maurischat, S., Wang, Y., Wu, C., & Schwarz, S. (2022). Metabolic Characteristics of Porcine LA-MRSA CC398 and CC9 Isolates from Germany and China via Biolog Phenotype MicroArrayTM. Microorganisms, 10(11), 2116. https://doi.org/10.3390/microorganisms10112116







