Super Shedding in Enteric Pathogens: A Review
Abstract
1. Introduction
2. What Is a Super Shedder?
2.1. Super Shedding and Infectiousness
2.2. Super Shedders as a Distinct Category: The 80/20 Rule
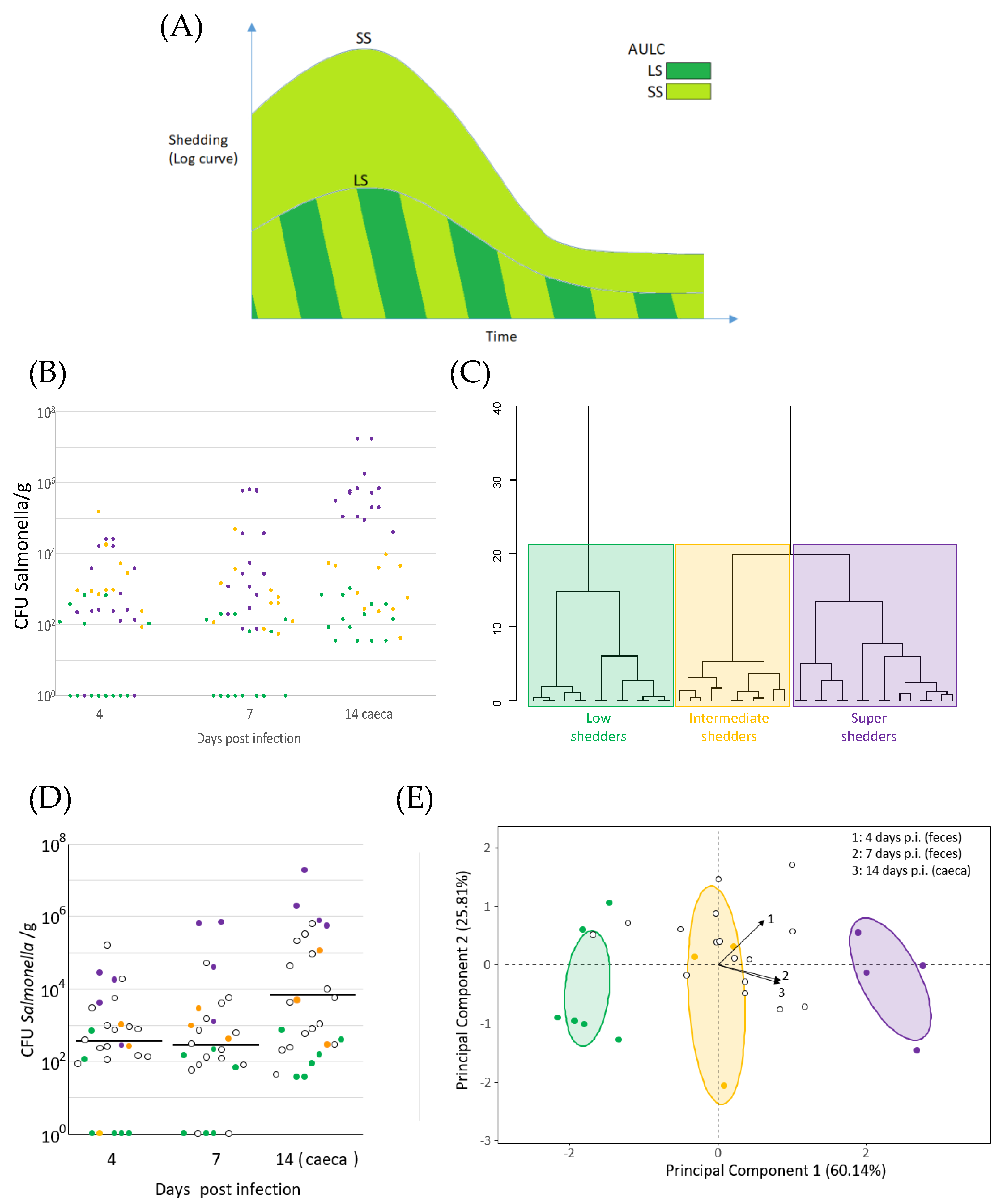
2.3. Temporal Shedding Patterns
2.4. Delineation of the Super Shedding Category
3. Enteric Bacteria and Super Shedding
3.1. Escherichia coli
3.2. Salmonella
3.3. Mycobacterium avium subspecies paratuberculosis
3.4. Campylobacter spp.
4. What Causes Super Shedding?
4.1. Environmental Factors
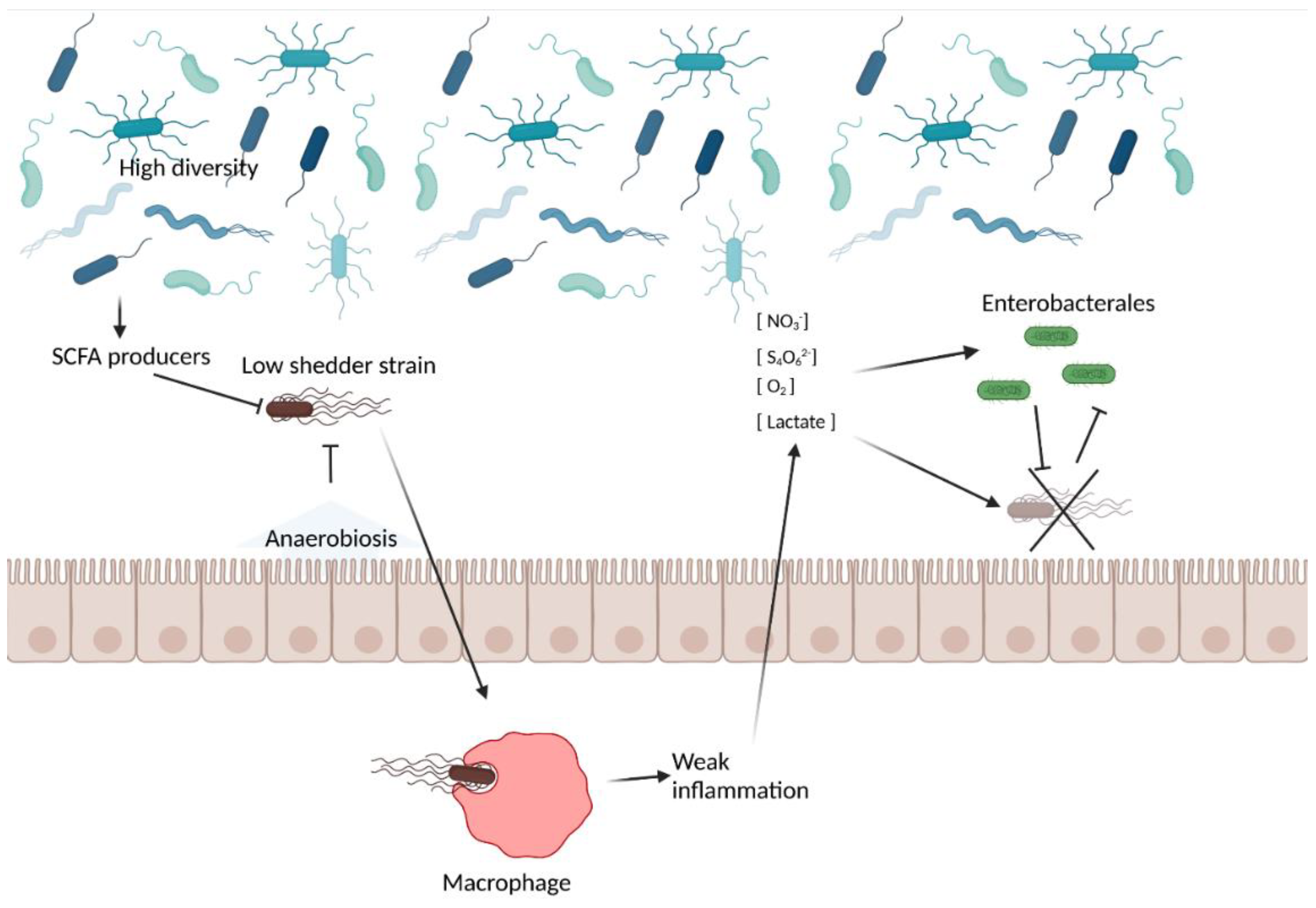
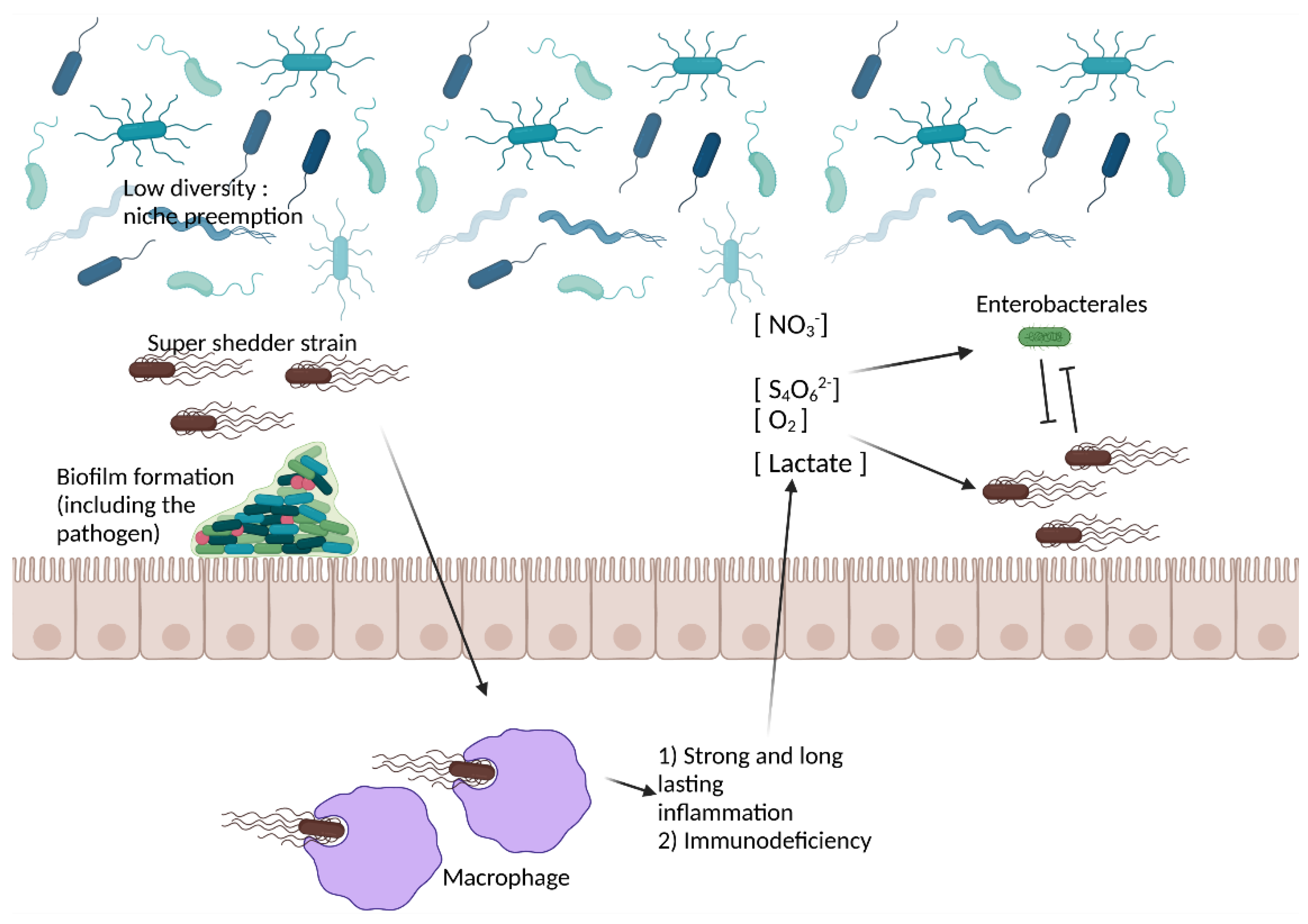
4.2. Gut Microbiota-Related Factors
4.3. Bacterial Genetics/Polymorphism
4.4. Host Related Factors: Age and Sex
4.5. Host Related Factors: Immunity
4.6. The Interplay between the Host, the Pathogen, and the Gut Microbiota
5. Control Strategies
5.1. Husbandry Practices
5.2. Vaccines
5.3. Bacteriophages
5.4. Feed Additives
6. Concluding Remarks
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lau, L.L.; Ip, D.K.; Nishiura, H.; Fang, V.J.; Chan, K.H.; Peiris, J.S.; Leung, G.M.; Cowling, B.J. Heterogeneity in viral shedding among individuals with medically attended influenza a virus infection. J. Infect. Dis. 2013, 207, 1281–1285. [Google Scholar] [CrossRef] [PubMed]
- Kucharski, A.J.; Althaus, C.L. The role of superspreading in Middle East respiratory syndrome coronavirus (MERS-CoV) transmission. Eurosurveillance 2015, 20, 14–18. [Google Scholar] [CrossRef] [PubMed]
- Al-Tawfiq, J.A.; Rodriguez-Morales, A.J. Super-spreading events and contribution to transmission of MERS, SARS, and SARS-CoV-2 (COVID-19). J. Hosp. Infect. 2020, 105, 111–112. [Google Scholar] [CrossRef]
- Chase-Topping, M.; Gally, D.; Low, C.; Matthews, L.; Woolhouse, M. Super-shedding and the link between human infection and livestock carriage of Escherichia coli O157. Nat. Rev. Microbiol. 2008, 6, 904–912. [Google Scholar] [CrossRef] [PubMed]
- Porten, K.; Rissland, J.; Tigges, A.; Broll, S.; Hopp, W.; Lunemann, M.; van Treeck, U.; Kimmig, P.; Brockmann, S.O.; Wagner-Wiening, C.; et al. A super-spreading ewe infects hundreds with Q fever at a farmers’ market in Germany. BMC Infect. Dis. 2006, 6, 147. [Google Scholar] [CrossRef]
- Young, T.M.; Bray, A.S.; Nagpal, R.K.; Caudell, D.L.; Yadav, H.; Zafar, M.A. Animal model to study Klebsiella pneumoniae gastrointestinal colonization and host-to-host transmission. Infect. Immun. 2020, 88, e00071-20. [Google Scholar] [CrossRef]
- Lawley, T.D.; Bouley, D.M.; Hoy, Y.E.; Gerke, C.; Relman, D.A.; Monack, D.M. Host transmission of Salmonella enterica serovar Typhimurium is controlled by virulence factors and indigenous intestinal microbiota. Infect. Immun. 2008, 76, 403–416. [Google Scholar] [CrossRef]
- Achen, M.; Morishita, T.Y.; Ley, E.C. Shedding and colonization of Campylobacter jejuni in broilers from day-of-hatch to slaughter age. Avian Dis. 1998, 42, 732–737. [Google Scholar] [CrossRef]
- Matthews, L.; Low, J.C.; Gally, D.L.; Pearce, M.C.; Mellor, D.J.; Heesterbeek, J.A.; Chase-Topping, M.; Naylor, S.W.; Shaw, D.J.; Reid, S.W.; et al. Heterogeneous shedding of Escherichia coli O157 in cattle and its implications for control. Proc. Natl. Acad. Sci. USA 2006, 103, 547–552. [Google Scholar] [CrossRef]
- Woolhouse, M.E.; Dye, C.; Etard, J.F.; Smith, T.; Charlwood, J.D.; Garnett, G.P.; Hagan, P.; Hii, J.L.; Ndhlovu, P.D.; Quinnell, R.J.; et al. Heterogeneities in the transmission of infectious agents: Implications for the design of control programs. Proc. Natl. Acad. Sci. USA 1997, 94, 338–342. [Google Scholar] [CrossRef]
- Anderson, R.M.; May, R.M. Infectious Diseases of Humans: Dynamics and Control; Oxford University Press: Oxford, UK, 1992. [Google Scholar]
- VanderWaal, K.L.; Ezenwa, V.O. Heterogeneity in pathogen transmission. Funct. Ecol. 2016, 30, 1606–1622. [Google Scholar] [CrossRef]
- Lloyd-Smith, J.O.; Schreiber, S.J.; Kopp, P.E.; Getz, W.M. Superspreading and the effect of individual variation on disease emergence. Nature 2005, 438, 355–359. [Google Scholar] [CrossRef] [PubMed]
- Spencer, S.E.; Besser, T.E.; Cobbold, R.N.; French, N.P. ‘Super’ or just ‘above average’? Supershedders and the transmission of Escherichia coli O157:H7 among feedlot cattle. J. R. Soc. Interface 2015, 12, 20150446. [Google Scholar] [CrossRef] [PubMed]
- Slater, N.; Mitchell, R.M.; Whitlock, R.H.; Fyock, T.; Pradhan, A.K.; Knupfer, E.; Schukken, Y.H.; Louzoun, Y. Impact of the shedding level on transmission of persistent infections in Mycobacterium avium subspecies paratuberculosis (MAP). Vet. Res. 2016, 47, 38. [Google Scholar] [CrossRef]
- Mitchell, R.M.; Whitlock, R.H.; Gröhn, Y.T.; Schukken, Y.H. Back to the real world: Connecting models with data. Prev. Vet. Med. 2015, 118, 215–225. [Google Scholar] [CrossRef]
- Omisakin, F.; MacRae, M.; Ogden, I.D.; Strachan, N.J. Concentration and prevalence of Escherichia coli O157 in cattle feces at slaughter. Appl. Environ. Microbiol. 2003, 69, 2444–2447. [Google Scholar] [CrossRef]
- Lanzas, C.; Brien, S.; Ivanek, R.; Lo, Y.; Chapagain, P.P.; Ray, K.A.; Ayscue, P.; Warnick, L.D.; Gröhn, Y.T. The effect of heterogeneous infectious period and contagiousness on the dynamics of Salmonella transmission in dairy cattle. Epidemiol. Infect. 2008, 136, 1496–1510. [Google Scholar] [CrossRef]
- Gopinath, S.; Carden, S.; Monack, D. Shedding light on Salmonella carriers. Trends Microbiol. 2012, 20, 320–327. [Google Scholar] [CrossRef]
- Robinson, S.E.; Wright, E.J.; Hart, C.A.; Bennett, M.; French, N.P. Intermittent and persistent shedding of Escherichia coli O157 in cohorts of naturally infected calves. J. Appl. Microbiol. 2004, 97, 1045–1053. [Google Scholar] [CrossRef]
- Mitchell, R.M.; Schukken, Y.; Koets, A.; Weber, M.; Bakker, D.; Stabel, J.; Whitlock, R.H.; Louzoun, Y. Differences in intermittent and continuous fecal shedding patterns between natural and experimental Mycobacterium avium subspecies paratuberculosis infections in cattle. Vet. Res. 2015, 46, 66. [Google Scholar] [CrossRef]
- Robinson, S.; Brown, P.; Wright, E.; Hart, C.; French, N. Quantifying within-and between-animal variation and uncertainty associated with counts of Escherichia coli O157 occurring in naturally infected cattle faeces. J. R. Soc. Interface 2009, 6, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Munns, K.D.; Selinger, L.; Stanford, K.; Selinger, L.B.; McAllister, T.A. Are super-shedder feedlot cattle really super? Foodborne Pathog. Dis. 2014, 11, 329–331. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.R. Vaccination of Cattle against Escherichia coli O157:H7. Microbiol Spectr 2014, 2, EHEC-0006-2013. [Google Scholar] [CrossRef] [PubMed]
- Kempf, F.; Menanteau, P.; Rychlik, I.; Kubasová, T.; Trotereau, J.; Virlogeux-Payant, I.; Schaeffer, S.; Schouler, C.; Drumo, R.; Guitton, E.; et al. Gut microbiota composition before infection determines the Salmonella super- and low-shedder phenotypes in chicken. Microb. Biotechnol. 2020, 13, 1611–1630. [Google Scholar] [CrossRef] [PubMed]
- Velge, P.; Menanteau, P.; Chaumeil, T.; Barilleau, E.; Trotereau, J.; Virlogeux-Payant, I. Two in vivo models to study Salmonella asymptomatic carrier state in chicks. Methods Mol. Biol. 2022, 2427, 249–264. [Google Scholar] [CrossRef] [PubMed]
- Lahodny, G.E.; Gautam, R.; Ivanek, R. Understanding the effects of intermittent shedding on the transmission of infectious diseases: Example of salmonellosis in pigs. J. Biol. Dyn. 2017, 11, 436–460. [Google Scholar] [CrossRef]
- Aly, S.S.; Anderson, R.J.; Whitlock, R.H.; Fyock, T.L.; McAdams, S.C.; Byrem, T.M.; Jiang, J.; Adaska, J.M.; Gardner, I.A. Cost-effectiveness of diagnostic strategies to identify Mycobacterium avium subspecies paratuberculosis super-shedder cows in a large dairy herd using antibody enzyme-linked immunosorbent assays, quantitative real-time polymerase chain reaction, and bacterial culture. J. Vet. Diagn. Investig. 2012, 24, 821–832. [Google Scholar] [CrossRef]
- Arthur, T.M.; Keen, J.E.; Bosilevac, J.M.; Brichta-Harhay, D.M.; Kalchayanand, N.; Shackelford, S.D.; Wheeler, T.L.; Nou, X.; Koohmaraie, M. Longitudinal study of Escherichia coli O157:H7 in a beef cattle feedlot and role of high-level shedders in hide contamination. Appl. Environ. Microbiol. 2009, 75, 6515–6523. [Google Scholar] [CrossRef]
- Chase-Topping, M.E.; McKendrick, I.J.; Pearce, M.C.; MacDonald, P.; Matthews, L.; Halliday, J.; Allison, L.; Fenlon, D.; Low, J.C.; Gunn, G.; et al. Risk factors for the presence of high-level shedders of Escherichia coli O157 on Scottish farms. J. Clin. Microbiol. 2007, 45, 1594–1603. [Google Scholar] [CrossRef][Green Version]
- Cobbold, R.N.; Hancock, D.D.; Rice, D.H.; Berg, J.; Stilborn, R.; Hovde, C.J.; Besser, T.E. Rectoanal junction colonization of feedlot cattle by Escherichia coli O157:H7 and its association with supershedders and excretion dynamics. Appl. Environ. Microbiol. 2007, 73, 1563–1568. [Google Scholar] [CrossRef]
- Bearson, S.M.; Allen, H.K.; Bearson, B.L.; Looft, T.; Brunelle, B.W.; Kich, J.D.; Tuggle, C.K.; Bayles, D.O.; Alt, D.; Levine, U.Y.; et al. Profiling the gastrointestinal microbiota in response to Salmonella: Low versus high Salmonella shedding in the natural porcine host. Infect. Genet. Evol. 2013, 16, 330–340. [Google Scholar] [CrossRef] [PubMed]
- Menanteau, P.; Kempf, F.; Trotereau, J.; Virlogeux-Payant, I.; Gitton, E.; Dalifard, J.; Gabriel, I.; Rychlik, I.; Velge, P. Role of systemic infection, cross contaminations and super-shedders in Salmonella carrier state in chicken. Environ. Microbiol. 2018, 20, 3246–3260. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.H.; Uthe, J.J.; Bearson, S.M.; Demirkale, C.Y.; Nettleton, D.; Knetter, S.; Christian, C.; Ramer-Tait, A.E.; Wannemuehler, M.J.; Tuggle, C.K. Distinct peripheral blood RNA responses to Salmonella in pigs differing in Salmonella shedding levels: Intersection of IFNG, TLR and miRNA pathways. PLoS ONE 2011, 6, e28768. [Google Scholar] [CrossRef] [PubMed]
- Knetter, S.M.; Bearson, S.M.; Huang, T.H.; Kurkiewicz, D.; Schroyen, M.; Nettleton, D.; Berman, D.; Cohen, V.; Lunney, J.K.; Ramer-Tait, A.E.; et al. Salmonella enterica serovar Typhimurium-infected pigs with different shedding levels exhibit distinct clinical, peripheral cytokine and transcriptomic immune response phenotypes. Innate Immun. 2015, 21, 227–241. [Google Scholar] [CrossRef]
- Kommadath, A.; Bao, H.; Arantes, A.S.; Plastow, G.S.; Tuggle, C.K.; Bearson, S.M.; Guan, L.L.; Stothard, P. Gene co-expression network analysis identifies porcine genes associated with variation in Salmonella shedding. BMC Genom. 2014, 15, 452. [Google Scholar] [CrossRef]
- Denamur, E.; Clermont, O.; Bonacorsi, S.; Gordon, D. The population genetics of pathogenic Escherichia coli. Nat. Rev. Microbiol. 2021, 19, 37–54. [Google Scholar] [CrossRef]
- European Food Safety Authority and European Centre for Disease Prevention and Control. The European Union one health 2019 zoonoses report. EFSA J. 2021, 19, e06406. [Google Scholar] [CrossRef]
- Munns, K.D.; Selinger, L.B.; Stanford, K.; Guan, L.; Callaway, T.R.; McAllister, T.A. Perspectives on super-shedding of Escherichia coli O157:H7 by cattle. Foodborne Pathog. Dis. 2015, 12, 89–103. [Google Scholar] [CrossRef]
- Stephens, T.P.; McAllister, T.A.; Stanford, K. Development of an experimental model to assess the ability of Escherichia coli O157:H7-inoculated fecal pats to mimic a super shedder within a feedlot environment. J. Food Prot. 2008, 71, 648–652. [Google Scholar] [CrossRef]
- Ogden, I.D.; MacRae, M.; Strachan, N.J. Is the prevalence and shedding concentrations of E. coli O157 in beef cattle in Scotland seasonal? FEMS Microbiol. Lett. 2004, 233, 297–300. [Google Scholar] [CrossRef] [PubMed]
- La Ragione, R.M.; Best, A.; Sprigings, K.; Liebana, E.; Woodward, G.R.; Sayers, A.R.; Woodward, M.J. Variable and strain dependent colonisation of chickens by Escherichia coli O157. Vet. Microbiol. 2005, 107, 103–113. [Google Scholar] [CrossRef] [PubMed]
- McCabe, E.; Burgess, C.M.; Lawal, D.; Whyte, P.; Duffy, G. An investigation of shedding and super-shedding of Shiga toxigenic Escherichia coli O157 and E. coli O26 in cattle presented for slaughter in the Republic of Ireland. Zoonoses Public Health 2019, 66, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Geenen, P.L.; Van der Meulen, J.; Bouma, A.; Engel, B.; Heesterbeek, J.A.; De Jong, M.C. Classification of temporal profiles of F4+ E. coli shedding and faecal dry matter in experimental post-weaning diarrhoea of pigs. Epidemiol. Infect. 2007, 135, 1001–1009. [Google Scholar] [CrossRef] [PubMed]
- Pollock, J.; Gally, D.L.; Glendinning, L.; Tiwari, R.; Hutchings, M.R.; Houdijk, J.G.M. Analysis of temporal fecal microbiota dynamics in weaner pigs with and without exposure to enterotoxigenic Escherichia coli. J. Anim. Sci. 2018, 96, 3777–3790. [Google Scholar] [CrossRef]
- Parry, C.M.; Hien, T.T.; Dougan, G.; White, N.J.; Farrar, J.J. Typhoid fever. N. Engl. J. Med. 2002, 347, 1770–1782. [Google Scholar] [CrossRef]
- McAloon, C.G.; Roche, S.; Ritter, C.; Barkema, H.W.; Whyte, P.; More, S.J.; O’Grady, L.; Green, M.J.; Doherty, M.L. A review of paratuberculosis in dairy herds—Part 1: Epidemiology. Vet. J. 2019, 246, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Ott, S.L.; Wells, S.J.; Wagner, B.A. Herd-level economic losses associated with Johne’s disease on US dairy operations. Prev. Vet. Med. 1999, 40, 179–192. [Google Scholar] [CrossRef]
- Crossley, B.M.; Zagmutt-Vergara, F.J.; Fyock, T.L.; Whitlock, R.H.; Gardner, I.A. Fecal shedding of Mycobacterium avium subsp. paratuberculosis by dairy cows. Vet. Microbiol. 2005, 107, 257–263. [Google Scholar] [CrossRef]
- Whitlock, R.; Sweeney, R.; Fyock, T. MAP super-shedders. In Proceedings of the American Association of Bovine Practitioners, Annual Conference, Salt Lake City, UT, USA, 24–26 September 2005; pp. 193–194. [Google Scholar]
- Coker, A.O.; Isokpehi, R.D.; Thomas, B.N.; Amisu, K.O.; Obi, C.L. Human campylobacteriosis in developing countries. Emerg. Infect. Dis. 2002, 8, 237–244. [Google Scholar] [CrossRef]
- Zhang, Q.; Sahin, O. Campylobacteriosis. In Diseases of Poultry, 14th ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2020; pp. 754–769. [Google Scholar]
- Rawson, T.; Paton, R.S.; Colles, F.M.; Maiden, M.C.; Dawkins, M.S.; Bonsall, M.B. A mathematical modeling approach to uncover factors influencing the spread of Campylobacter in a flock of broiler-breeder chickens. Front. Microbiol. 2020, 11, 576646. [Google Scholar] [CrossRef]
- Rapp, D.; Ross, C.M.; Pleydell, E.J.; Muirhead, R.W. Differences in the fecal concentrations and genetic diversities of Campylobacter jejuni populations among individual cows in two dairy herds. Appl. Environ. Microbiol. 2012, 78, 7564–7571. [Google Scholar] [CrossRef] [PubMed]
- Money, P.; Kelly, A.F.; Gould, S.W.; Denholm-Price, J.; Threlfall, E.J.; Fielder, M.D. Cattle, weather and water: Mapping Escherichia coli O157:H7 infections in humans in England and Scotland. Environ. Microbiol. 2010, 12, 2633–2644. [Google Scholar] [CrossRef] [PubMed]
- Williams, K.J.; Ward, M.P.; Dhungyel, O.P.; Hall, E.J. Risk factors for Escherichia coli O157 shedding and super-shedding by dairy heifers at pasture. Epidemiol. Infect. 2015, 143, 1004–1015. [Google Scholar] [CrossRef] [PubMed]
- Venegas-Vargas, C.; Henderson, S.; Khare, A.; Mosci, R.E.; Lehnert, J.D.; Singh, P.; Ouellette, L.M.; Norby, B.; Funk, J.A.; Rust, S.; et al. Factors associated with shiga toxin-producing Escherichia coli shedding by dairy and beef cattle. Appl. Environ. Microbiol. 2016, 82, 5049–5056. [Google Scholar] [CrossRef]
- Edrington, T.S.; Callaway, T.R.; Ives, S.E.; Engler, M.J.; Looper, M.L.; Anderson, R.C.; Nisbet, D.J. Seasonal shedding of Escherichia coli O157:H7 in ruminants: A new hypothesis. Foodborne Pathog. Dis. 2006, 3, 413–421. [Google Scholar] [CrossRef]
- Sheng, H.; Shringi, S.; Baker, K.N.; Minnich, S.A.; Hovde, C.J.; Besser, T.E. Standardized Escherichia coli O157:H7 exposure studies in cattle provide evidence that bovine factors do not drive increased summertime colonization. Appl. Environ. Microbiol. 2016, 82, 964–971. [Google Scholar] [CrossRef]
- Stenkamp-Strahm, C.; McConnel, C.; Rao, S.; Magnuson, R.; Hyatt, D.R.; Linke, L. Climate, lactation, and treatment factors influence faecal shedding of Escherichia coli O157 pathotypes in dairy cows. Epidemiol. Infect. 2017, 145, 115–125. [Google Scholar] [CrossRef]
- Gunn, G.J.; McKendrick, I.J.; Ternent, H.E.; Thomson-Carter, F.; Foster, G.; Synge, B.A. An investigation of factors associated with the prevalence of verocytotoxin producing Escherichia coli O157 shedding in Scottish beef cattle. Vet. J. 2007, 174, 554–564. [Google Scholar] [CrossRef]
- Jacob, M.E.; Callaway, T.R.; Nagaraja, T.G. Dietary interactions and interventions affecting Escherichia coli O157 colonization and shedding in cattle. Foodborne Pathog. Dis. 2009, 6, 785–792. [Google Scholar] [CrossRef]
- Traub-Dargatz, J.L.; Ladely, S.R.; Dargatz, D.A.; Fedorka-Cray, P.J. Impact of heat stress on the fecal shedding patterns of Salmonella enterica Typhimurium DT104 and Salmonella enterica Infantis by 5-week-old male broilers. Foodborne Pathog. Dis. 2006, 3, 178–183. [Google Scholar] [CrossRef]
- Rogers, A.W.L.; Tsolis, R.M.; Bäumler, A.J. Salmonella versus the Microbiome. Microbiol. Mol. Biol. Rev. 2021, 85, e00027-19. [Google Scholar] [CrossRef] [PubMed]
- Argüello, H.; Estellé, J.; Leonard, F.C.; Crispie, F.; Cotter, P.D.; O’Sullivan, O.; Lynch, H.; Walia, K.; Duffy, G.; Lawlor, P.G.; et al. Influence of the intestinal microbiota on colonization resistance to Salmonella and the shedding pattern of naturally exposed pigs. mSystems 2019, 4, e00021-19. [Google Scholar] [CrossRef] [PubMed]
- Pedroso, A.A.; Lee, M.D.; Maurer, J.J. Strength lies in diversity: How community diversity limits. Front. Microbiol. 2021, 12, 694215. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Tyler, P.J.; Starnes, J.; Bratcher, C.L.; Rankins, D.; McCaskey, T.A.; Wang, L. Correlation analysis of Shiga toxin-producing Escherichia coli shedding and faecal bacterial composition in beef cattle. J. Appl. Microbiol. 2013, 115, 591–603. [Google Scholar] [CrossRef] [PubMed]
- Stenkamp-Strahm, C.; McConnel, C.; Magzamen, S.; Abdo, Z.; Reynolds, S. Associations between Escherichia coli O157 shedding and the faecal microbiota of dairy cows. J. Appl. Microbiol. 2018, 124, 881–898. [Google Scholar] [CrossRef]
- Xu, Y.; Dugat-Bony, E.; Zaheer, R.; Selinger, L.; Barbieri, R.; Munns, K.; McAllister, T.A.; Selinger, L.B. Escherichia coli O157:H7 super-shedder and non-shedder feedlot steers harbour distinct fecal bacterial communities. PLoS ONE 2014, 9, e98115. [Google Scholar] [CrossRef]
- Zaheer, R.; Dugat-Bony, E.; Holman, D.B.; Cousteix, E.; Xu, Y.; Munns, K.; Selinger, L.J.; Barbieri, R.; Alexander, T.; McAllister, T.A.; et al. Changes in bacterial community composition of Escherichia coli O157:H7 super-shedder cattle occur in the lower intestine. PLoS ONE 2017, 12, e0170050. [Google Scholar] [CrossRef]
- Bibbal, D.; Ruiz, P.; Sapountzis, P.; Mazuy-Cruchaudet, C.; Loukiadis, E.; Auvray, F.; Forano, E.; Brugère, H. Persistent circulation of enterohemorrhagic Escherichia coli (EHEC) O157: H7 in cattle farms: Characterization of enterohemorrhagic Escherichia coli O157: H7 strains and fecal microbial communities of bovine shedders and non-shedders. Front. Vet. Sci. 2022, 9, 852475. [Google Scholar] [CrossRef]
- Sofka, D.; Pfeifer, A.; Gleiss, B.; Paulsen, P.; Hilbert, F. Changes within the intestinal flora of broilers by colonisation with Campylobacter jejuni. Berl. Münch. Tierärztl. Wochenschr. 2015, 128, 104–110. [Google Scholar]
- Kaevska, M.; Videnska, P.; Sedlar, K.; Bartejsova, I.; Kralova, A.; Slana, I. Faecal bacterial composition in dairy cows shedding Mycobacterium avium subsp. paratuberculosis in faeces in comparison with nonshedding cows. Can. J. Microbiol. 2016, 62, 538–541. [Google Scholar] [CrossRef]
- Wang, O.; McAllister, T.A.; Plastow, G.; Stanford, K.; Selinger, B.; Guan, L.L. Interactions of the hindgut mucosa-associated microbiome with its host regulate shedding of Escherichia coli O157:H7 by cattle. Appl. Environ. Microbiol. 2018, 84, e01738-17. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Cong, G.; Zhang, Q.; Yao, H.; Wang, Z.; Kang, K.; He, X.; Shi, S. Infection heterogeneity and microbiota differences in chicks infected by Salmonella enteritidis. Microorganisms 2021, 9, 1705. [Google Scholar] [CrossRef]
- Stevens, E.J.; Bates, K.A.; King, K.C. Host microbiota can facilitate pathogen infection. PLoS Pathog. 2021, 17, e1009514. [Google Scholar] [CrossRef] [PubMed]
- Hallewell, J.; Niu, Y.D.; Munns, K.; McAllister, T.A.; Johnson, R.P.; Ackermann, H.W.; Thomas, J.E.; Stanford, K. Differing populations of endemic bacteriophages in cattle shedding high and low numbers of Escherichia coli O157:H7 bacteria in feces. Appl. Environ. Microbiol. 2014, 80, 3819–3825. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, M.K.; Maurice, C.F. Ménage à trois in the human gut: Interactions between host, bacteria and phages. Nat. Rev. Microbiol. 2017, 15, 397–408. [Google Scholar] [CrossRef]
- Munns, K.D.; Zaheer, R.; Xu, Y.; Stanford, K.; Laing, C.R.; Gannon, V.P.; Selinger, L.B.; McAllister, T.A. Comparative genomic analysis of Escherichia coli O157:H7 isolated from super-shedder and low-shedder cattle. PLoS ONE 2016, 11, e0151673. [Google Scholar] [CrossRef][Green Version]
- Katani, R.; Cote, R.; Kudva, I.T.; DebRoy, C.; Arthur, T.M.; Kapur, V. Comparative genomics of two super-shedder isolates of Escherichia coli O157:H7. PLoS ONE 2017, 12, e0182940. [Google Scholar] [CrossRef]
- Cote, R.; Katani, R.; Moreau, M.R.; Kudva, I.T.; Arthur, T.M.; DebRoy, C.; Mwangi, M.M.; Albert, I.; Raygoza Garay, J.A.; Li, L.; et al. Comparative analysis of super-shedder strains of Escherichia coli O157:H7 reveals distinctive genomic features and a strongly aggregative adherent phenotype on bovine rectoanal junction squamous epithelial cells. PLoS ONE 2015, 10, e0116743. [Google Scholar] [CrossRef]
- Leitão, J.H. Microbial virulence factors. Int. J. Mol. Sci. 2020, 21, 5320. [Google Scholar] [CrossRef]
- Teng, L.; Lee, S.; Park, D.; Jeong, K.C. Genetic and functional analyses of virulence potential of an Escherichia coli O157:H7 strain isolated from super-shedder cattle. Front. Cell. Infect. Microbiol. 2020, 10, 271. [Google Scholar] [CrossRef]
- Castro, V.S.; Figueiredo, E.; McAllister, T.; Stanford, K. Farm to fork impacts of super-shedders and high-event periods on food safety. Trends Food Sci. Technol. 2022, 127, 129–142. [Google Scholar] [CrossRef]
- Raza, A.; Sarwar, Y.; Ali, A.; Jamil, A.; Haque, A. Effect of biofilm formation on the excretion of Salmonella enterica serovar Typhi in feces. Int. J. Infect. Dis. 2011, 15, e747–e752. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, R.M.; Medley, G.F.; Collins, M.T.; Schukken, Y.H. A meta-analysis of the effect of dose and age at exposure on shedding of Mycobacterium avium subspecies paratuberculosis (MAP) in experimentally infected calves and cows. Epidemiol. Infect. 2012, 140, 231–246. [Google Scholar] [CrossRef] [PubMed]
- Mir, R.A.; Weppelmann, T.A.; Elzo, M.; Ahn, S.; Driver, J.D.; Jeong, K.C. Colonization of beef cattle by shiga toxin-producing Escherichia coli during the first year of life: A cohort study. PLoS ONE 2016, 11, e0148518. [Google Scholar] [CrossRef] [PubMed]
- Antaki-Zukoski, E.M.; Li, X.; Hoar, B.; Adaska, J.M.; Byrne, B.A.; Atwill, E.R. Understanding the transmission dynamics of Escherichia coli O157: H7 super-shedding infections in feedlot cattle. PeerJ 2021, 9, e12524. [Google Scholar] [CrossRef]
- Wang, O.; Liang, G.; McAllister, T.A.; Plastow, G.; Stanford, K.; Selinger, B.; Guan, L.L. Comparative transcriptomic analysis of rectal tissue from beef steers revealed reduced host immunity in Escherichia coli O157:H7 super-shedders. PLoS ONE 2016, 11, e0151284. [Google Scholar] [CrossRef][Green Version]
- Wang, O.; McAllister, T.A.; Plastow, G.; Stanford, K.; Selinger, B.; Guan, L.L. Host mechanisms involved in cattle Escherichia coli O157 shedding: A fundamental understanding for reducing foodborne pathogen in food animal production. Sci. Rep. 2017, 7, 7630. [Google Scholar] [CrossRef]
- Wang, O.; Zhou, M.; Chen, Y.; McAllister, T.A.; Plastow, G.; Stanford, K.; Selinger, B.; Guan, L.L. MicroRNAomes of cattle intestinal tissues revealed possible miRNA regulated mechanisms involved in Escherichia coli O157 fecal shedding. Front. Cell. Infect. Microbiol. 2021, 11, 634505. [Google Scholar] [CrossRef]
- Uthe, J.J.; Wang, Y.; Qu, L.; Nettleton, D.; Tuggle, C.K.; Bearson, S.M. Correlating blood immune parameters and a CCT7 genetic variant with the shedding of Salmonella enterica serovar Typhimurium in swine. Vet. Microbiol. 2009, 135, 384–388. [Google Scholar] [CrossRef]
- Cazals, A.; Estellé, J.; Bruneau, N.; Coville, J.L.; Menanteau, P.; Rossignol, M.N.; Jardet, D.; Bevilacqua, C.; Rau, A.; Bed’Hom, B.; et al. Differences in caecal microbiota composition and Salmonella carriage between experimentally infected inbred lines of chickens. Genet. Sel. Evol. 2022, 54, 7. [Google Scholar] [CrossRef]
- Ciarlo, E.; Heinonen, T.; Théroude, C.; Asgari, F.; Le Roy, D.; Netea, M.G.; Roger, T. Trained immunity confers broad-spectrum protection against bacterial infections. J. Infect. Dis. 2020, 222, 1869–1881. [Google Scholar] [CrossRef]
- Byrne, K.A.; Loving, C.L.; McGill, J.L. Innate immunomodulation in food animals: Evidence for trained immunity? Front. Immunol. 2020, 11, 1099. [Google Scholar] [CrossRef]
- Litvak, Y.; Bäumler, A.J. Microbiota-nourishing immunity: A guide to understanding our microbial self. Immunity 2019, 51, 214–224. [Google Scholar] [CrossRef]
- LeJeune, J.T.; Wetzel, A.N. Preharvest control of Escherichia coli O157 in cattle. J. Anim Sci 2007, 85, E73–E80. [Google Scholar] [CrossRef]
- De Cort, W.; Ducatelle, R.; Van Immerseel, F. Preharvest measures to improve the safety of eggs. In Producing Safe Eggs; Elsevier: Amsterdam, The Netherlands, 2017; pp. 259–280. [Google Scholar]
- Pessoa, J.; Rodrigues da Costa, M.; Nesbakken, T.; Meemken, D. Assessment of the effectiveness of pre-harvest meat safety interventions to control foodborne pathogens in broilers: A systematic review. Curr. Clin. Microbiol. Rep. 2021, 8, 21–30. [Google Scholar] [CrossRef]
- Rodrigues da Costa, M.; Pessoa, J.; Meemken, D.; Nesbakken, T. A systematic review on the effectiveness of pre-harvest meat safety interventions in pig herds to control Salmonella and other foodborne pathogens. Microorganisms 2021, 9, 1825. [Google Scholar] [CrossRef] [PubMed]
- Ellis-Iversen, J.; Smith, R.P.; Van Winden, S.; Paiba, G.A.; Watson, E.; Snow, L.C.; Cook, A.J. Farm practices to control E. coli O157 in young cattle—A randomised controlled trial. Vet. Res. 2008, 39, 3. [Google Scholar] [CrossRef][Green Version]
- Lu, Z.; Mitchell, R.M.; Smith, R.L.; Van Kessel, J.S.; Chapagain, P.P.; Schukken, Y.H.; Grohn, Y.T. The importance of culling in Johne’s disease control. J. Biol. 2008, 254, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Berriman, A.D.; Clancy, D.; Clough, H.E.; Armstrong, D.; Christley, R.M. Effectiveness of simulated interventions in reducing the estimated prevalence of Salmonella in UK pig herds. PLoS ONE 2013, 8, e66054. [Google Scholar] [CrossRef]
- Humphrey, T. Are happy chickens safer chickens? Poultry welfare and disease susceptibility. Br. Poult. Sci 2006, 47, 379–391. [Google Scholar] [CrossRef] [PubMed]
- Bannantine, J.P.; Hines, M.E.; Bermudez, L.E.; Talaat, A.M.; Sreevatsan, S.; Stabel, J.R.; Chang, Y.F.; Coussens, P.M.; Barletta, R.G.; Davis, W.C.; et al. A rational framework for evaluating the next generation of vaccines against Mycobacterium avium subspecies paratuberculosis. Front. Cell. Infect. Microbiol. 2014, 4, 126. [Google Scholar] [CrossRef] [PubMed]
- Wales, A.D.; Davies, R.H. Salmonella vaccination in pigs: A review. Zoonoses Public Health 2017, 64, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Desin, T.S.; Köster, W.; Potter, A.A. Salmonella vaccines in poultry: Past, present and future. Expert Rev. Vaccines 2013, 12, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Van Immerseel, F.; Methner, U.; Rychlik, I.; Nagy, B.; Velge, P.; Martin, G.; Foster, N.; Ducatelle, R.; Barrow, P.A. Vaccination and early protection against non-host-specific Salmonella serotypes in poultry: Exploitation of innate immunity and microbial activity. Epidemiol. Infect. 2005, 133, 959–978. [Google Scholar] [CrossRef] [PubMed]
- Endersen, L.; Coffey, A. The use of bacteriophages for food safety. Curr. Opin. Food Sci. 2020, 36, 1–8. [Google Scholar] [CrossRef]
- Lamy-Besnier, Q.; Chaffringeon, L.; Lourenço, M.; Payne, R.B.; Trinh, J.T.; Schwartz, J.A.; Sulakvelidze, A.; Debarbieux, L. Prophylactic administration of a bacteriophage cocktail is safe and effective in reducing Salmonella enterica serovar Typhimurium burden in Vivo. Microbiol. Spectr. 2021, 9, e0049721. [Google Scholar] [CrossRef]
- Tanji, Y.; Shimada, T.; Fukudomi, H.; Miyanaga, K.; Nakai, Y.; Unno, H. Therapeutic use of phage cocktail for controlling Escherichia coli O157:H7 in gastrointestinal tract of mice. J. Biosci. Bioeng. 2005, 100, 280–287. [Google Scholar] [CrossRef]
- Callaway, T.R.; Edrington, T.S.; Brabban, A.D.; Anderson, R.C.; Rossman, M.L.; Engler, M.J.; Carr, M.A.; Genovese, K.J.; Keen, J.E.; Looper, M.L.; et al. Bacteriophage isolated from feedlot cattle can reduce Escherichia coli O157:H7 populations in ruminant gastrointestinal tracts. Foodborne Pathog. Dis. 2008, 5, 183–191. [Google Scholar] [CrossRef]
- Carvalho, C.M.; Gannon, B.W.; Halfhide, D.E.; Santos, S.B.; Hayes, C.M.; Roe, J.M.; Azeredo, J. The in vivo efficacy of two administration routes of a phage cocktail to reduce numbers of Campylobacter coli and Campylobacter jejuni in chickens. BMC Microbiol. 2010, 10, 232. [Google Scholar] [CrossRef]
- Bardina, C.; Spricigo, D.A.; Cortés, P.; Llagostera, M. Significance of the bacteriophage treatment schedule in reducing Salmonella colonization of poultry. Appl. Environ. Microbiol. 2012, 78, 6600–6607. [Google Scholar] [CrossRef]
- Torres-Barceló, C.; Turner, P.E.; Buckling, A. Mitigation of evolved bacterial resistance to phage therapy. Curr. Opin. Virol. 2022, 53, 101201. [Google Scholar] [CrossRef] [PubMed]
- Collins, J.W.; La Ragione, R.M.; Woodward, M.J.; Searle, L.E. Application of prebiotics and probiotics in livestock. In Prebiotics and Probiotics Science and Technology; Springer: New York, NY, USA, 2009; pp. 1123–1192. [Google Scholar]
- Madsen, K. Probiotics and the immune response. J. Clin. Gastroenterol. 2006, 40, 232–234. [Google Scholar] [CrossRef] [PubMed]
- Larsen, M.H.; Dalmasso, M.; Ingmer, H.; Langsrud, S.; Malakauskas, M.; Mader, A.; Møretrø, T.; Možina, S.S.; Rychli, K.; Wagner, M. Persistence of foodborne pathogens and their control in primary and secondary food production chains. Food Control 2014, 44, 92–109. [Google Scholar] [CrossRef]
- Zhao, T.; Doyle, M.P.; Harmon, B.G.; Brown, C.A.; Mueller, P.O.; Parks, A.H. Reduction of carriage of enterohemorrhagic Escherichia coli O157:H7 in cattle by inoculation with probiotic bacteria. J. Clin. Microbiol. 1998, 36, 641–647. [Google Scholar] [CrossRef]
- Lema, M.; Williams, L.; Rao, D.R. Reduction of fecal shedding of enterohemorrhagic Escherichia coli O157:H7 in lambs by feeding microbial feed supplement. Small Rumin. Res. 2001, 39, 31–39. [Google Scholar] [CrossRef]
- Brashears, M.M.; Galyean, M.L.; Loneragan, G.H.; Mann, J.E.; Killinger-Mann, K. Prevalence of Escherichia coli O157:H7 and performance by beef feedlot cattle given Lactobacillus direct-fed microbials. J. Food Prot. 2003, 66, 748–754. [Google Scholar] [CrossRef]
- Younts-Dahl, S.M.; Osborn, G.D.; Galyean, M.L.; Rivera, J.D.; Loneragan, G.H.; Brashears, M.M. Reduction of Escherichia coli O157 in finishing beef cattle by various doses of Lactobacillus acidophilus in direct-fed microbials. J. Food Prot. 2005, 68, 6–10. [Google Scholar] [CrossRef]
- Wisener, L.V.; Sargeant, J.M.; O’Connor, A.M.; Faires, M.C.; Glass-Kaastra, S.K. The use of direct-fed microbials to reduce shedding of Escherichia coli O157 in beef cattle: A systematic review and meta-analysis. Zoonoses Public Health 2015, 62, 75–89. [Google Scholar] [CrossRef]
- Sargeant, J.M.; Amezcua, M.R.; Rajic, A.; Waddell, L. Pre-harvest interventions to reduce the shedding of E. coli O157 in the faeces of weaned domestic ruminants: A systematic review. Zoonoses Public Health 2007, 54, 260–277. [Google Scholar] [CrossRef]
- Casey, P.G.; Gardiner, G.E.; Casey, G.; Bradshaw, B.; Lawlor, P.G.; Lynch, P.B.; Leonard, F.C.; Stanton, C.; Ross, R.P.; Fitzgerald, G.F.; et al. A five-strain probiotic combination reduces pathogen shedding and alleviates disease signs in pigs challenged with Salmonella enterica Serovar Typhimurium. Appl. Environ. Microbiol. 2007, 73, 1858–1863. [Google Scholar] [CrossRef]
- Carter, A.; Adams, M.; La Ragione, R.M.; Woodward, M.J. Colonisation of poultry by Salmonella Enteritidis S1400 is reduced by combined administration of Lactobacillus salivarius 59 and Enterococcus faecium PXN-33. Vet. Microbiol. 2017, 199, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Zenner, C.; Hitch, T.C.A.; Riedel, T.; Wortmann, E.; Tiede, S.; Buhl, E.M.; Abt, B.; Neuhaus, K.; Velge, P.; Overmann, J.; et al. Early-life immune system maturation in chickens using a synthetic community of cultured gut bacteria. mSystems 2021, 6, e01300-20. [Google Scholar] [CrossRef] [PubMed]
- Brugiroux, S.; Beutler, M.; Pfann, C.; Garzetti, D.; Ruscheweyh, H.J.; Ring, D.; Diehl, M.; Herp, S.; Lötscher, Y.; Hussain, S.; et al. Genome-guided design of a defined mouse microbiota that confers colonization resistance against Salmonella enterica serovar Typhimurium. Nat. Microbiol. 2016, 2, 16215. [Google Scholar] [CrossRef]
- Edrington, T.; Callaway, T.; Anderson, R.; Genovese, K.; Jung, Y.S.; McReynolds, J.; Bischoff, K.; Nisbet, D. Reduction of E. coli O157: H7 populations in sheep by supplementation of an experimental sodium chlorate product. Small Rumin. Res. 2003, 49, 173–181. [Google Scholar] [CrossRef]
- Oyofo, B.A.; DeLoach, J.R.; Corrier, D.E.; Norman, J.O.; Ziprin, R.L.; Mollenhauer, H.H. Prevention of Salmonella typhimurium colonization of broilers with D-mannose. Poult. Sci. 1989, 68, 1357–1360. [Google Scholar] [CrossRef]
- Lema, M.; Williams, L.; Walker, L.; Rao, D. Effect of dietary fiber on E. coli O157: H7 shedding in lambs. Small Rumin. Res. 2002, 43, 249–255. [Google Scholar] [CrossRef]
- Fernandez, F.; Hinton, M.; Van Gils, B. Dietary mannan-oligosaccharides and their effect on chicken caecal microflora in relation to Salmonella Enteritidis colonization. Avian Pathol. 2002, 31, 49–58. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kempf, F.; La Ragione, R.; Chirullo, B.; Schouler, C.; Velge, P. Super Shedding in Enteric Pathogens: A Review. Microorganisms 2022, 10, 2101. https://doi.org/10.3390/microorganisms10112101
Kempf F, La Ragione R, Chirullo B, Schouler C, Velge P. Super Shedding in Enteric Pathogens: A Review. Microorganisms. 2022; 10(11):2101. https://doi.org/10.3390/microorganisms10112101
Chicago/Turabian StyleKempf, Florent, Roberto La Ragione, Barbara Chirullo, Catherine Schouler, and Philippe Velge. 2022. "Super Shedding in Enteric Pathogens: A Review" Microorganisms 10, no. 11: 2101. https://doi.org/10.3390/microorganisms10112101
APA StyleKempf, F., La Ragione, R., Chirullo, B., Schouler, C., & Velge, P. (2022). Super Shedding in Enteric Pathogens: A Review. Microorganisms, 10(11), 2101. https://doi.org/10.3390/microorganisms10112101





