Epigenetic Regulation of Human T-Cell Leukemia Virus Gene Expression
Abstract
:1. Epigenetic Dysregulation in Virus-Associated Cancers
2. HTLV and ATLL
3. Tax and HBZ
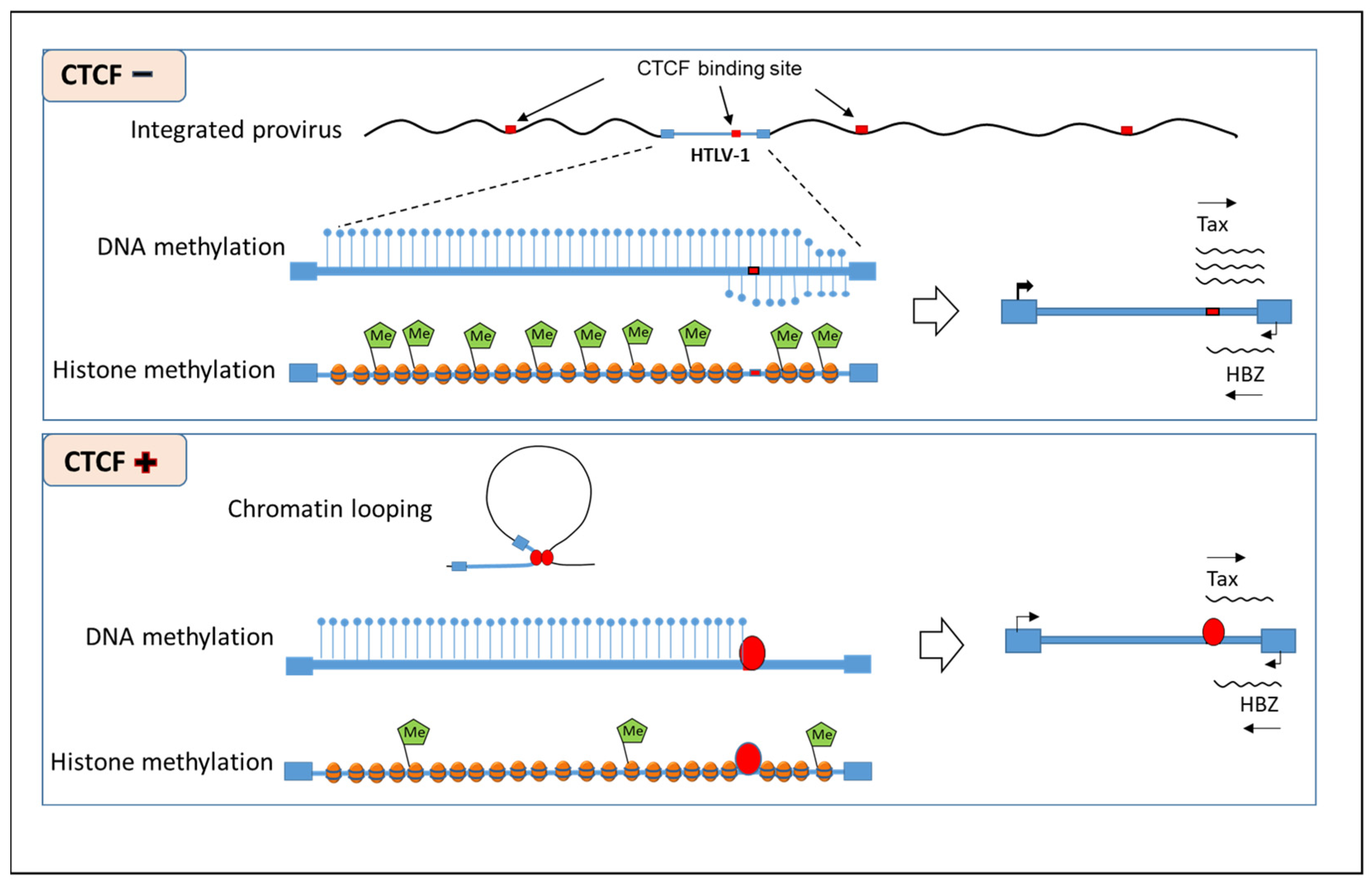
4. HTLV-1 DNA Methylation
5. HTLV-1 Chromatin-Associated Histone Modifications
6. Nucleosome Positioning along the HTLV-1 Provirus
7. Topology-Associated Domains in HTLV-1-Infected Cells
8. Future Queries
9. Summary
Funding
Conflicts of Interest
References
- Rutherford, A. Beware the pseudo gene geneis. Guardian, 19 July 2015. [Google Scholar]
- Bird, A. Perceptions of epigenetics. Nature 2007, 447, 396–398. [Google Scholar] [CrossRef]
- Berger, S.L.; Kousarides, T.; Shiekhattar, R.; Shilatifard, A. An operational definition of epigenetics. Genes Dev. 2009, 23, 781–783. [Google Scholar] [CrossRef] [Green Version]
- Waddington, C.H. The epigenotype. Endeavor 1942, 1, 18–20. [Google Scholar] [CrossRef] [Green Version]
- Holliday, R. DNA methylation and epigenetic inheritance. Phillosophical Trans. R. Soc. Lond. Ser. B Biol. Sci. 1990, 326, 329–338. [Google Scholar] [CrossRef]
- Minarovits, J.; Demcsak, A.; Banati, F.; Niller, H.H. Epigenetic dysregulation in virus-associated neoplasms. Adv. Exp. Med. Biol. 2016, 879, 71–90. [Google Scholar]
- Proietti, F.A.; Carneiro-Proietti, A.B.F.; Catalan-Soares, B.C.; Murphy, E.L. Global epidemiology of HTLV-I and associated diseases. Oncogene 2005, 24, 6058–6068. [Google Scholar] [CrossRef] [Green Version]
- Miyazato, P.; Matsuoka, M. Human T-cell leukemia virus type 1 and Foxp3 expression: Viral strategy in vivo. Int. Immunol. 2014, 26, 419–425. [Google Scholar] [CrossRef] [Green Version]
- Gasparini, C.; Celeghini, C.; Monasta, L.; Zauli, G. NF-kappaB pathways in hematological malignancies. Cell Mol. Life Sci. 2014, 71, 2083–2102. [Google Scholar] [CrossRef]
- Gessain, A.; Mahieux, R. Tropical spastic paraparesis and HTLV-1 associated myelopathy: Clinical, epidemiological, virological, and therapeutic aspects. Rev. Neurol. 2012, 168, 257–269. [Google Scholar] [CrossRef]
- Rosadas, C.; Taylor, G.P. Mother-to-child HTLV-1 transmission: Unmet research needs. Front. Microbiol. 2019, 10, 999. [Google Scholar] [CrossRef] [Green Version]
- Lairmore, M.D.; Anupam, R.; Bowden, N.; Haines, R.; Haynes, R.A.; Ratner, L.; Green, P.L. Molecular determinants of human T-lymphotropic virus type 1 transmission and spread. Viruses 2011, 3, 1131–1165. [Google Scholar] [CrossRef] [Green Version]
- Edwards, D.; Fenizia, C.; Gold, H.; Castro-Amarante, M.F.d.; Buchmann, C.; Pise-Masison, C.A.; Franchini, G. Orf-1 and orf-II encoded proteins in HTLV-1 infection and persistence. Viruses 2011, 3, 861–865. [Google Scholar] [CrossRef]
- Matsuoka, M.; Jeang, K.-T. Human T-cell leukemia virus type 1 (HTLV-1) and leukemic transformation: Viral infectivity, Tax, HBZ, and therapy. Oncogene 2011, 30, 1379–1389. [Google Scholar] [CrossRef] [Green Version]
- Harhaj, E.W.; Giam, C.Z. NF-kB signaling mechanisms in HTLV-1-induced adult T-cell leukemia/lymphoma. FEBS J. 2018, 285, 3324–3336. [Google Scholar] [CrossRef] [Green Version]
- Giam, C.Z.; Semmes, O.J. HTLV-1 infection and adult T cell leukemia/lymphoma—A tale of two proteins: Tax and HBZ. Viruses 2016, 8, 161. [Google Scholar] [CrossRef]
- Harrod, R.; Tang, Y.; Nicot, C.; Lu, H.; Vasillve, A.; Nakatani, Y.; Giam, C.-Z. An exposed KID-like domain in human T-cell lymphotropic virus type 1 Tax is responsible for the recruitment of coactivators CBP/p300. Molecular Cell Biology 1998, 18, 5052–5061. [Google Scholar] [CrossRef] [Green Version]
- Bernal-Mizrachi, L.; Lovly, C.M.; Ratner, L. The role of nuclear factor kB-1 and -2-mediated resistance to apoptosis in lymphomas. Proc. Natl. Acad. Sci. USA 2006, 103, 9220–9225. [Google Scholar] [CrossRef] [Green Version]
- Iha, H.; Kibler, K.V.; Yedavalli, V.R.K.; Peloponese, J.-M.; Haller, K.; Miyazato, A.; Kasai, T.; Jeang, K.-T. Segregation of NF-kB activation through NEMO/IKKgamma by Tax and TNFalpha: Implications for stimulus-specific interruption of oncogenic signaling. Oncogene 2003, 22, 8912–8923. [Google Scholar] [CrossRef] [Green Version]
- Uhlik, M.; Good, L.; Xiao, G.; Harhaj, E.W.; Zandi, E.; Karin, M.; Sun, S.-C. NF-kB-inducing kinase and IkB kinase participate in human T-cell leukemia I Tax-mediated NF-kB activation. J. Biol. Chem. 1998, 273, 21132–21136. [Google Scholar] [CrossRef] [Green Version]
- Murakami, T.; Hirai, H.; Suzuki, T.; Fujisawa, J.; Yoshida, M. HTLV-1 Tax enhanced NF-kappa B2 expression and binds to the products p52 and p100, but does not suppress the inhibitory function of p100. Virology 1995, 206, 1066–1074. [Google Scholar] [CrossRef] [Green Version]
- Robek, M.D.; Ratner, L. Immortlaization of CD4(+) and CD8(+) T lymphocytes by human T-cell leukemia virus type 1 Tax mutants expressed in a functional molecular clone. J. Virol. 1999, 73, 4856–4865. [Google Scholar] [CrossRef] [Green Version]
- Matsuoka, M.; Jeang, K.-T. Human T-cell leukaemia virus type 1 (HTLV-1) infectivity and cellular transformation. Nat. Rev. Cancer 2007, 7, 270–280. [Google Scholar] [CrossRef]
- Kattan, T.; MacNamara, A.; Rowan, A.G.; Nose, H.; Mosley, A.J.; Tanaka, Y.; Taylor, G.P.; Asquith, B.; Bangham, C.R.M. The avidity and lytic efficiency of the CTL response to HTLV-1. J. Immunol. 2009, 182, 5723–5729. [Google Scholar] [CrossRef] [Green Version]
- Koiwa, T.; Hamano-Usami, A.; Ishida, T.; Okayama, A.; Tamaguchi, K.; Kamihira, S.; Watanabe, T. 5′Long terminal repeat-selective CpG methylation of latent human T cell leukemia virus type 1 provirus in vitro and in vivo. J. Virol. 2002, 76, 9389–9397. [Google Scholar] [CrossRef] [Green Version]
- Cook, L.B.; Elemans, M.; Rowan, A.G.; Asquith, B. HTLV-1: Persistence and pathogenesis. Virology 2013, 435, 131–140. [Google Scholar] [CrossRef] [Green Version]
- Matsuoka, M.; Green, P.L. The HBZ gene, a key player in HTLV-1 pathogenesis. Retrovirology 2009, 6, 71. [Google Scholar] [CrossRef] [Green Version]
- Matsuoka, M.; Mesnard, J.-M. HTLV-1 bZIP factor; the key viral gene for pathogenesis. Retrovirology 2020, 17, 2. [Google Scholar] [CrossRef]
- Borchiellini, M.; Ummarino, S.; DiRuscio, A. The bright and dark side of DNA methylation: A matter of balance. Cells 2019, 12, 1243. [Google Scholar] [CrossRef] [Green Version]
- Deaton, A.M.; Bird, A. CpG islands and the regulation of transcription. Genes Dev. 2011, 25, 1010–1022. [Google Scholar] [CrossRef] [Green Version]
- Siegried, Z.; Eden, S.; Mendelsohn, M.; Feng, X.; Tsuberi, B.-Z.; Cedar, H. DNA methylation represses transcription in vivo. Nat. Genet. 1999, 22, 203–206. [Google Scholar] [CrossRef]
- Datta, S.; Kothari, N.H.; Fan, H. Induction of Tax I expression in MT-4 cells by 5-azacytidine leads to protein binding in the HTLV-1 LTR in vivo. Virology 2001, 283, 207–214. [Google Scholar] [CrossRef] [Green Version]
- Verma, M. Viral genes and methylation. Ann. N. Y. Acad. Sci. 2003, 983, 170–180. [Google Scholar] [CrossRef]
- Baylin, S.B.; Herman, J.G.; Graff, J.R.; Vertino, P.M.; Issa, J.P. Alterations in DNA methylation: A fundamental aspect of neoplasia. Adv. Cancer Res. 1998, 72, 141–196. [Google Scholar]
- Taniguchi, Y.; Nosaka, K.; Yaunaga, J.-i.; Maeda, M.; Mueler, N.; Okayama, A.; Matsuoka, M. Silencing of human T-cell leukemia virus type I gene transcription by epigenetic mechanisms. Retrovirolgy 2005, 2, 64. [Google Scholar] [CrossRef] [Green Version]
- Kulkarni, A.; Bangham, C.R.M. HTLV-1: Regulating the balance between proviral latency and reactivation. Front. Microbiol. 2018, 9, 449. [Google Scholar] [CrossRef]
- Kataoka, S.; Shiraishi, Y.; Takeda, Y.; Sakata, S.; Matsumoto, M.; Nagano, S.; Maeda, T.; Nagata, Y.; Ktanaka, A.; Mizuno, S.; et al. Aberrant PD-L1- expression through 3′-UTR disruption in multiple cancers. Nature 2016, 534, 402–406. [Google Scholar] [CrossRef]
- Yeh, C.-H.; Bai, X.T.; Moles, R.; Ratner, L.; Waldmann, T.A.; Tonsiki, W.; Nicot, C. Mutation of epigenetic regulators TET2 and MLL3 in patinets with HTLV-I-induced acute adult T-cell leukemia. Mol. Cancer 2016, 15, 15. [Google Scholar] [CrossRef] [Green Version]
- Kataoka, K.; Iwanaga, M.; Yasunaga, J.-i.; Nagata, Y.; Kitanaka, A.; Kameda, T.; Yoshimitsu, M.; Shiraishi, Y.; Sato-Otsubo, A.; Sanada, M.; et al. Prognostic relevance of integrated genetic profiling in adult T-cell leukemia/lymphoma. Blood 2018, 131, 215–225. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, T.; Yamashita, S.; Ureshino, H.; Kamachi, K.; Kurahashi, Y.; Fukuda-Kurahashi, Y.; Yoshida, N.; Hattori, N.; Nakamura, H.; Sato, A.; et al. Targeting aberrant DNA hypermethylation as a driver of ATL leukemogenesis by using the new oral demethylating agent OR-2100. Blood 2020, 136, 871–884. [Google Scholar] [CrossRef]
- Taniguchi, A.; Nemoto, Y.; Yokoyama, A.; Kotani, N.; Imagi, S.; Shuin, T.; Daibata, M. Promoter methylation of the bone morphogenetic protein-6 gene in association with adult T-cell leukemia. Int. J. Cancer 2008, 123, 1824–1831. [Google Scholar] [CrossRef] [Green Version]
- Nosaka, K.; Maeda, M.; Tamiya, S.; Sakai, T.; Mitsuya, H.; Matsuoka, M. Increasing methylation of the CDKN2A gene is associated with the progression of adult T-cell leukemia. Cancer Res. 2000, 60, 1043–1048. [Google Scholar]
- Matsusaka, K.; Kaneda, A.; Nagae, G.; Ushiku, T.; Kikuchi, Y.; Hino, R.; Uozaki, H.; Seto, Y.; Takada, K.; Aburatani, H.; et al. Classification of Epstein-Barr virus-positive gastric cancer by definition of DNA methylation epigenotypes. Cancer Res. 2011, 71, 7187–7197. [Google Scholar] [CrossRef] [Green Version]
- Lieras, R.A.; Smith, R.V.; Adrien, L.R.; Schlecht, N.F.; Burk, R.D.; Harris, T.M.; Childs, G.; Prystowsky, M.B.; Belbiri, T.J. Unique DNA methyulation loci distinguish anatomic site and HPV status in head and neck squamous cell carcinoma. Clin. Cancer Res. 2013, 19, 5444–5455. [Google Scholar] [CrossRef] [Green Version]
- Biktasova, A.; Hajek, M.; Sewell, A.; Gary, C.; Bellinger, G.; Deshpande, H.A.; Bhatia, A.; Burtness, B.; Judson, B.; Mehra, S.; et al. Demethylation therapy as a targeted treatment for human papillomavirus-associated head and neck cancer. Clin. Cancer Res. 2017, 23, 7276–7287. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, M.; Nishikawa, J.; Saito, M.; Sakai, K.; Sasaki, S.; Hashimoto, S.; Okamoto, T.; Suehiro, Y.; Yamasaki, T.; Sakaida, I. Decitabine inhibits tumor cell proliferation and up-regulates e-cadherin expression in Epstein-Barr virus-associated gastric cancer. J. Med. Virol. 2016, 89, 508–517. [Google Scholar] [CrossRef]
- Tropberger, P.; Schneider, R. Scratching the (lateral) surface of chromatin regulation by histone modifications. Nat. Struct. Mol. Biol. 2013, 20, 657–661. [Google Scholar] [CrossRef]
- Bernstein, B.E.; Karral, M.; Lindblad-Toh, K.; Bekiranov, S.; Bailey, D.K.; Huebert, D.J.; McMahon, S.; Karlsson, E.K.; Kulbokas, E.J.; Gingeras, T.R.; et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 2005, 120, 169–181. [Google Scholar] [CrossRef] [Green Version]
- Hsin, J.-P.; Manley, J.L. The RNA polymerase II CTD coordinates transcrition and RNA processing. Genes Dev. 2012, 26, 2119–2137. [Google Scholar] [CrossRef] [Green Version]
- Kang, H.; Lieberman, P.M. Mechanism of glycyrrhizic acid inhibition of Kaposi’s sarcoma-associated herpesvirus: Disruption of CTCF-cohesin-mediated RNA polymerase II pausing and sister chromatin cohesion. J. Virol. 2011, 85, 11159–11169. [Google Scholar] [CrossRef] [Green Version]
- Nyborg, J.K.; Egan, D.; Sharmna, N. The HTLV-1 Tax protein: Revealing mechanisms of transcriptional activation through histone acetylation and nucleosome disassembly. Biochim. Biophys. Acta 2010, 1799, 266–274. [Google Scholar] [CrossRef]
- Georges, S.A.; Kraus, W.L.; Luger, K.; Nyborg, J.K.; Laybourn, P.J. p300-mediated tax transcactivation from recombinant chromatin: Histone tail deletion mimics coactivator function. Mol. Cell. Biol. 2002, 22, 127–137. [Google Scholar] [CrossRef] [Green Version]
- Canettieri, G.; Morantte, I.; Guzman, E.; Asahara, H.; Herzi, S.; Anderson, S.D.; Yates, J.R.; Montminy, M. Attenuation of a phosphorylation-dependent activator by an HDAC-PP1 complex. Nat. Struct. Biol. 2003, 10, 175–181. [Google Scholar] [CrossRef]
- Lu, H.; Pise-Masison, C.A.; Linton, R.; Park, H.U.; Schiltz, R.L.; Sartorelli, V.; Brady, J.N. Tax relieves transcriptional repression by promoting histone deacetylase 1 release from the human T-cell leukemia virus type 1 long terminal repeat. J. Virol. 2004, 78, 6735–6743. [Google Scholar] [CrossRef] [Green Version]
- Tang, H.M.; Gao, W.W.; Chan, C.P.; Cheng, Y.; Deng, J.J.; Yuen, K.S.; Iha, H.; Jin, D.Y. SIRT1 suppresses human T-cell leukemia virus type 1 transcription. J. Virol. 2015, 89, 8623–8631. [Google Scholar] [CrossRef] [Green Version]
- Lezin, A.; Gillet, N.; Olindo, S.; Signate, A.; Grandvaux, N.; Verlaeten, O.; Belrose, G.; Bittencourt, M.C.; Hiscott, J.; Asquith, B.; et al. Histone deacetylase mediated transcriptional activation reduces proviral loads in HTLV-1 associated myelopathy/tropical spastic paraparesis patients. Blood 2007, 110, 3722–3728. [Google Scholar] [CrossRef] [Green Version]
- Lezin, A.; Olindo, S.; Belrose, G.; Signate, A.; Cesaire, R.; Smadja, D.; Macallan, D.; Asquith, B.; Bangham, C.; Bouzar, A.; et al. Gene activation therapy: From the BLV model to HAM/TSP patients. Front. Biosci. 2009, S1, 205–215. [Google Scholar] [CrossRef]
- Yamamoto, K.; Ishida, T.; Yamagishi, K.N.; Yamochi, T.; Tanaka, Y.; Furukawa, Y.; Nakamura, Y.; Watanabe, T. SMYD3 interacts with HTLV-1 Tax and regulates subcellular localization of Tax. Cancer Sci. 2011, 102, 260–266. [Google Scholar] [CrossRef]
- Kamoi, K.; Yamamoto, K.; Misawa, A.; Miyake, A.; Ishida, T.; Tanaka, Y.; Mochizuki, M.; Watanabe, T. SUV39H1 interacts wtih HTLV-1 Tax and abrogates Tax transcativation of HTLV-1 LTR. Retrovirology 2006, 3. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Wang, G.Z.; Cingoz, O.; Goff, S.P. NP220 mediated silencing of unintegrated retroviral DNA. Nature 2018, 264, 278–282. [Google Scholar] [CrossRef]
- Arbuckle, J.H.; Kristie, T.M. Epigenetic repression of herpes simplex virus infection by the nucleosome remodeler CHD3. mBio 2014, 5, e01027-13. [Google Scholar] [CrossRef] [Green Version]
- Rai, T.S.; Glass, M.; Cole, J.J.; Rather, M.I.; Marsden, M.; Neilson, M.; Brock, C.; Humphreys, I.R.; Everett, R.D.; Adams, P.D. Histone chaperone HIRA deposits histone H3.3 onto foreign viral DNA and contributes to anti-viral intrinsic immunity. Nucleic Acids Res. 2017, 45, 11673–11683. [Google Scholar] [CrossRef] [Green Version]
- Schreiner, S.; Burck, C.; Glass, M.; Groitl, P.; Wimmer, P.; Kinkley, S.; Mund, A.; Everett, R.D.; Dobner, T. Control of human adenovirus type 5 gene expression by cellular Daxx/ATRX chromatin-associated complexes. Nucleic Acids Res. 2013, 41, 3532–3550. [Google Scholar] [CrossRef] [Green Version]
- Okada, T.; Uchibori, R.; Iwata-Okada, M.; Takahashi, M.; Nomoto, T.; Nonaka-Sarukawa, M.; Ito, T.; Liu, Y.; Mizukami, H.; Kume, A.; et al. A histone deacetylase inhibitor enhances recombinant adeno-assoicated virus-mediated gene expression in tumor cells. Mol. Ther. 2006, 13, 738–746. [Google Scholar] [CrossRef]
- Geis, F.K.; Goff, S.P. Unintegrated HIV-1 DNAs are loaded with core and linker histones and transcreiptionally silenced. Proc. Natl. Acad. Sci. USA 2019, 116, 23735–23742. [Google Scholar] [CrossRef]
- Poon, B.; Chen, I.S. Human immunodeficiency virus type 1 (HIV-1) Vpr enhances expression from unintegration HIV-1 DNA. J. Viral Entry 2003, 77, 3962–3972. [Google Scholar]
- Laguette, N.; Bregnard, C.; Hue, P.; Basbous, J.; Ytaim, A.; Larroque, M.; Kirchhoff, F.; Constantinou, A.; Sobhian, B.; Benikirane, M. Premature activation of the SLX4 coplex by Vpr promotes G2/M arrest and escape from innate immune sensing. Cell 2014, 156, 134–145. [Google Scholar] [CrossRef] [Green Version]
- Irwan, I.D.; Cullen, B.R. Tax induces the recruitment of NF-kB to unintegrated HIV-1 DNA to rescue viral gene expression and replication. J. Virol. 2021, in press. [Google Scholar] [CrossRef]
- Dupont, L.; Bloor, S.; Williamson, J.C.; Cuesta, S.M.; Shah, R.; Teixeira-Silva, A.; Naarmati, A.; Greenwood, E.J.D.; Sarafianos, S.G.; Matheson, N.J.; et al. The SMC5/6 complex compacts and silences unintegrated HIV-1 DNA and is antagnoized by Vpr. Cell Host Microbe 2021, 29, 792–805. [Google Scholar] [CrossRef]
- Makita, C.; Tobinai, K. Targeting EZH@ with tazometostat. Lancet Oncol. 2018, 19, 586–587. [Google Scholar] [CrossRef]
- Iwanaga, M.; Watanabe, T.; Utsonomiya, A.; Okayama, A.; Uchimaru, K.; Koh, K.R.; Ogata, M.; Kikuchi, H.; Sagara, Y.; Uozumi, K.; et al. Human T-cell leukemia virus type 1 (HTLV-1) proviral load and disease progression in a symptomatic HTLV-1 cariers: A nationwide prospective study in Japan. Blood 2010, 116, 1211–1219. [Google Scholar] [CrossRef] [Green Version]
- Katsuya, H.; Yamanka, T.; Ishitsuka, K.; Utsonomiya, A.; Sasaki, H.; Hanada, S.; Eto, T.; Moriuchi, Y.; Saburi, Y.; Miyahara, M.; et al. Prognostic index for acute- and lymphoma-type adult T-cell leukemia/lymphoma. J. Clin. Oncol. 2012, 30, 1635–1640. [Google Scholar] [CrossRef]
- Song, Z.; Wu, W.; Chen, M.; Cheng, W.; Yu, J.; Fang, J.; Xu, L.; Yasunaga, J.-I.; Matsuoka, M.; Zhao, T. Long noncoding RNA ANRIL supports proliferation of adult T-cell leukemia cells through cooperation with EZH2. J. Virol. 2018, 92, e00909–e00918. [Google Scholar] [CrossRef] [Green Version]
- Yamagishi, M.; Hori, M.; Fujikawa, D.; Ohsugi, T.; Honma, D.; Adachi, N.; Katano, H.; Hishima, T.; Kobayashi, S.; Nakano, K.; et al. Targeting excessive EZH1 and EZH2 activities for abnormal histone methylation and transcription network in malignant lymphomas. Cell Rep. 2019, 29, 2321–2337. [Google Scholar] [CrossRef]
- Morishima, S.; Ishitsuka, K.; Izutsu, K.; Kusumoto, S.; Makiyama, J.; Utsunomiya, A.; Nosaka, K.; Ishida, T.; Imaizumi, Y.; Yamauchi, N.; et al. First-in-human study of hte EZH1/2 dual inhibitor valemetostat in relapsed or refractory non-Hodgkin lymphoma (NHL)—Updates results focusing on adult T-cell leukemia-lymphoma (ATL). Blood 2019, 134 (Suppl. 1), 4025. [Google Scholar] [CrossRef]
- Kulkarni, A.; Taylor, G.P.; Klose, R.J.; Schofield, C.J.; Bangham, C.R. Histone H2A monoubiquitylation and p38-MAPKs regulated immediate-early gene-like reactivation of latent retroviurs HTLV-1. JCI Insight 2018, 3, e123196. [Google Scholar] [CrossRef]
- Alberts, B. Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002. [Google Scholar]
- Zhou, Y.B.; Gerchman, S.E.; Ramakrishnan, V.; Travers, A.; Muyldermans, S. Position and oreintation of the globular domain of linker histone H5 on the nucleosome. Nature 1998, 395, 402–405. [Google Scholar] [CrossRef]
- Lemasson, I.; Polakowski, N.J.; Laybourn, P.J.; Nyborg, J.K. Tax-dependent displacement of nucleosomes during transcriptional activation of human T-cell leukemia virus type 1. J. Biol. Chem. 2006, 281, 13075–13082. [Google Scholar] [CrossRef] [Green Version]
- Sharma, N.; Nyborg, J.K. The coactivators of CBP/p300 and the histone chaperone NAP1 promote transcription-independent nucleosome eviction at the HTLV-1 promoter. Proc. Natl. Acad. Sci. USA 2008, 105, 7959–7963. [Google Scholar] [CrossRef] [Green Version]
- Wu, K.; Bottazzi, M.E.; delaFuente, C.; Deng, L.; Gitlin, S.D.; Maddukuri, A.; Dadgar, S.; Li, H.; Vertes, A.; Pumfery, A.; et al. Protein profile of tax-associated complexes. J. Biol. Chem. 2004, 279, 495–508. [Google Scholar] [CrossRef] [Green Version]
- Alasiri, A.; Guerr, J.A.; Hall, W.W.; Sheehy, N. Novel interactions between the human T-cell leukemia virus type 1 antisense protein HBZ and the SWI/SNF chromatin remodeling family: Implications for viral life cycle. J. Virol. 2019, 93, e00412–e00419. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Liu, M.; Merling, R.; Giam, C.-Z. Versatile reporter systems show that transactivation by human T-cell leukemia virus type 1 Tax occurs independently of chromatin remodeling factor BRG1. J. Virol. 2006, 80, 7459–7468. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.; Santoso, N.; Power, D.; Simposn, S.; Dieringer, M.; Miao, H.; Gurova, K.; Giam, C.-Z.; Elledge, S.J.; Zhu, J. FACT proteins, SUPT16H and SSRP1, are transcriptional suppressor of HIV-1 and HTLV-1 that facilitate viral latency. J. Biol. Chem. 2015, 290, 27297–27310. [Google Scholar] [CrossRef] [Green Version]
- Aliya, N.; Rahman, S.; Khan, Z.K.; Jain, P. Corranscriptional chromatin remodeling by small RNA species: An HTLV-1 perspective. Leuk. Res. Treat. 2012, 2012, 984754. [Google Scholar]
- Wang, J.; Lawry, S.T.; Cohen, A.L.; Jia, S. Chromosome boundary elements and regulation of heterochromatin spreading. Cell. Mol. Life Sci. 2014, 71, 4841–4852. [Google Scholar] [CrossRef] [Green Version]
- Valenzuela, L.; Kamakaka, R.T. Chromatin insulators. Annu. Rev. Genet. 2006, 40, 107–138. [Google Scholar] [CrossRef] [Green Version]
- Guelen, L.; Pagie, L.; Brasset, E.; Meuleman, W.; Faza, M.B.; Talhout, W.; Eussen, B.H.; de Klein, A.; Wessels, L.; de Laat, W.; et al. Domain organization of human choromosomes revealed by mapping of nuclear lamina interactions. Nature 2008, 453, 948–951. [Google Scholar] [CrossRef]
- Nakahashi, H.; Kwon, K.-R.K.; Resch, W.; Vian, L.; Dose, M.; Stavreva, D.; Hakim, O.; Pruett, N.; Nelson, S.; Yamane, A.; et al. A genome-wide map of CTCF multivalency redefines the CTCF code. Cell Rep. 2013, 3, 1678–1689. [Google Scholar] [CrossRef] [Green Version]
- Schoenfelder, S.; Sugar, R.; Dimond, A.; Javierre, B.M.; Armstrong, H.; Mifsud, B.; Dimitrova, E.; Matheson, L.; Tavares-Cadete, F.; Furlan-Magani, M.; et al. Polycomb repressive complex PRC1 spatially constrains the mouse mebryonic stem cell genome. Nat. Genet. 2015, 47, 1179–1186. [Google Scholar] [CrossRef]
- Phanstiel, D.H.; Van Bortle, K.; Spacek, D.; Hess, G.T.; Shamim, M.S.; Machol, I.; Love, M.I.; Aiden, E.L.; Bassik, M.C.; Snyder, M.P. Static and dynamic DNA loops from AP-1 bound activation hubs during macrophage development. Mol. Cell 2017, 67, 1037–1048. [Google Scholar] [CrossRef] [Green Version]
- Beagan, J.A.; Duong, M.T.; Titus, K.R.; Zhou, L.; Cao, Z.; Ma, J.; Lachanski, C.V.; Gillis, D.R.; Phillips-Cremins, J.E. YY1 and CTCF orchestrate a 3D chromatin looping switch during early neural lineage commitment. Genome Res. 2017, 27, 1139–1152. [Google Scholar] [CrossRef] [Green Version]
- Bartman, C.R.; Hsu, S.C.; Hsiung, C.C.; Raj, A.; Blobel, G.A. Enhancer regulation of transcriptional bursting parameters revealed by forced chromatin looping. Mol. Cell 2016, 62, 237–247. [Google Scholar] [CrossRef] [Green Version]
- Corces, M.R.; Corces, V.G. The three-dimensional cancer genome. Curr. Opin. Genet. Dev. 2016, 36, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Lupianez, D.G.; Kraft, K.; Heinrich, V.; Krawitz, P.; Brancati, F.; Klopocki, E.; Horn, D.; Kaysenli, H.; Opitz, J.M.; Laxova, R.; et al. Disruption of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 2015, 161, 1012–1025. [Google Scholar] [CrossRef] [Green Version]
- Clarkson, D.T.; Deeks, E.A.; Samarista, R.; Mamayusupova, H.; Zhurkin, V.B.; Teif, V.B. CTCF-dependent chromatin boundaries formed by asymmetric nucleosome arrays wtih decreased linker length. Nucleic Acids Res. 2019, 47, 11181–11196. [Google Scholar] [CrossRef]
- Tempera, I.; Lieberman, P.M. Epigenetic regulation of EBV persistence. Semin. Cancer Biol. 2014, 26, 22–29. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.-S.; Martin, K.A.; Lu, F.; Lupey, L.N.; Mueller, J.M.; Lieberman, P.M.; Tempera, I. Epigenetic deregulation of the LMP1/LMP2 locus of Epstein-Barr virus by mutation of a single CTCF-cohesin binding site. J. Virol. 2014, 88, 1703–1713. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.-S.; Wikramasinghe, P.; Showe, L.; Lieberman, P.M. Cohesins repress Kaposi’s sarcoma-associated herpesvirus immediate early gene transcription during latency. J. Virol. 2012, 86, 9454–9464. [Google Scholar] [CrossRef] [Green Version]
- Bangham, C.R.M.; Cook, L.B.; Melamed, A. HTLV-1 clonality in adult T-cell leukaemia and non-malignant HTLV-1 infection. Semin. Cancer Biol. 2014, 26, 89–98. [Google Scholar] [CrossRef] [Green Version]
- Kang, H.; Cho, H.; Sung, G.-H.; Lieberman, P.M. CTCF regulates Kaposi’s sarcoma-associated herpesvirus latency transcription by nucleosome displacement and RNA polymerase programming. J. Virol. 2013, 87, 1789–1799. [Google Scholar] [CrossRef] [Green Version]
- Martinez, F.P.; Cruz, R.; Lu, F.; Plasschaert, R.; Deng, Z.; Rivera-Molina, Y.A.; Bartolomei, M.S.; Lieberman, P.M.; Tang, Q. CTCF binding of the first intron of the major immediate early (MIE) gene of human cytomegalovirus (HCMV) negatively regulates MIE gene expression and HCMV replication. J. Virol. 2014, 88, 7389–7401. [Google Scholar] [CrossRef] [Green Version]
- Komatsu, T.; Sekiya, T.; Nagata, K. DNA replication-dependent binding of CTCF plays a critical role in adenovirus genome functions. Sci. Rep. 2013, 3, 2187. [Google Scholar] [CrossRef] [Green Version]
- Rebollo, R.; Miceli-Royer, K.; Zhang, Y.; Farivar, S.; Gagnier, L.; Mager, D.L. Epigenetic interplay between mouse endogenous retroviruses and host genes. Genome Biol. 2012, 13, R89. [Google Scholar] [CrossRef] [Green Version]
- Satou, Y.; Miyazato, P.; Ishihara, K.; Yaguchi, H.; Melamed, A.; Miura, M.; Fukuda, A.; Nosaka, K.; Watanabe, T.; Rowan, A.G.; et al. The retrovirus HTLV-1 inserts an ectopic CTCF-binding site into the human genome. Proc. Natl. Acad. Sci. USA 2016, 113, 3054–3059. [Google Scholar] [CrossRef] [Green Version]
- Miura, M.; Miyazato, P.; Satou, Y.; Tanaka, Y.; Bangham, C.R.M. Epigenetic changes around the pX region and spontaneous HTLV-1 transcription are CTCF-independent. Wellcome Open Res. 2018, 3, 105. [Google Scholar] [CrossRef]
- Cheng, X.; Joseph, A.; Castro, V.; Chien-Liaw, A.; Skidmore, Z.; Ueno, T.; Fujisawa, J.-i.; Rauch, D.A.; Challen, G.A.; Martinez, M.P.; et al. Epigenomic regulation of human T-cell leukemia virus by chromatin-insulator CTCF. PLoS Pathog. 2021, 17, e1009577. [Google Scholar] [CrossRef]
- Melamed, A.; Yaguchi, H.; Miura, M.; Witkover, A.; Fitzgeerald, T.W.; Birney, E.; Bangham, C.R. The human leukemia virus HTLV-1 alters the structure and transcription of host chromatin in cis. Elife 2018, 7, e36245. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ratner, L. Epigenetic Regulation of Human T-Cell Leukemia Virus Gene Expression. Microorganisms 2022, 10, 84. https://doi.org/10.3390/microorganisms10010084
Ratner L. Epigenetic Regulation of Human T-Cell Leukemia Virus Gene Expression. Microorganisms. 2022; 10(1):84. https://doi.org/10.3390/microorganisms10010084
Chicago/Turabian StyleRatner, Lee. 2022. "Epigenetic Regulation of Human T-Cell Leukemia Virus Gene Expression" Microorganisms 10, no. 1: 84. https://doi.org/10.3390/microorganisms10010084
APA StyleRatner, L. (2022). Epigenetic Regulation of Human T-Cell Leukemia Virus Gene Expression. Microorganisms, 10(1), 84. https://doi.org/10.3390/microorganisms10010084




