An Evaluation of the Pathogenic Potential, and the Antimicrobial Resistance, of Salmonella Strains Isolated from Mussels
Abstract
1. Introduction
2. Materials and Methods
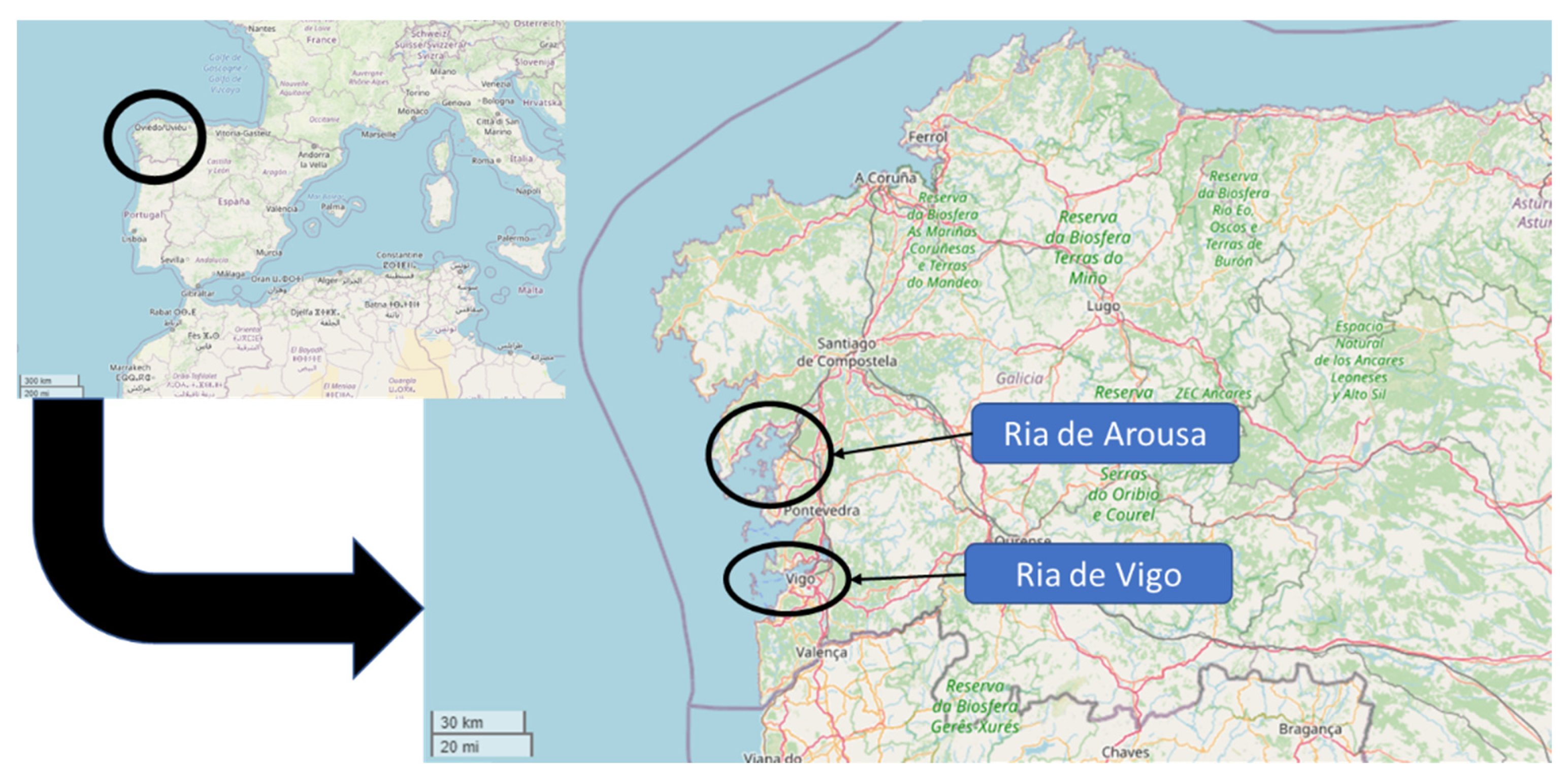
2.1. Sampling
2.2. DNA Extraction Protocol
2.3. Screening of Salmonella spp. by qPCR in Mussel Samples
2.4. Genetic Characterization of the Isolates
2.4.1. Screening for Virulence Genes
2.4.2. Screening for Antibiotic Resistance Genes
2.5. PCR Tests for Isolate Characterization
2.6. Antimicrobial Resistance Test
3. Results and Discussion
3.1. Prevalence and Serotypes of Salmonella enterica
3.2. Genetic Characterization of the Isolates
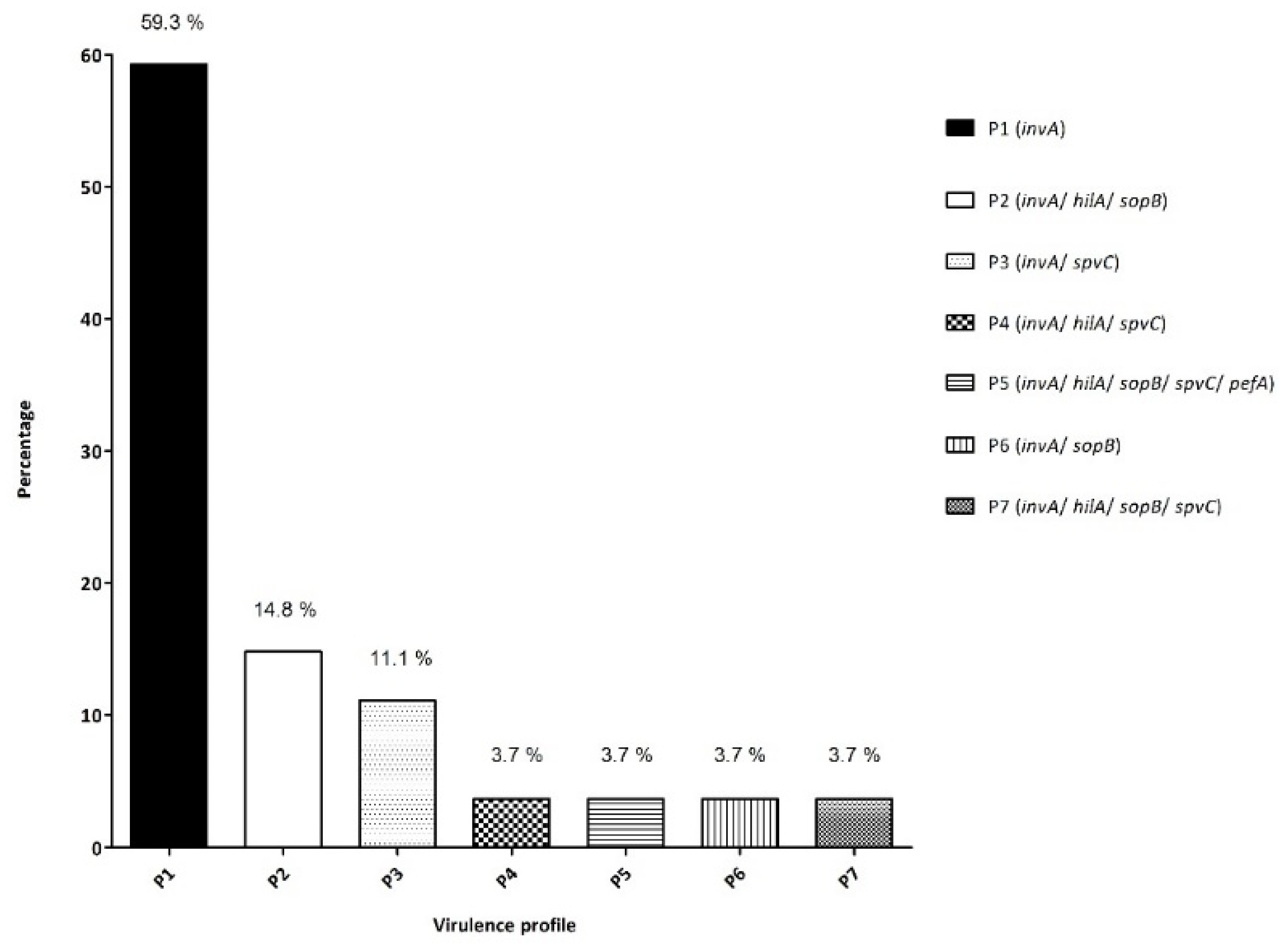
3.2.1. Screening for Virulence Genes
3.2.2. Screening for Antimicrobial Resistance Genes
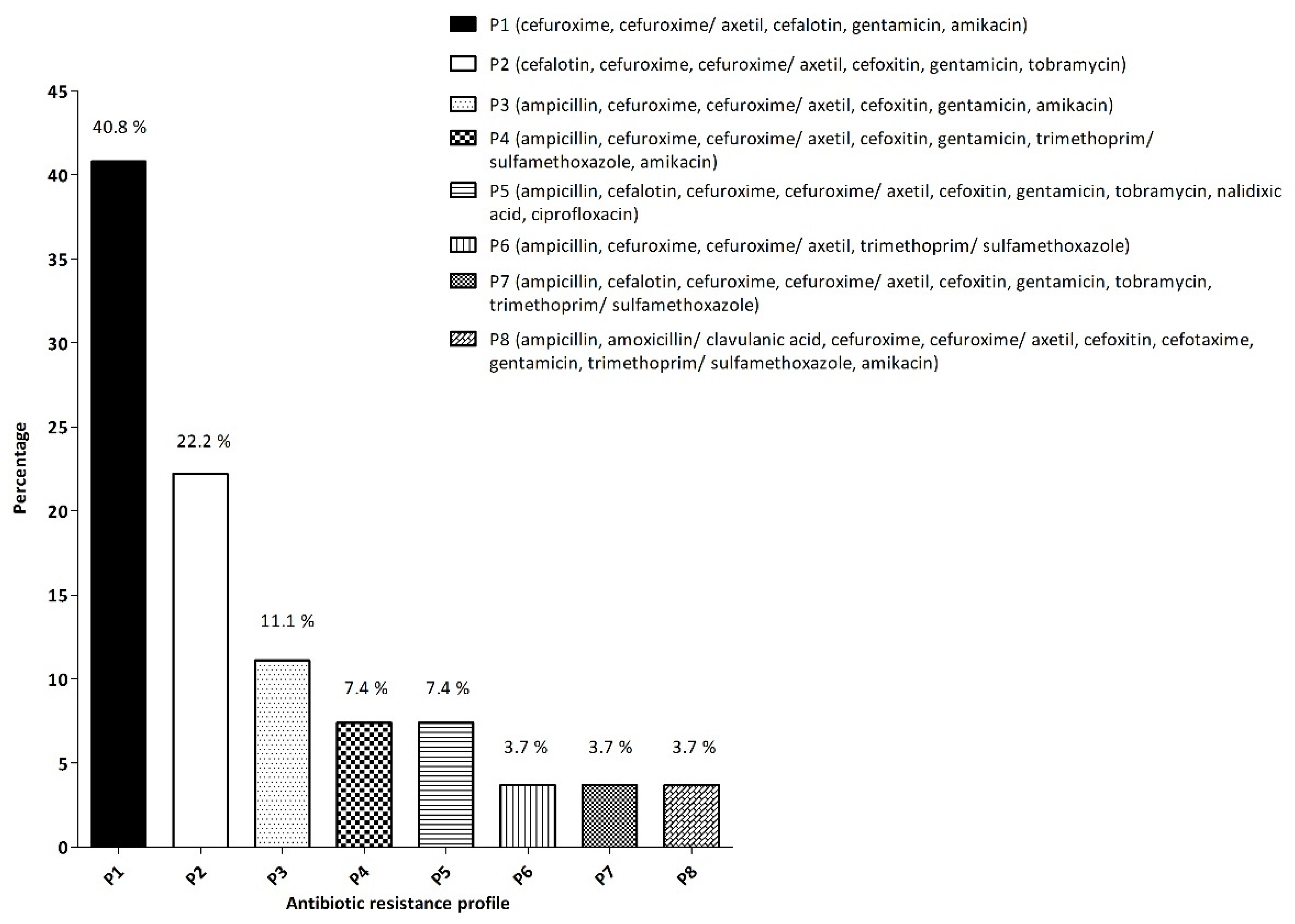
3.3. Antimicrobial Resistance
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- EFSA; ECDC. The European Union One Health 2020 Zoonoses Report. EFSA J. 2021, 19, e6971. [Google Scholar]
- European Commission Regulation (EC) No 2073/2005. Microbiological Criteria for Foodstuffs; European Union: Brussels, Belgium, 2005.
- EFSA. Foodborne Outbreaks Reported in 2019. Available online: https://app.powerbi.com/view?r=eyJrIjoiY2FmNmUzYWEtZjg4Zi00ODJiLTkwMDMtOGUzYTY0YzYxMWY5IiwidCI6ImM0ODdkZDVhLTM3NjktNDQyYy1hYjc3LTI5MTkwODFkODVmYyIsImMiOjl9 (accessed on 7 December 2021).
- Mir, R.A.; Weppelmann, T.A.; Johnson, J.A.; Archer, D.; Morris, J.G.; Jeong, K.C.C. Identification and characterization of cefotaxime resistant bacteria in beef cattle. PLoS ONE 2016, 11, e0163279. [Google Scholar] [CrossRef] [PubMed]
- Mir, R.A.; Weppelmann, T.A.; Teng, L.; Kirpich, A.; Elzo, M.A.; Driver, J.D.; Jeong, K.C. Colonization Dynamics of Cefotaxime Resistant Bacteria in Beef Cattle Raised Without Cephalosporin Antibiotics. Front. Microbiol. 2018, 9, 500. [Google Scholar] [CrossRef]
- Chen, S.; Zhao, S.; White, D.G.; Schroeder, C.M.; Lu, R.; Yang, H.; McDermott, P.F.; Ayers, S.; Meng, J. Characterization of Multiple-Antimicrobial-Resistant Salmonella Serovars Isolated from Retail Meats. Appl. Environ. Microbiol. 2004, 70, 1–7. [Google Scholar] [CrossRef]
- EFSA; ECDC (European Food Safety Authority and European Centre for Disease Prevention and Control). The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria. EFSA J. 2021, 18. [Google Scholar]
- Ferreira, M.; Lago, J.; Vieites, J.M.; Cabado, A.G. Chapter 8: World Production of Bivalve Mollusks and Socioeconomic Facts Related to Teh Impact of Marine Biotoxins, in Seafood and Freshwater Toxins: Pharmacology, Physiology, and Detection, 3rd ed.; Botana, L.M., Ed.; Taylor & Francis Group: Abingdon, UK, 2014; ISBN 978-1-4665-0514-8. [Google Scholar]
- Caballero Miguez, G.; Garza Gil, M.D.; Varela Lafuente, M.M. The institutional foundations of economic performance of mussel production: The Spanish case of the Galician floating raft culture. Mar. Policy 2009, 33, 288–296. [Google Scholar] [CrossRef]
- Caballero-Miguez, G.; Garza-Gil, M.D.; Varela-Lafuente, M.M. Legal change, property rights system and institutional stability: The case of the floating raft culture in the Galician mussel sector. Ocean Coast. Manag. 2012, 55, 84–93. [Google Scholar] [CrossRef]
- Costas-Rodríguez, M.; Lavilla, I.; Bendicho, C. Classification of cultivated mussels from Galicia (Northwest Spain) with European protected designation of origin using trace element fingerprint and chemometric analysis. Anal. Chim. Acta 2010, 664, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Souto, R.R.; Garrido-Maestu, A.; Pastoriza-Fontan, A.; Lozano-Leon, A. Investigation and characterization of Shiga toxin-producing Escherichia coli present in mussels from harvesting areas in Galician southern Rias (NW Spain). J. Food Saf. 2017, 37, e12367. [Google Scholar] [CrossRef]
- Garrido-Maestu, A.; Lozano-León, A.; Rodríguez-Souto, R.R.; Vieites-Maneiro, R.; Chapela, M.-J.; Cabado, A.G. Presence of pathogenic Vibrio species in fresh mussels harvested in the southern Rias of Galicia (NW Spain). Food Control 2016, 59, 759–765. [Google Scholar] [CrossRef]
- Lozano-León, A.; Rodríguez-Souto, R.R.; González-Escalona, N.; Llovo-Taboada, J.; Iglesias-Canle, J.; Álvarez-Castro, A.; Garrido-Maestu, A. Detection, molecular characterization, and antimicrobial susceptibility, of Campylobacter spp. isolated from shellfish. Microb. Risk Anal. 2021, 18, 100176. [Google Scholar] [CrossRef]
- Martinez-Urtaza, J.; Saco, M.; Hernandez-Cordova, G.; Lozano, A.; Garcia-Martin, O.; Espinosa, J. Identification of Salmonella Serovars Isolated from Live Molluscan Shellfish and Their Significance in the Marine Environment. J. Food Prot. 2003, 66, 226–232. [Google Scholar] [CrossRef]
- Martinez-Urtaza, J.; Liebana, E.; Garcia-Migura, L.; Perez-Pin, P.; Perez-Piñeiro, P.; Saco, M. Characterization of Salmonella enterica serovar Typhimurium from marine environments in coastal waters of Galicia (Spain). Appl. Environ. Microbiol. 2004, 70, 4030–4034. [Google Scholar] [CrossRef] [PubMed]
- Van Asten, A.J.A.M.; Van Dijk, J.E. Distribution of “classic” virulence factors among Salmonella spp. FEMS Immunol. Med. Microbiol. 2005, 44, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Swamy, S.C.; Barnhart, H.M.; Lee, M.D.; Dreesen, D.W. Virulence determinants invA and spvC in Salmonellae isolated from poultry products, wastewater, and human sources. Appl. Environ. Microbiol. 1996, 62, 3768–3771. [Google Scholar] [CrossRef]
- Rahn, K.; De Grandis, S.A.; Clarke, R.C.; McEwen, S.A.; Galan, J.E.; Ginocchio, C.; Curtiss, R.; Gyles, C.L. Amplification of an invA gene sequence of Salmonella Typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol. Cell. Probes 1992, 6, 271–279. [Google Scholar] [CrossRef]
- Murugkar, H.V.; Rahman, H.; Dutta, P.K. Distribution of virulence genes in Salmonella serovars isolated from man & animals. Indian J. Med. Res. 2003, 117, 66–70. [Google Scholar]
- Cardona-Castro, N.; Restrepo-Pineda, E.; Correa-Ochoa, M. Detection of hilA gene sequences in serovars of Salmonella enterica subspecies enterica. Mem. Inst. Oswaldo Cruz 2002, 97, 1153–1156. [Google Scholar] [CrossRef][Green Version]
- Rahman, H. Prevalence & phenotypic expression of sopB gene among clinical isolates of Salmonella enterica. Indian J. Med. Res. 2006, 123, 83. [Google Scholar]
- Fritsche, T.R.; Castanheira, M.; Miller, G.H.; Jones, R.N.; Armstrong, E.S. Detection of methyltransferases conferring high-level resistance to aminoglycosides in Enterobacteriaceae from Europe, North America, and Latin America. Antimicrob. Agents Chemother. 2008, 52, 1843–1845. [Google Scholar] [CrossRef]
- Leflon-Guibout, V.; Jurand, C.; Bonacorsi, S.; Espinasse, F.; Guelfi, M.C.; Duportail, F.; Heym, B.; Bingen, E.; Nicolas-Chanoine, M.H. Emergence and spread, of three clonally related virulent isolates of CTX-M-15-producing Escherichia coli with variable resistance to aminoglycosides and tetracycline in a French geriatric hospital. Antimicrob. Agents Chemother. 2004, 48, 3736–3742. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 9th ed.; CLSI: Pittsburgh, PA, USA, 2012; Volume 32, ISBN 1562387839.
- Martínez, O.; Rodríguez-Calleja, J.M.; Santos, J.A.; Otero, A.; García-López, M.L. Foodborne and indicator bacteria in farmed molluscan shellfish before and after depuration. J. Food Prot. 2009, 72, 1443–1449. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Leon, A.; Garcia-Omil, C.; Dalama, J.; Rodriguez-Souto, R.; Martinez-Urtaza, J.; Gonzalez-Escalona, N. Detection of colistin resistance mcr-1 gene in Salmonella enterica serovar Rissen isolated from mussels, Spain, 2012 to 2016. Eurosurveillance 2019, 24, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Zahli, R.; Soliveri, J.; Abrini, J.; Copa-Patiño, J.L.; Nadia, A.; Scheu, A.-K.; Nadia, S.S. Prevalence, typing and antimicrobial resistance of Salmonella isolates from commercial shellfish in the North coast of Morocco. World J. Microbiol. Biotechnol. 2021, 37, 170. [Google Scholar] [CrossRef]
- Setti, I.; Rodriguez-Castro, A.; Pata, M.P.; Cadarso-Suarez, C.; Yacoubi, B.; Bensmael, L.; Moukrim, A.; Martinez-Urtaza, J. Characteristics and dynamics of Salmonella contamination along the coast of agadir, Morocco. Appl. Environ. Microbiol. 2009, 75, 7700–7709. [Google Scholar] [CrossRef] [PubMed]
- Mannas, H.; Mimouni, R.; Chaouqy, N.; Hamadi, F.; Martinez-Urtaza, J. Occurrence of Vibrio and Salmonella species in mussels (Mytilus galloprovincialis) collected along the Moroccan Atlantic coast. Springerplus 2014, 3, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Urtaza, J.; Peiteado, J.; Lozano-León, A.; Garcia-Martin, O. Detection of Salmonella Senftenberg Associated with High Saline Environments in Mussel Processing Facilities. J. Food Prot. 2004, 67, 256–263. [Google Scholar] [CrossRef]
- Martinez-Urtaza, J.; Saco, M.; De Novoa, J.; Perez-Piñeiro, P.; Peiteado, J.; Lozano-Leon, A.; Garcia-Martin, O. Influence of Environmental Factors and Human Activity on the Presence of Salmonella Serovars in a Marine Environment. Appl. Environ. Microbiol. 2004, 70, 2089–2097. [Google Scholar] [CrossRef] [PubMed]
- Obe, T.; Nannapaneni, R.; Sharma, C.S.; Kiess, A. Homologous stress adaptation, antibiotic resistance, and biofilm forming ability of Salmonella enterica serovar Heidelberg ATCC8326 on different food-contact surfaces following exposure to sublethal chlorine concentrations 1. Poult. Sci. 2018, 97, 951–961. [Google Scholar] [CrossRef]
- Soto-Varela, Z.E.; Rosado-Porto, D.; Bolívar-Anillo, H.J.; González, C.P.; Pantoja, B.G.; Alvarado, D.E.; Anfuso, G. Preliminary microbiological coastal water quality determination along the department of atlántico (Colombia): Relationships with beach characteristics. J. Mar. Sci. Eng. 2021, 9, 122. [Google Scholar] [CrossRef]
- EFSA; ECDC. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2016. EFSA J. 2017, 15, e05077. [Google Scholar] [CrossRef]
- Lamas, A.; Fernandez-No, I.C.; Miranda, J.M.; Vázquez, B.; Cepeda, A.; Franco, C.M. Prevalence, molecular characterization and antimicrobial resistance of Salmonella serovars isolated from northwestern Spanish broiler flocks (2011–2015). Poult. Sci. 2016, 95, 2097–2105. [Google Scholar] [CrossRef] [PubMed]
- Díez-García, M.; Capita, R.; Alonso-Calleja, C. Influence of serotype on the growth kinetics and the ability to form biofilms of Salmonella isolates from poultry. Food Microbiol. 2012, 31, 173–180. [Google Scholar] [CrossRef]
- Cheng, C.M.; Lin, W.; Van, K.T.; Phan, L.; Tran, N.N.; Farmer, D. Rapid Detection of Salmonella in Foods Using Real-Time PCR. J. Food Prot. 2008, 71, 2436–2441. [Google Scholar] [CrossRef] [PubMed]
- Hara-Kudo, Y.; Yoshino, M.; Kojima, T.; Ikedo, M. Loop-mediated isothermal amplification for the rapid detection of Salmonella. FEMS Microbiol. Lett. 2005, 253, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Malorny, B.; Cook, N.; D’Agostino, M.; De Medici, D.; Croci, L.; Abdulmawjood, A.; Fach, P.; Karpiskova, R.; Aymerich, T.; Kwaitek, K.; et al. Multicenter validation of PCR-based method for detection of Salmonella in chicken and pig samples. J. AOAC Int. 2004, 87, 861–866. [Google Scholar] [CrossRef]
- Boddicker, J.D.; Knosp, B.M.; Jones, B.D. Transcription of the Salmonella invasion gene activator, hilA, requires HilD activation in the absence of negative regulators. J. Bacteriol. 2003, 185, 525–533. [Google Scholar] [CrossRef]
- Campioni, F.; Moratto Bergamini, A.M.; Falcão, J.P. Genetic diversity, virulence genes and antimicrobial resistance of Salmonella Enteritidis isolated from food and humans over a 24-year period in Brazil. Food Microbiol. 2012, 32, 254–264. [Google Scholar] [CrossRef]
- Libby, S.J.; Lesnick, M.; Hasegawa, P.; Kurth, M.; Belcher, C.; Fierer, J.; Guiney, D.G. Characterization of the spv locus in Salmonella enterica serovar Arizona. Infect. Immun. 2002, 70, 3290–3294. [Google Scholar] [CrossRef][Green Version]
- Ammar, A.M.; Mohamed, A.A.; El-Hamid, M.I.A.; El-Azzouny, M.M. Virulence genotypes of clinical Salmonella serovars from broilers in Egypt. J. Infect. Dev. Ctries. 2016, 10, 337–346. [Google Scholar] [CrossRef]
- Gharieb, R.M.; Tartor, Y.H.; Khedr, M.H.E. Non-Typhoidal Salmonella in poultry meat and diarrhoeic patients: Prevalence, antibiogram, virulotyping, molecular detection and sequencing of class I integrons in multidrug resistant strains. Gut Pathog. 2015, 7, 34. [Google Scholar] [CrossRef]
- Ho, P.L.; Wong, R.C.; Lo, S.W.; Chow, K.H.; Wong, S.S.; Que, T.L. Genetic identity of aminoglycoside-resistance genes in Escherichia coli isolates from human and animal sources. J. Med. Microbiol. 2010, 59, 702–707. [Google Scholar] [CrossRef] [PubMed]
- Cloeckaert, A.; Praud, K.; Doublet, B.; Bertini, A.; Carattoli, A.; Butaye, P.; Imberechts, H.; Bertrand, S.; Collard, J.M.; Arlet, G.; et al. Dissemination of an extended-spectrum-β-lactamase blaTEM-52 gene-carrying IncI1 plasmid in various Salmonella enterica serovars isolated from poultry and humans in Belgium and France between 2001 and 2005. Antimicrob. Agents Chemother. 2007, 51, 1872–1875. [Google Scholar] [CrossRef]
- Wachino, J.I.; Shibayama, K.; Kurokawa, H.; Kimura, K.; Yamane, K.; Suzuki, S.; Shibata, N.; Ike, Y.; Arakawa, Y. Novel plasmid-mediated 16S rRNA m1A1408 methyltransferase, NpmA, found in a clinically isolated Escherichia coli strain resistant to structurally diverse aminoglycosides. Antimicrob. Agents Chemother. 2007, 51, 4401–4409. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Urtaza, J.; Liebana, E. Use of pulsed-field gel electrophoresis to characterize the genetic diversity and clonal persistence of Salmonella Senftenberg in mussel processing facilities. Int. J. Food Microbiol. 2005, 105, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Michael, G.B.; Schwarz, S. Antimicrobial resistance in zoonotic nontyphoidal Salmonella: An alarming trend? Clin. Microbiol. Infect. 2016, 22, 968–974. [Google Scholar] [CrossRef] [PubMed]
- Giacometti, F.; Pezzi, A.; Galletti, G.; Tamba, M.; Merialdi, G.; Piva, S.; Serraino, A.; Rubini, S. Antimicrobial resistance patterns in Salmonella enterica subsp. enterica and Escherichia coli isolated from bivalve molluscs and marine environment. Food Control 2021, 121, 107590. [Google Scholar] [CrossRef]
- Álvarez-Fernández, E.; Alonso-Calleja, C.; García-Fernández, C.; Capita, R. Prevalence and antimicrobial resistance of Salmonella serotypes isolated from poultry in Spain: Comparison between 1993 and 2006. Int. J. Food Microbiol. 2012, 153, 281–287. [Google Scholar] [CrossRef]
- EFSA. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2015. EFSA J. 2017, 15, e04694. [Google Scholar] [CrossRef]
- Prioritarias, T. Boletín Epidemiolóxico de Galicia; Galician Healthcare Service: Galicia, Spain, 2018; Volume 27, pp. 1–23. [Google Scholar]
- European Medicines Agency. Sales of Veterinary Antimicrobial Agents in 31 European Countries in 2018: Trends from 2010–2018; European Medicines Agency: Amsterdam, The Netherlands, 2019; ISBN 9789291550685.
- Collignon, P.C.; Conly, J.M.; Andremont, A.; McEwen, S.A.; Aidara-Kane, A.; Griffin, P.M.; Agerso, Y.; Dang Ninh, T.; Donado-Godoy, P.; Fedorka-Cray, P.; et al. World Health Organization Ranking of Antimicrobials According to Their Importance in Human Medicine: A Critical Step for Developing Risk Management Strategies to Control Antimicrobial Resistance from Food Animal Production. Clin. Infect. Dis. 2016, 63, 1087–1093. [Google Scholar] [CrossRef]
| Primers | Target Gene | Sequence (5′-3′) | Reference |
|---|---|---|---|
| invA-f | invA | GTG AAA TTA TCG CCA CGT TCG GGC AA | [19] |
| invA-r | TCA TCG CAC CGT CAA AGG AAC C | ||
| spvC-1 | spvC | CGG AAA TAC CAT CTA CAA ATA | [18] |
| spvC-2 | CCC AAA CCC ATA CTT ACT CTG | ||
| pefA1 | pefA | TGT TTC CGG GCT TGT GCT | [20] |
| pefA2 | CAG GGC ATT TGC TGA TTC TTC C | ||
| hilA DS | hilA | CGG AAG CTT ATT TGC GCC ATG CTG AGG TAG | [21] |
| hilA US | GCA TGG ATC CCC GCC GGC GAG ATT GTG | ||
| sopB PRSB1 | sopB | CCA CCG TTC TGG GTA AAC AAG AC | [22] |
| sopB PRSB2 | AGG ATT GAG CTC CTC TGG CGA T | ||
| armA-f | armA | TAT GGG GGT CTT ACT ATT CTG CCTAT | [23] |
| armA-r | TCT TCC ATT CCC TTC TCC TTT | ||
| rmtA-f | rmtA | CTA GCG TCC ATC CTT TCC TC | |
| rmtA-r | TTT GCT TCC ATG CCC TTG CC | ||
| rmtB-f | rmtB | TCA ACG ATG CCC TCA CCT C | |
| rmtB-r | GCA GGG CAA AGG TAA AAT CC | ||
| rmtC-f | rmtC | GCC AAA GTA CTC ACA AGT GG | |
| rmtC-r | CTC AGA TCT GAC CCA AC AAG | ||
| rmtD-f | rmtD | CTG TTT GAA GCC AGC GGA ACG C | |
| rmtD-r | GCG CCT CCA TCC ATT CGG AAT AG | ||
| npmA-f | npmA | CTC AAA GGA ACA AAG ACG G | |
| npmA-r | GAA ACA TGG CCA GAA ACT C | ||
| CTX-M-15-F1 | blaCTX-M-15 | ATA AAA CCG GCA GCG GTG | [24] |
| CTX-M-15-F2 | GAA TTT TGA CGA TCG GGG |
| Gene | Hot-Start | Cycles | Denaturalization | Hybridization | Extension | Final Extension | Fragment Size (bp) |
|---|---|---|---|---|---|---|---|
| invA | 94 °C/5 min | 30 | 93 °C/1 min | 42 °C/1 min | 72 °C/2 min | 72 °C/4 min | 284 |
| spvC | 669 | ||||||
| pefA | 94 °C/5 min | 25 | 94 °C/55 s | 55 °C/55 s | 72 °C/55 s | 72 °C/10 min | 700 |
| hilA | 94 °C/3 min | 30 | 94 °C/1 min | 65 °C/1 min | 72 °C/1 min | 72 °C/10 min | 854 |
| sopB | 94 °C/5 min | 30 | 94 °C/1 min | 55 °C/1 min | 72 °C/2 min | 72 °C/10 min | 1348 |
| armA | 94 °C/5 min | 30 | 94 °C/1 min | 55 °C/1 min | 72 °C/2 min | 72 °C/10 min | 514 |
| rmtA | 635 | ||||||
| rmtB | 459 | ||||||
| rmtC | 752 | ||||||
| rmtD | 375 | ||||||
| npmA | 641 | ||||||
| blaCTX-M-15 | 483 |
| Code | Strain | Year of Isolation | Origin |
|---|---|---|---|
| AMC 28 | S. montevideo * | 2012 | Ria de Arousa |
| AMC 90 | S. rissen * | 2014 | Ria de Arousa |
| AMC 92 | Salmonella spp. | 2014 | Ria de Arousa |
| AMC 93 | Salmonella spp. | 2014 | Ria de Arousa |
| AMC 200 | S. wentworth * | 2014 | Ria de Arousa |
| AMC 238 | S. typhimurium * | 2015 | Ria de Arousa |
| AMC 239 | S. rissen * | 2015 | Ria de Arousa |
| AMC 240 | S. rissen * | 2015 | Ria de Arousa |
| AMC 256 | Salmonella spp. | 2015 | Ria de Arousa |
| AMC 257 | S. offa * | 2015 | Ria de Arousa |
| AMC 265 | S. montevideo * | 2015 | Ria de Arousa |
| AMC 266 | S. senftenberg * | 2015 | Ria de Arousa |
| AMC 267 | S. senftenberg * | 2015 | Ria de Arousa |
| AMC 268 | S. typhimurium * | 2015 | Ria de Vigo |
| AMC 270 | S. agona * | 2015 | Ria de Vigo |
| AMC 281 | Salmonella spp. | 2015 | Ria de Arousa |
| AMC 287 | Salmonella spp. | 2015 | Ria de Arousa |
| AMC 288 | Salmonella spp. | 2015 | Ria de Arousa |
| AMC 289 | S. senftenberg * | 2015 | Ria de Arousa |
| AMC 290 | Salmonella spp. | 2015 | Ria de Arousa |
| AMC 291 | S. typhimurium * | 2015 | Ria de Arousa |
| AMC 294 | S. typhimurium * | 2015 | Ria de Arousa |
| AMC 299 | S. typhimurium * | 2015 | Ria de Arousa |
| AMC 300 | Salmonella spp. | 2015 | Ria de Vigo |
| AMC 301 | S. bredeney * | 2015 | Ria de Arousa |
| AMC 303 | Salmonella spp. | 2016 | Ria de Vigo |
| AMC 327 | S. liverpool * | 2016 | Ria de Vigo |
| Antibiotics | Virulence Genes | |||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain | AMC Code | Area | AMP | AMC | CEF | CXM | CXM/axetil | FOX | CDN | CTX | CAZ | FEP | ETP | IPM | GEN | TOB | NAL | CIP | FOF | NIT | SXT | AMK | TGC | TZP | invA | spvC | pefA | hilA | sopB | armA | rmtA | rmtB | rmtC | rmtD | npmA | blaCTX-M-15 |
| S. montevideo * | 28 | A | S | S | R | R | R | R | S | S | S | S | S | S | R | R | S | S | S | S | S | N | N | N | + | |||||||||||
| S. rissen * | 90 | A | R | S | R | R | R | R | S | S | S | S | S | S | R | R | S | S | S | S | R | N | N | N | + | |||||||||||
| Salmonella spp. | 92 | A | R | R | N | R | R | R | N | R | I | I | S | S | R | N | S | S | N | N | R | R | S | I | + | |||||||||||
| Salmonella spp. | 93 | A | R | I | N | R | R | R | N | S | S | S | S | S | R | N | S | S | N | N | R | R | S | S | + | |||||||||||
| S. wentworth * | 200 | A | S | S | R | R | R | R | S | S | S | S | S | S | R | R | S | S | S | S | S | N | N | N | + | |||||||||||
| S. typhimurium * | 238 | A | R | S | N | R | R | R | N | S | S | S | S | S | R | N | S | S | N | N | S | R | S | S | + | |||||||||||
| S. rissen * | 239 | A | R | S | N | R | R | R | N | S | S | S | S | S | R | N | S | S | N | N | S | R | S | S | + | |||||||||||
| S. rissen * | 240 | A | R | S | N | R | R | R | N | S | S | S | S | S | R | N | S | S | N | N | S | R | S | S | + | |||||||||||
| Salmonella spp. | 256 | A | R | S | N | R | R | R | N | S | S | S | S | S | R | N | S | S | N | N | R | R | S | S | + | + | + | |||||||||
| S. offa * | 257 | A | S | S | N | R | R | R | N | S | S | S | S | S | R | N | S | S | N | N | S | R | S | S | + | |||||||||||
| S. montevideo * | 265 | A | S | S | N | R | R | R | N | S | S | S | S | S | R | N | S | S | N | N | S | R | S | S | + | |||||||||||
| S. senftenberg * | 266 | A | S | S | R | R | R | R | S | S | S | S | S | S | R | R | S | S | S | S | S | N | N | N | + | + | + | |||||||||
| S. senftenberg * | 267 | A | S | S | R | R | R | R | S | S | S | S | S | S | R | R | S | S | S | S | S | N | N | N | + | + | + | |||||||||
| S. typhimurium * | 268 | V | S | S | R | R | R | R | S | S | S | S | S | S | R | R | S | S | S | S | S | N | N | N | + | + | + | + | + | |||||||
| S. agona * | 270 | V | R | S | R | R | R | R | S | S | S | S | S | S | R | R | R | R | S | I | S | N | N | N | + | + | ||||||||||
| Salmonella spp. | 281 | A | R | S | R | R | R | R | S | S | S | S | S | S | R | R | R | R | S | I | S | N | N | N | + | + | + | + | ||||||||
| Salmonella spp. | 287 | A | S | S | N | R | R | R | N | S | S | S | S | S | R | N | S | S | N | N | S | R | S | S | + | + | ||||||||||
| Salmonella spp. | 288 | A | S | S | N | R | R | R | N | S | S | S | S | S | R | N | S | S | N | N | S | R | S | S | + | |||||||||||
| S. senftenberg * | 289 | A | S | S | N | R | R | R | N | S | S | S | S | S | R | N | S | S | N | N | S | R | S | S | + | + | ||||||||||
| Salmonella spp. | 290 | A | S | S | N | R | R | R | N | S | S | S | S | S | R | N | S | S | N | N | S | R | S | S | + | + | + | |||||||||
| S. typhimurium * | 291 | V | S | S | R | R | R | R | S | S | S | S | S | S | R | R | S | S | S | S | S | N | N | N | + | + | ||||||||||
| S. typhimurium * | 294 | A | R | I | N | R | R | S | N | S | S | S | S | S | S | N | S | S | N | N | R | S | S | I | + | |||||||||||
| S. typhimurium * | 299 | A | S | S | N | R | R | R | N | S | S | S | S | S | R | N | S | S | N | N | S | R | S | S | + | |||||||||||
| Salmonella spp. | 300 | V | S | S | N | R | R | R | N | S | S | S | S | S | R | N | S | S | N | N | S | R | S | S | + | |||||||||||
| S. bredeney * | 301 | A | S | S | N | R | R | R | N | S | S | S | S | S | R | N | S | S | N | N | S | R | S | S | + | |||||||||||
| Salmonella spp. | 303 | V | S | S | N | R | R | R | N | S | S | S | S | S | R | N | S | S | N | N | S | R | S | S | + | + | + | |||||||||
| S. liverpool * | 327 | V | S | S | N | R | R | R | N | S | S | S | S | S | R | N | S | S | N | N | S | R | S | S | + | |||||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lozano-León, A.; García-Omil, C.; Rodríguez-Souto, R.R.; Lamas, A.; Garrido-Maestu, A. An Evaluation of the Pathogenic Potential, and the Antimicrobial Resistance, of Salmonella Strains Isolated from Mussels. Microorganisms 2022, 10, 126. https://doi.org/10.3390/microorganisms10010126
Lozano-León A, García-Omil C, Rodríguez-Souto RR, Lamas A, Garrido-Maestu A. An Evaluation of the Pathogenic Potential, and the Antimicrobial Resistance, of Salmonella Strains Isolated from Mussels. Microorganisms. 2022; 10(1):126. https://doi.org/10.3390/microorganisms10010126
Chicago/Turabian StyleLozano-León, Antonio, Carlos García-Omil, Rafael R. Rodríguez-Souto, Alexandre Lamas, and Alejandro Garrido-Maestu. 2022. "An Evaluation of the Pathogenic Potential, and the Antimicrobial Resistance, of Salmonella Strains Isolated from Mussels" Microorganisms 10, no. 1: 126. https://doi.org/10.3390/microorganisms10010126
APA StyleLozano-León, A., García-Omil, C., Rodríguez-Souto, R. R., Lamas, A., & Garrido-Maestu, A. (2022). An Evaluation of the Pathogenic Potential, and the Antimicrobial Resistance, of Salmonella Strains Isolated from Mussels. Microorganisms, 10(1), 126. https://doi.org/10.3390/microorganisms10010126







