UV-Vis Spectrophotometric Analysis of DNA Retrieval for DNA Storage Applications
Abstract
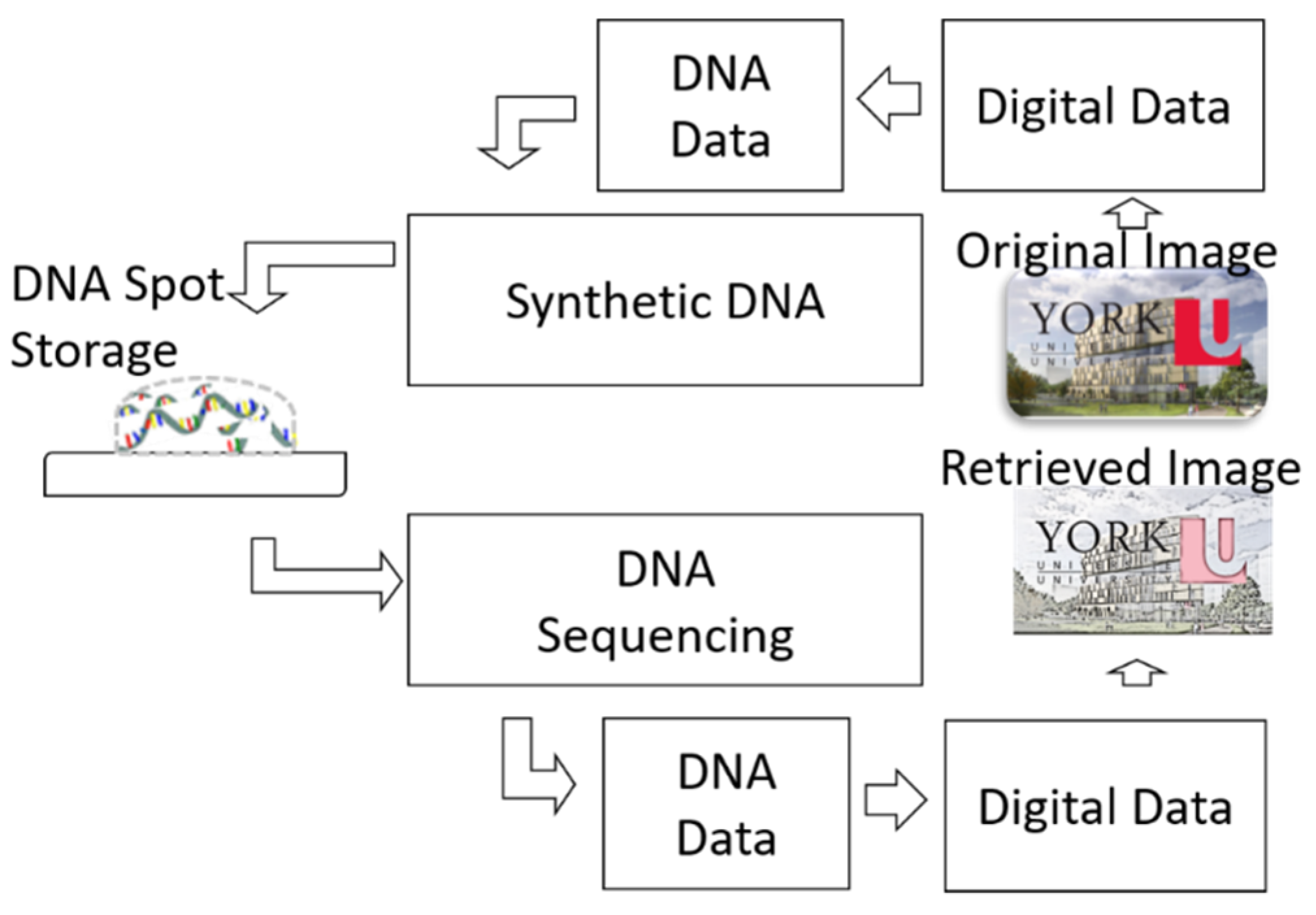
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation and Analysis
2.1.1. DNA Sample Preparation
2.1.2. NanoDrop Analysis
2.2. UV-Vis Spectrophotometer Setup
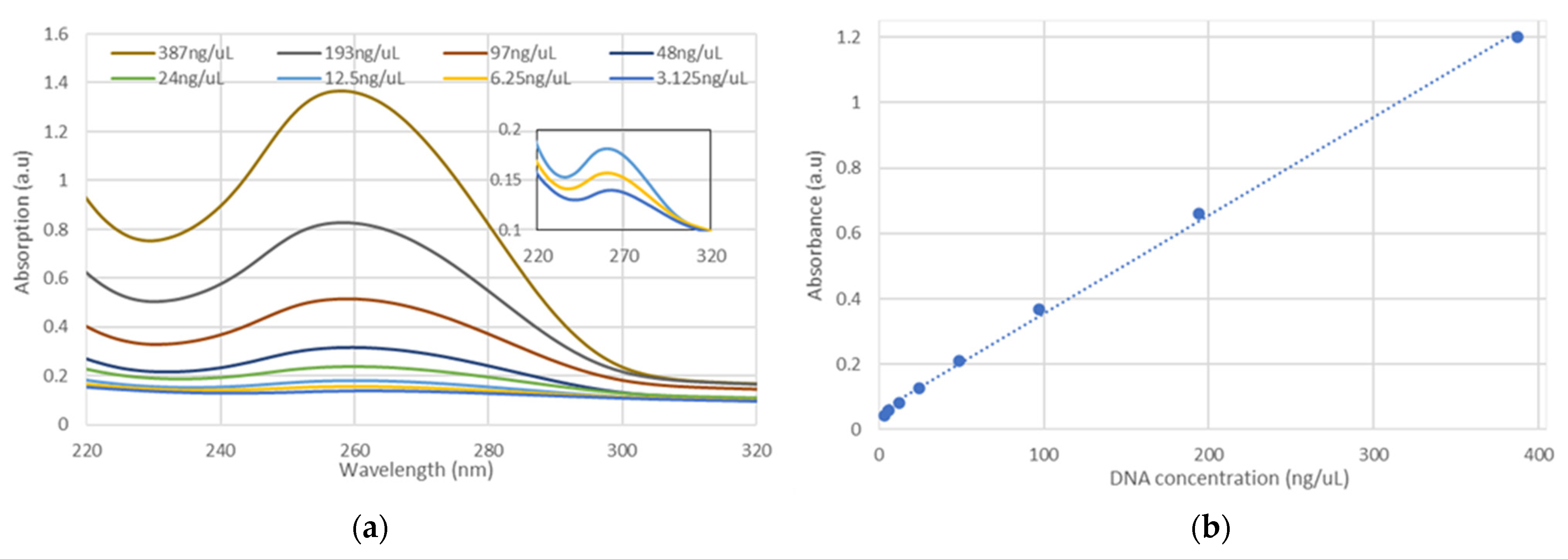
3. Results
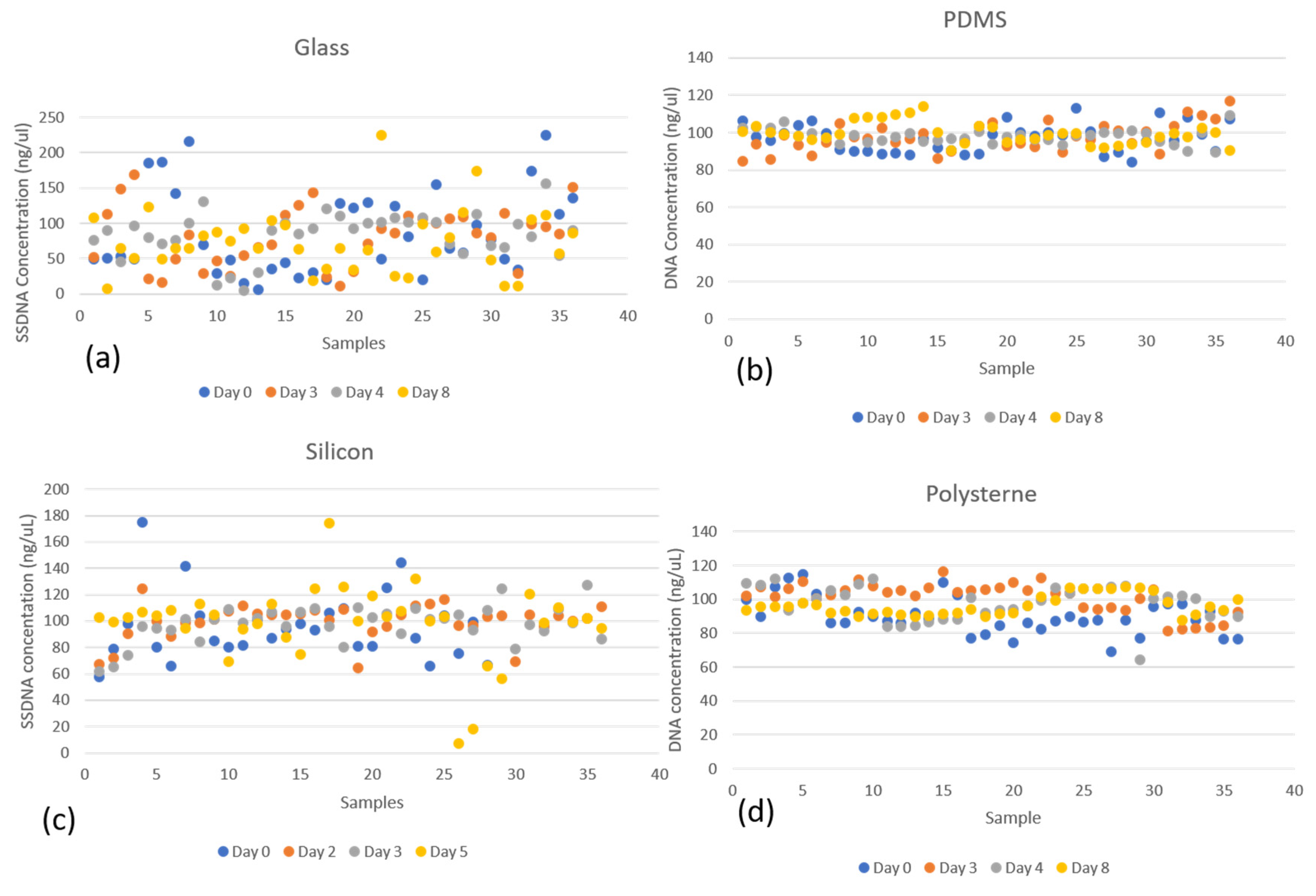
3.1. Wet-DNA Retrieval Assessment
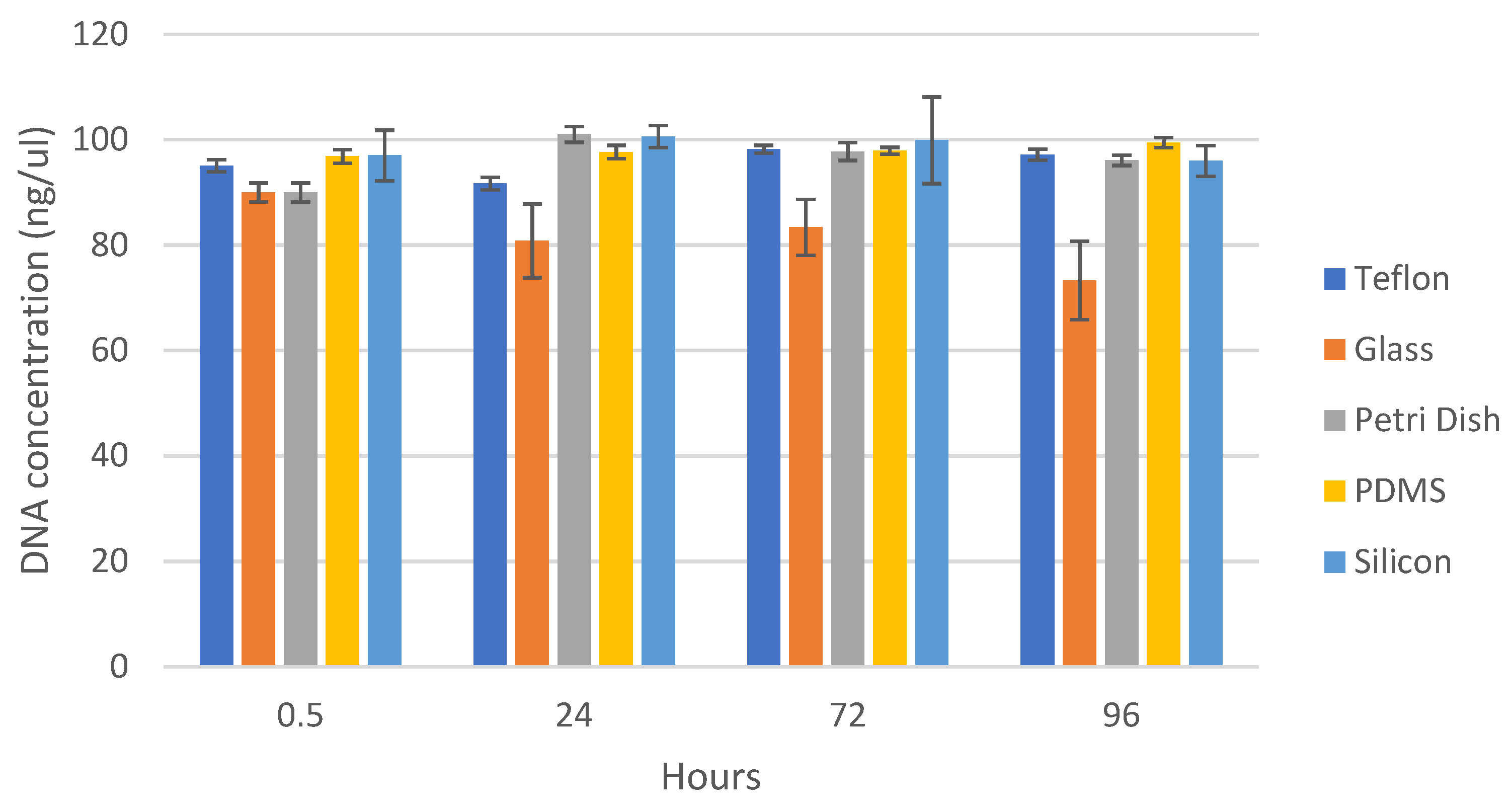
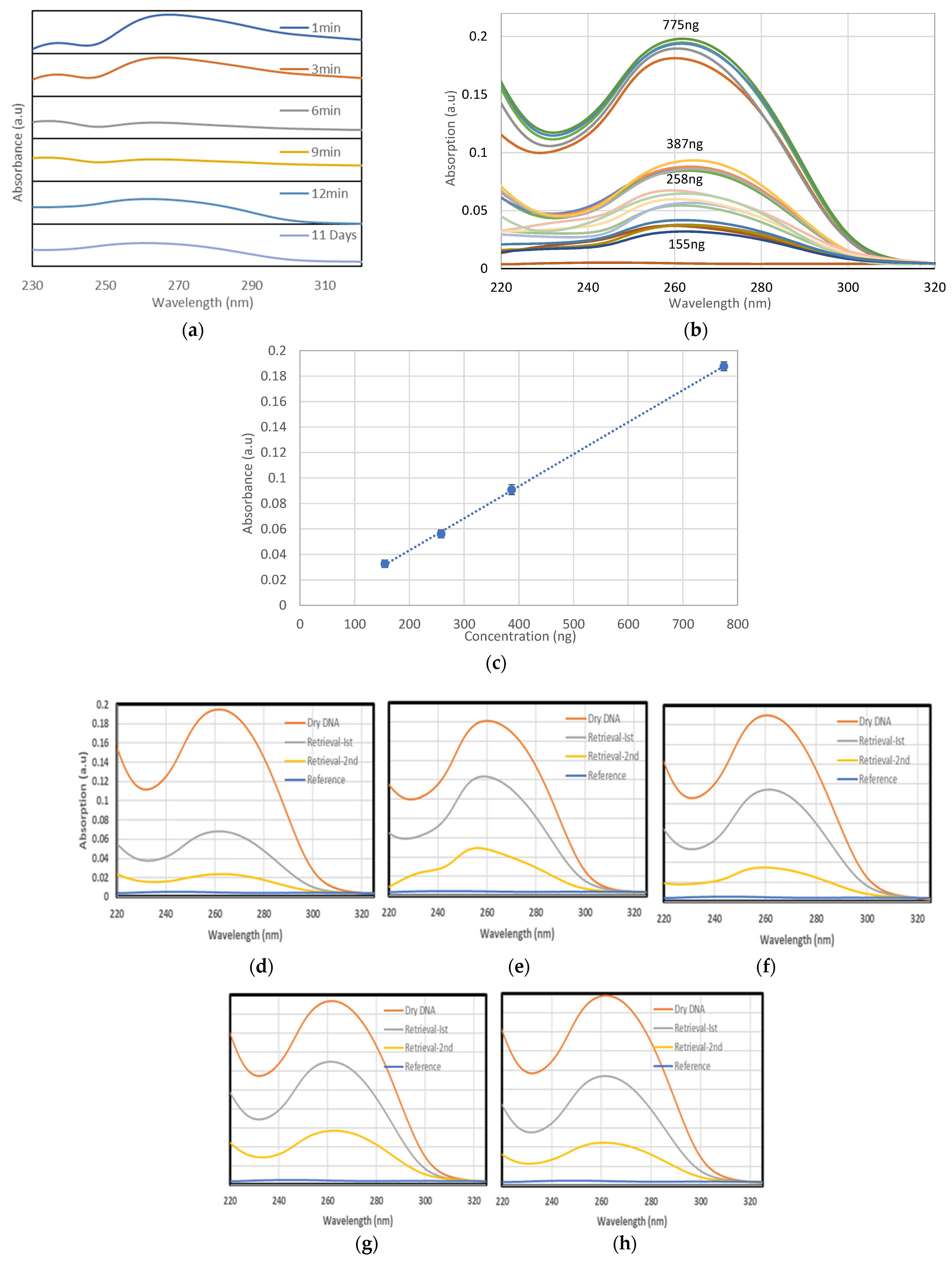
3.1.1. The Effect of Time on the Retrieved DNA from Teflon
3.1.2. Retrieved DNA from Various Surfaces after Different Times
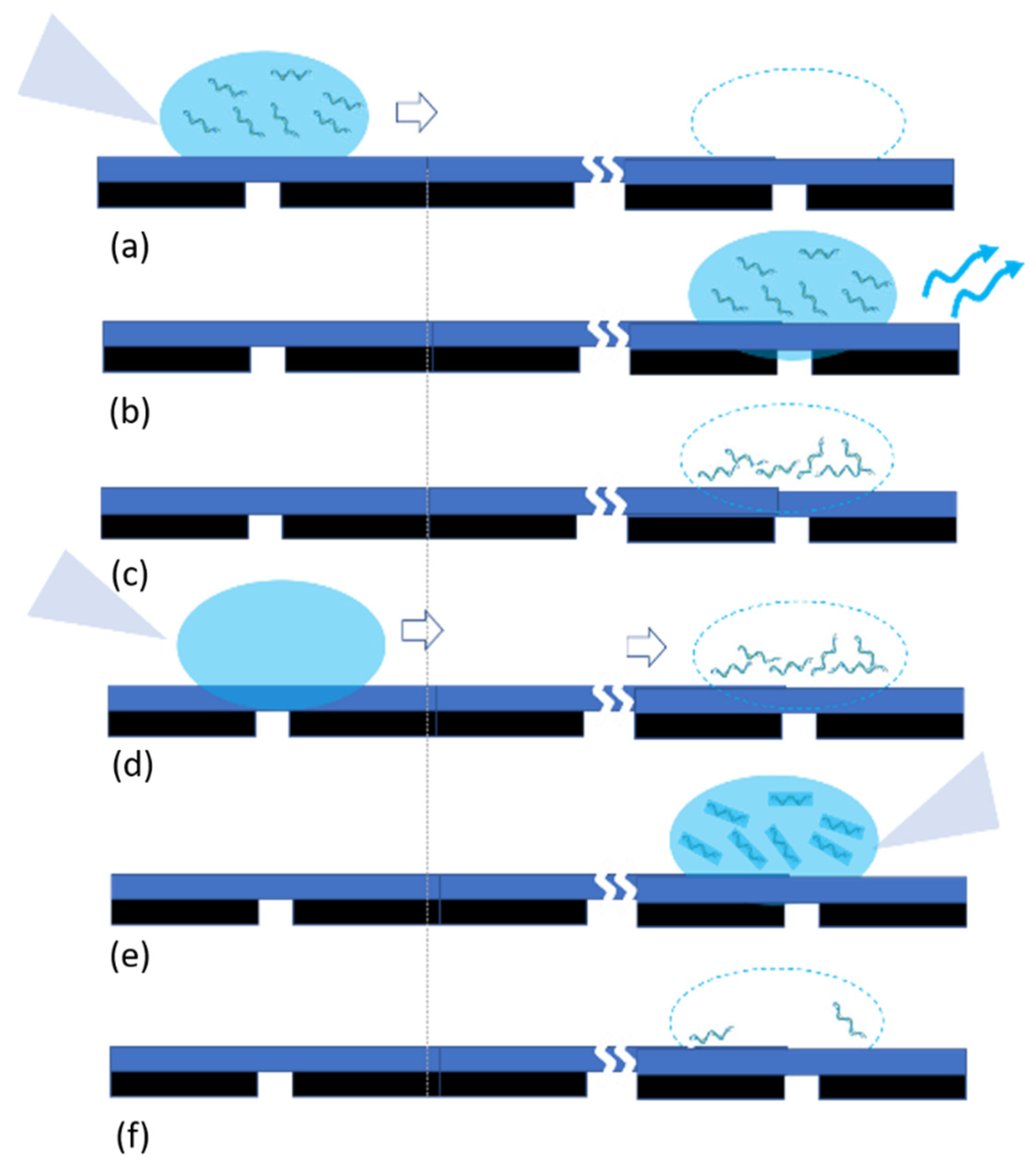
3.1.3. Pipetting Processes
3.2. Dry DNA UV Spectrophotometry
3.2.1. Liquid DNA in a Cuvette
3.2.2. Dry DNA
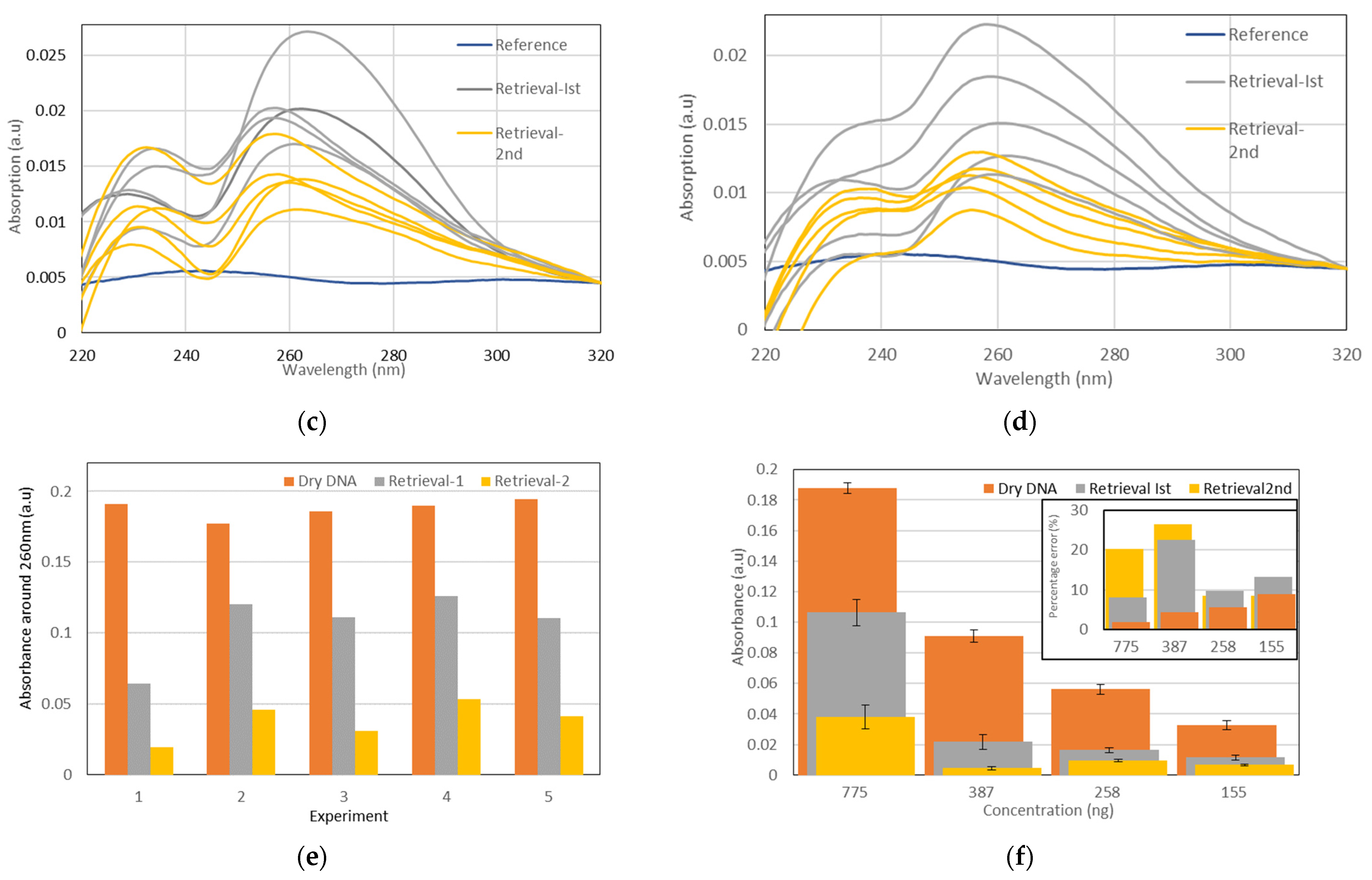
3.2.3. DNA Retrieval
4. Discussions
4.1. DNA Retrieval Assessment Using NanoDrop
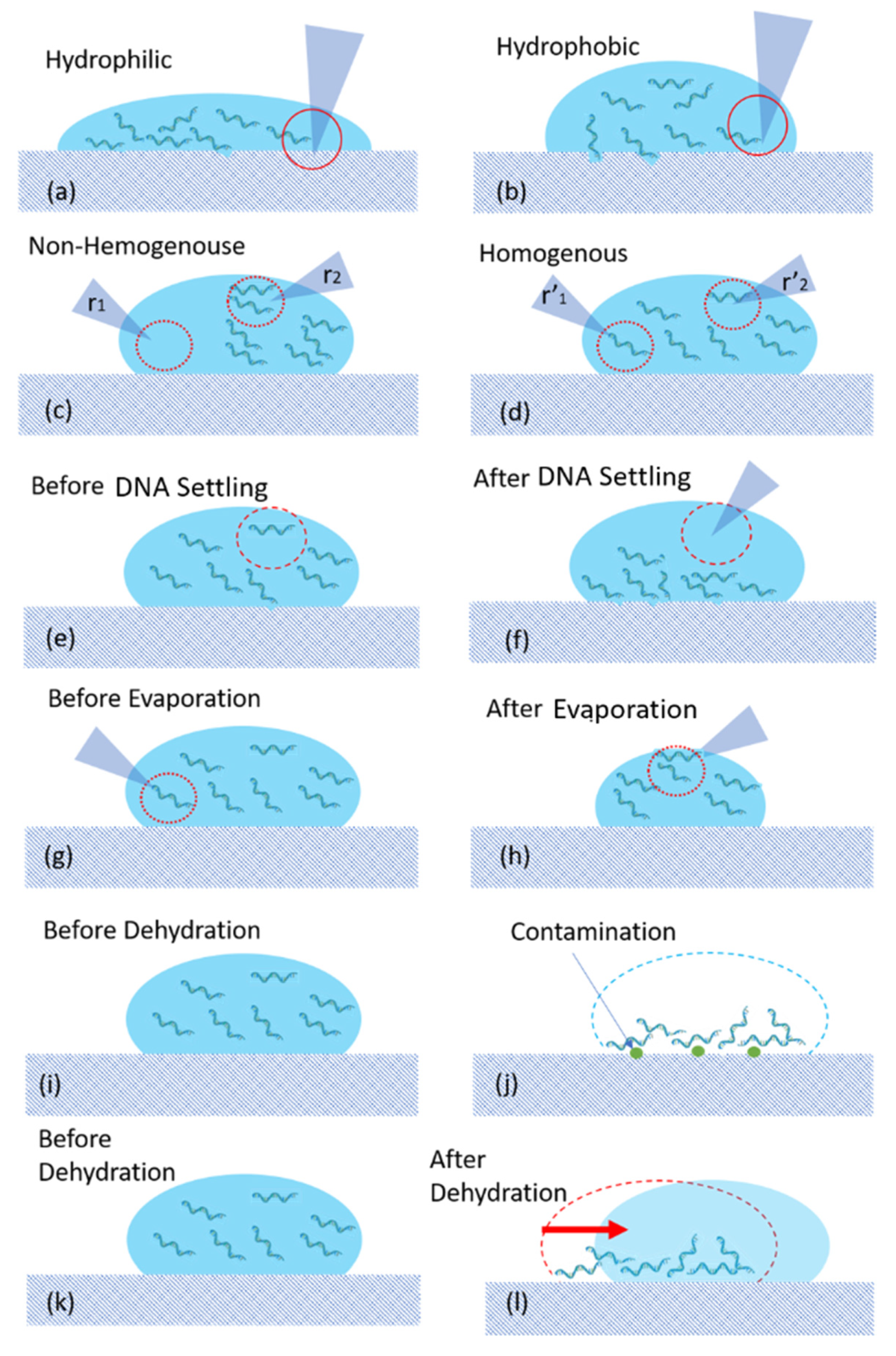
4.1.1. Source of Errors
- Hydrophilicity of surface
- 2.
- Non-Homogeneity
- 3.
- Contamination
- Dehydrated DNA spots are exposed to air containing dust. During the preparation of DNA spots, the experiments were performed under a biosafety cabinet and then were placed on the racks with a lid. Then, the DNA spot must be carried to another room and the samples were in direct contact with air for the duration time of the experiments (~1 h).
- As described in the last subsections and shown in Figure 10, there is always a risk of direct contact of the tip of the pipette with the substrate, which may increase the risk of contamination.
- The surface of substrates including Teflon tape will be in direct contact with the water and DNA. One may argue the risk of the removal of nanogram material from the surface of the substrate or the possibility of nanogram DNA residual. This can potentially increase the risk of contamination as well.
- 4.
- Misalignment
4.1.2. DNA Retrieval
- DNA storage time
- Dispersion of measurement results
4.2. Dry DNA Retrieval Assessment
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Polystyrene | Teflon | |||
|---|---|---|---|---|
| 260/230 | 260/280 | 260/230 | 260/280 | |
| 1 | 2.29 | 1.72 | 2.12 | 2.29 |
| 2 | 2.44 | 1.72 | 2.16 | 2.44 |
| 3 | 2.69 | 1.67 | 2.18 | 2.69 |
| 4 | 2.7 | 1.83 | 1.96 | 2.7 |
| 5 | 2.7 | 1.74 | 2.18 | 2.7 |
| 6 | 2.65 | 1.78 | 2.2 | 2.65 |
| 7 | 2.85 | 1.82 | 2.21 | 2.85 |
| 8 | 2.95 | 1.75 | 2.26 | 2.95 |
| 9 | 2.89 | 1.84 | 2.27 | 2.89 |
| 10 | 2.85 | 1.89 | 2.17 | 2.85 |
| 11 | 2.95 | 1.77 | 2.09 | 2.95 |
| 12 | 2.9 | 1.64 | 1.95 | 2.9 |
| 13 | 2.85 | 1.67 | 1.05 | 2.85 |
| 14 | 2.79 | 1.56 | 2.21 | 2.79 |
| 15 | 2.48 | 1.68 | 2.27 | 2.48 |
| 16 | 2.54 | 1.28 | 2.01 | 2.54 |
| 17 | 2.99 | 1.68 | 2.29 | 2.99 |
| 18 | 2.79 | 1.56 | 2.01 | 2.79 |
| 19 | 3.18 | 1.87 | 2.25 | 3.18 |
| 20 | 3.04 | 1.89 | 2.23 | 3.04 |
| 21 | 2.96 | 1.89 | 2.01 | 2.96 |
| 23 | 3.02 | 1.28 | 2.28 | 3.02 |
| 24 | 2.91 | 1.9 | 2.23 | 2.91 |
| 25 | 2.89 | 1.93 | 2.44 | 2.89 |
| 26 | 3 | 1.26 | 2.54 | 3 |
| 27 | 3.12 | 1.96 | 2.34 | 3.12 |
| 28 | 2.89 | 1.87 | 2.47 | 2.89 |
| 29 | 2.94 | 1.26 | 2.26 | 2.94 |
| 30 | 2.92 | 1.86 | 2.19 | 2.92 |
| 31 | 2.92 | 1.59 | 2.29 | 2.92 |
| 32 | 2.91 | 1.15 | 2.3 | 2.91 |
| 33 | 2.9 | 1.23 | 2.32 | 2.9 |
| 34 | 2.89 | 2.1 | 2.43 | 2.89 |
References
- IDC: Expect 175 Zettabytes of Data Worldwide by 2025|Network World. Available online: https://www.networkworld.com/article/3325397/idc-expect-175-zettabytes-of-data-worldwide-by-2025.html (accessed on 4 May 2021).
- What Causes Hard Drive Data Corruption. Available online: https://www.securedatarecovery.com/services/hard-drive-recovery/what-causes-hard-drive-data-corruption (accessed on 17 January 2021).
- Lunt, B.M. How long is long-term data storage? In Archiving Conference; Society for Imaging Science and Technology: Springfield, VA, USA, 2011; pp. 29–33. [Google Scholar]
- Matange, K.; Tuck, J.M.; Keung, A.J. DNA stability: A central design consideration for DNA data storage systems. Nat. Commun. 2021, 12, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Heckel, R.; Shomorony, I.; Ramchandran, K.; David, N. Fundamental limits of DNA storage systems. In Proceedings of the 2017 IEEE International Symposium on Information Theory (ISIT), Aachen, Germany, 25–30 June 2017; pp. 3130–3134. [Google Scholar]
- Ceze, L.; Nivala, J.; Strauss, K. Molecular digital data storage using DNA. Nat. Rev. Genet. 2019, 20, 456–466. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.K.; Nirantar, S.; Yew, W.S.; Poh, C.L. Novel modalities in DNA data storage. Trends Biotechnol. 2021, 39, 990–1003. [Google Scholar] [CrossRef] [PubMed]
- Yazdi, S.H.T.; Kiah, H.M.; Garcia-Ruiz, E.; Ma, J.; Zhao, H.; Milenkovic, O. DNA-based storage: Trends and methods. IEEE Trans. Mol. Biol. Multi-Scale Commun. 2015, 1, 230–248. [Google Scholar] [CrossRef] [Green Version]
- Regaladoarchive, A. Microsoft Has a Plan to Add DNA Data Storage to Its Cloud. Available online: https://www.technologyreview.com/2017/05/22/68387/microsoft-has-a-plan-to-add-dna-data-storage-to-its-cloud/ (accessed on 22 May 2017).
- Demidov, V.V. Hiding and Storing Messages and Data in DNA. In DNA Beyond Genes: From Data Storage and Computing to Nanobots, Nanomedicine, and Nanoelectronics; Springer International Publishing: Cham, Switzerlands, 2020; pp. 7–23. [Google Scholar]
- Organick, L.; Ang, S.D.; Chen, Y.-J.; Lopez, R.; Yekhanin, S.; Makarychev, K.; Racz, M.Z.; Kamath, G.; Gopalan, P.; Nguyen, B. Random access in large-scale DNA data storage. Nat. Biotechnol. 2018, 36, 242–248. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Ramteke, M. DNA Computing: Methodologies and Challenges. In DNA-and RNA-Based Computing Systems; Wiley-VCH: Weinheim, Germany, 2021; pp. 15–29. [Google Scholar]
- Martens, S.; Landuyt, A.; Espeel, P.; Devreese, B.; Dawyndt, P.; Du Prez, F. Multifunctional sequence-defined macromolecules for chemical data storage. Nat. Commun. 2018, 9, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newman, S.; Stephenson, A.P.; Willsey, M.; Nguyen, B.H.; Takahashi, C.N.; Strauss, K.; Ceze, L. High density DNA data storage library via dehydration with digital microfluidic retrieval. Nat. Commun. 2019, 10, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adhikari, P.R.; Tasneem, N.T.; Reid, R.C.; Mahbub, I. Electrode and electrolyte configurations for low frequency motion energy harvesting based on reverse electrowetting. Sci. Rep. 2021, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, K.; Mogensen, H.S.; Hedman, J.; Niederstätter, H.; Parson, W.; Morling, N. Comparison of five DNA quantification methods. Forensic Sci. Int. Genet. 2008, 2, 226–230. [Google Scholar] [CrossRef] [PubMed]
- Swinehart, D.F. The beer-lambert law. J. Chem. Educ. 1962, 39, 333–335. [Google Scholar] [CrossRef]
- Rani, N.; Sharma, S.; Sharma, M. Phytochemical analysis of Meizotropis pellita by FTIR and UV-VIS spectrophotometer. Indian J. Sci. Technol. 2016, 9, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Jovanović, N.; Bouchard, A.; Hofland, G.W.; Witkamp, G.-J.; Crommelin, D.J.; Jiskoot, W. Distinct effects of sucrose and trehalose on protein stability during supercritical fluid drying and freeze-drying. Eur. J. Pharm. Sci. 2006, 27, 336–345. [Google Scholar] [CrossRef] [PubMed]
- Weigmann, H.-J.; Lindemann, U.; Antoniou, C.; Tsikrikas, G.; Stratigos, A.; Katsambas, A.; Sterry, W.; Lademann, J. UV/VIS absorbance allows rapid, accurate, and reproducible mass determination of corneocytes removed by tape stripping. Skin Pharmacol. Physiol. 2003, 16, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Ren, K.; Ji, J.; Shen, J. Construction and enzymatic degradation of multilayered poly-L-lysine/DNA films. Biomaterials 2006, 27, 1152–1159. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, T.; Hieda, K.; Nikaido, O. Wavelength dependent formation of thymine dimers and (6-4) photoproducts in DNA by monochromatic ultraviolet light ranging from 150 to 365 nm. Photochem. Photobiol. 1991, 54, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-L.; Hu, Y.-J.; Mi, R.; Li, X.-Y.; Li, P.-Q.; Ouyang, Y. Spectroscopic exploring the affinities, characteristics, and mode of binding interaction of curcumin with DNA. Mol. Biol. Rep. 2013, 40, 4405–4413. [Google Scholar] [CrossRef] [PubMed]
- Faghihnejad, A.; Zeng, H. Hydrophobic interactions between polymer surfaces: Using polystyrene as a model system. Soft Matter 2012, 8, 2746–2759. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Minhas-Khan, A.; Ghafar-Zadeh, M.; Shaffaf, T.; Forouhi, S.; Scime, A.; Magierowski, S.; Ghafar-Zadeh, E. UV-Vis Spectrophotometric Analysis of DNA Retrieval for DNA Storage Applications. Actuators 2021, 10, 246. https://doi.org/10.3390/act10100246
Minhas-Khan A, Ghafar-Zadeh M, Shaffaf T, Forouhi S, Scime A, Magierowski S, Ghafar-Zadeh E. UV-Vis Spectrophotometric Analysis of DNA Retrieval for DNA Storage Applications. Actuators. 2021; 10(10):246. https://doi.org/10.3390/act10100246
Chicago/Turabian StyleMinhas-Khan, Aamir, Morteza Ghafar-Zadeh, Tina Shaffaf, Saghi Forouhi, Anthony Scime, Sebastian Magierowski, and Ebrahim Ghafar-Zadeh. 2021. "UV-Vis Spectrophotometric Analysis of DNA Retrieval for DNA Storage Applications" Actuators 10, no. 10: 246. https://doi.org/10.3390/act10100246
APA StyleMinhas-Khan, A., Ghafar-Zadeh, M., Shaffaf, T., Forouhi, S., Scime, A., Magierowski, S., & Ghafar-Zadeh, E. (2021). UV-Vis Spectrophotometric Analysis of DNA Retrieval for DNA Storage Applications. Actuators, 10(10), 246. https://doi.org/10.3390/act10100246







