Identification and Morphological Characterization of Biofilms Formed by Strains Causing Infection in Orthopedic Implants
Abstract
1. Introduction
2. Material and Methods
2.1. Isolation and Identification of the Strains
2.2. Antibiotic Sensitivity Tests
2.3. Biofilm Formation In Vitro
2.4. Counting of Colony Forming Units (CFU)
2.5. Metabolic Activity Tests
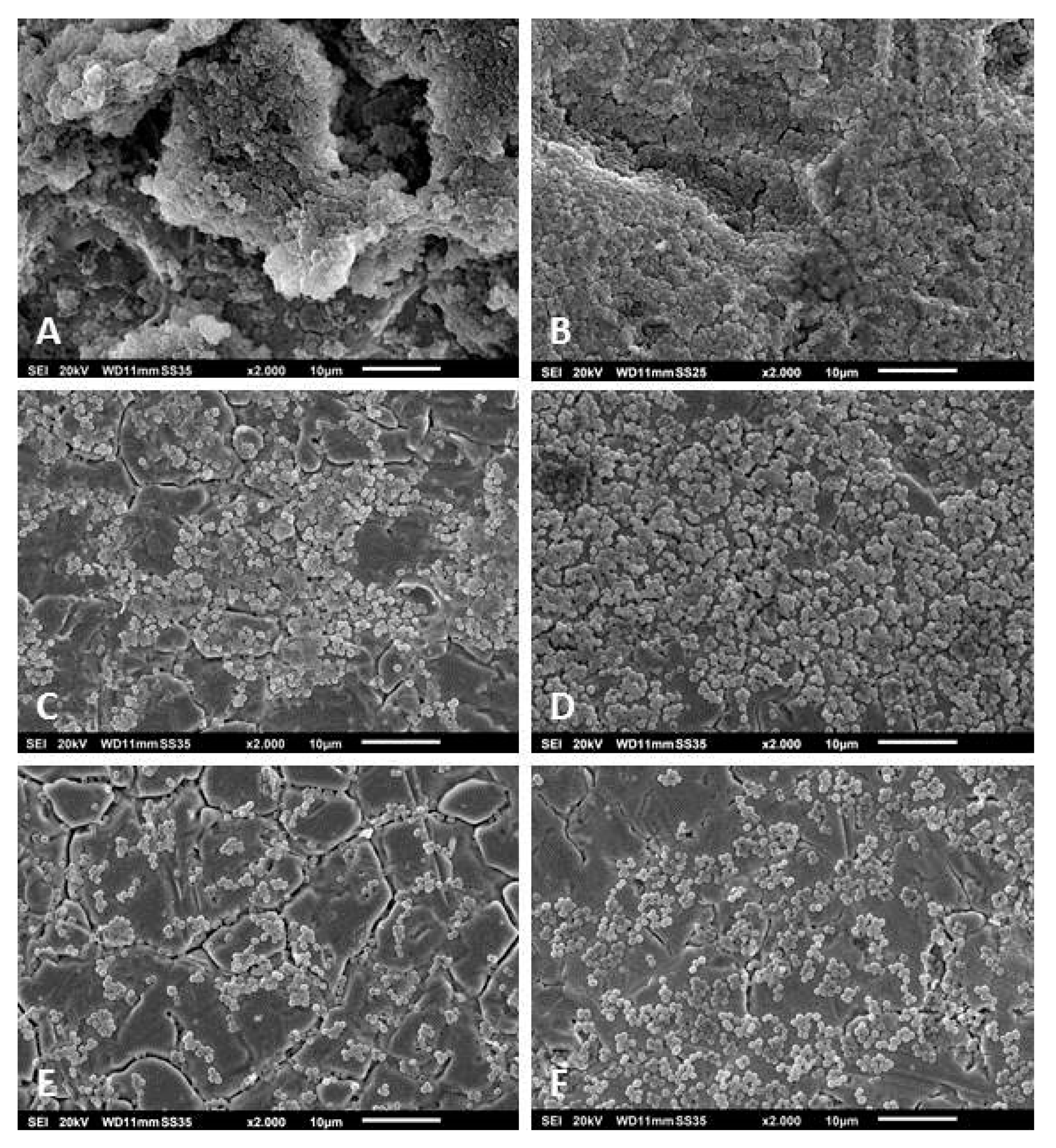
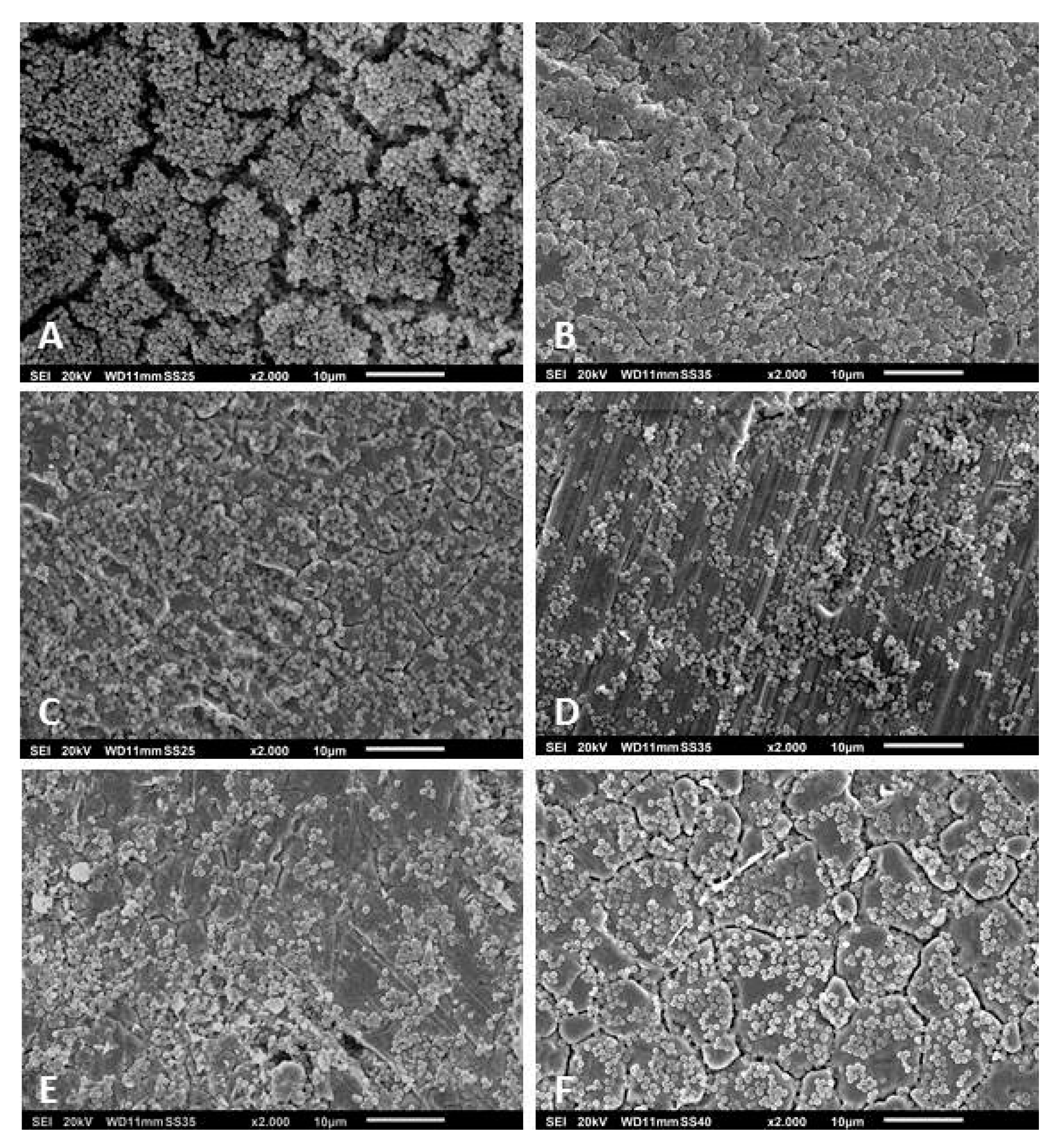
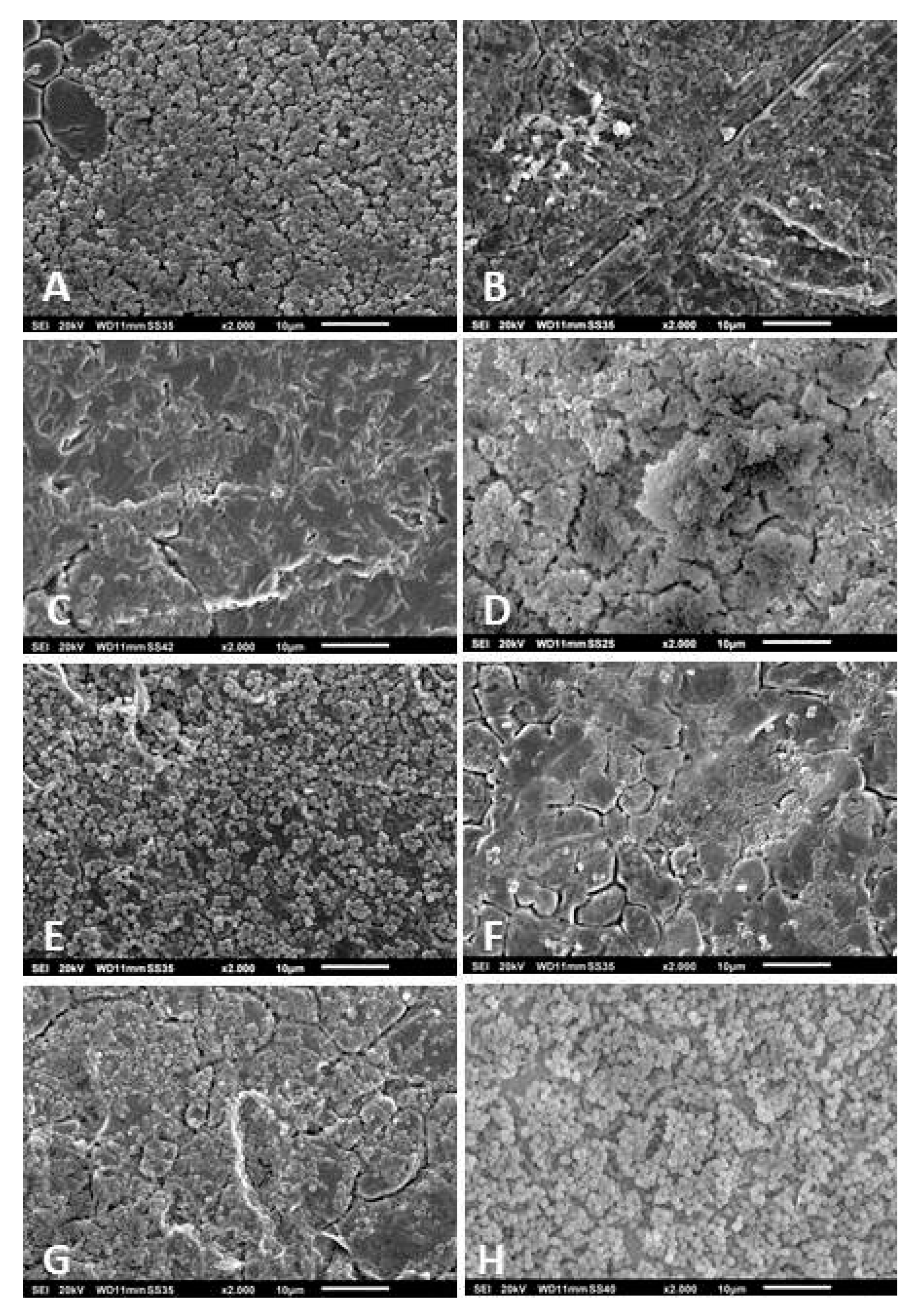
2.6. Scanning Electron Microscopy
2.7. Characterization of Biofilm Forming Capacity
2.8. Data Analysis
3. Results and Discussions
3.1. Colony Forming Units Counting
3.2. Metabolic Activity of Biofilm Cells
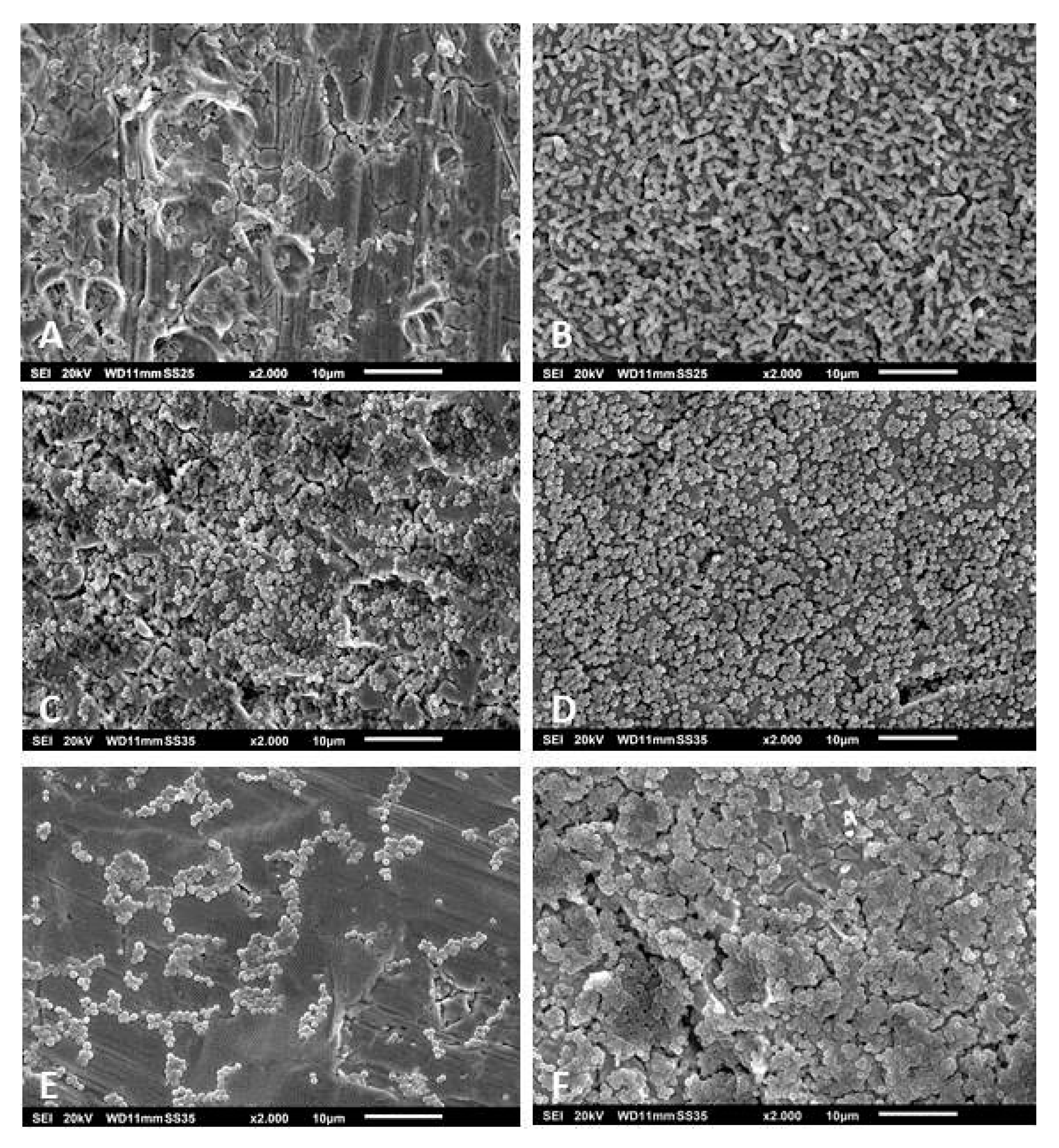
3.3. Scanning Electron Microscopy of the Biofilms
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kapadia, B.H.; Berg, R.A.; Daley, J.A.; Fritz, J.; Bhave, A.; Mont, M.A. Periprosthetic joint infection. Lancet 2016, 387, 386–394. [Google Scholar] [CrossRef]
- Vastag, B. Knee replacement underused, says panel: Useful option when nonsurgical therapies fail. JAMA 2004, 291, 413–414. [Google Scholar] [PubMed]
- Lamagni, T. Epidemiology and burden of prosthetic joint infections. J. Antimicrob. Chemother. 2014, 69, i5–i10. [Google Scholar] [CrossRef] [PubMed]
- Saeed, K.; McLaren, A.C.; Schwarz, E.M.; Antoci, V.; Arnold, W.V.; Chen, A.F.; Clauss, M.; Esteban, J.; Gant, V.; Hendershot, E.; et al. 2018 International consensus meeting on musculoskeletal infection: Summary from the biofilm workgroup and consensus on biofilm related musculoskeletal infections. J. Orthop. Res. 2019, 37, 1007–1017. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.J.; Engemann, J.J.; Harrell, L.J.; Carmeli, Y.; Reller, L.B.; Kaye, K.S. Predictors of mortality in patients with bloodstream infection due to ceftazidime-resistant Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2006, 50, 1715–1720. [Google Scholar] [CrossRef]
- Cosgrove, S.E.; Sakoulas, G.; Perencevich, E.N.; Schwaber, M.J.; Karchmer, A.W.; Carmeli, Y. Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus Bacteremia: A meta-analysis. Clin. Infect. Dis. 2003, 36, 53–59. [Google Scholar] [CrossRef]
- Roberts, R.R.; Hota, B.; Ahmad, I.; Scott, R.D.; Foster, S.D.; Abbasi, F.; Schabowski, S.; Kampe, L.M.; Ciavarella, G.G.; Supino, M.; et al. Hospital and societal costs of antimicrobial-resistant infections in a Chicago Teaching Hospital: Implications for antibiotic stewardship. Clin. Infect. Dis. 2009, 49, 1175–1184. [Google Scholar] [CrossRef]
- Ibrahim, E.H.; Sherman, G.; Ward, S.; Fraser, V.J.; Kollef, M.H. The influence of inadequate antimicrobial treatment of bloodstream infections on patient outcomes in the ICU setting. Chest 2000, 118, 146–155. [Google Scholar] [CrossRef]
- Ito, A.; Taniuchi, A.; May, T.; Kawata, K.; Okabe, S. Increased Antibiotic Resistance of Escherichia coli in mature biofilms. Appl. Environ. Microbiol. 2009, 75, 4093–4100. [Google Scholar] [CrossRef]
- Alhede, M.; Kragh, K.N.; Qvortrup, K.; Allesen-Holm, M.; van Gennip, M.; Christensen, L.D.; Jensen, P.Ø.; Nielsen, A.K.; Parsek, M.; Wozniak, D.; et al. Phenotypes of non-attached Pseudomonas aeruginosa aggregates resemble surface attached biofilm. PLoS ONE 2011, 6, e27943. [Google Scholar] [CrossRef]
- Bowler, L.L.; Zhanel, G.G.; Ball, T.B.; Saward, L.L. Mature Pseudomonas aeruginosa biofilms prevail compared to young biofilms in the presence of ceftazidime. Antimicrob. Agents Chemother. 2012, 56, 4976–4979. [Google Scholar] [CrossRef] [PubMed]
- Haaber, J.; Cohn, M.T.; Frees, D.; Andersen, T.J.; Ingmer, H. Planktonic aggregates of Staphylococcus aureus protect against common antibiotics. PLoS ONE 2012, 7, e41075. [Google Scholar] [CrossRef]
- Stewart, P.S. Antimicrobial tolerance in biofilms. In Microbiology Spectrum; Ghannoum, M., Parsek, M., Whiteley, M., Mukherjee, P.K., Eds.; American Society for Microbiology: Washington, DC, USA, 2015; p. 3. [Google Scholar]
- Hall, C.W.; Mah, T.-F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301. [Google Scholar] [CrossRef] [PubMed]
- Ceri, H.; Olson, M.E.; Stremick, C.; Read, R.R.; Morck, D.; Buret, A. The Calgary Biofilm Device: New technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. 1999, 37, 1771–1776. [Google Scholar] [CrossRef] [PubMed]
- Hall-Stoodley, L.; Rayner, J.C.; Stoodley, P.; Lappin-Scott, H.M. Establishment of experimental biofilms using the Modified Robbins Device and flow cells. In Methods in Biotechnology: Environmental Monitoring of Bacteria; Humana Press Inc.: Honoken, NJ, USA, 1999; pp. 307–319. [Google Scholar]
- Merritt, J.H.; Kadouri, D.E.; O’Toole, G.A. Growing and analyzing static biofilms. In Current Protocols in Microbiology; Coico, R., Kowalik, T., Quarles, J., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2005; pp. 1–17. [Google Scholar]
- Sternberg, C.; Tolker-Nielsen, T. Growing and analyzing biofilms in flow cells. In Current Protocols in Microbiology; Coico, R., Kowalik, T., Quarles, J., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2006; pp. 1–15. [Google Scholar]
- O’Toole, G.A. Microtiter dish biofilm formation assay. JoVE J. Vis. Exp. 2011, 47, e2437. [Google Scholar] [CrossRef] [PubMed]
- Peterson, S.B.; Irie, Y.; Borlee, B.R.; Murakami, K.; Harrison, J.J.; Colvin, K.M.; Parsek, M.R. Different methods for culturing biofilms in vitro. In Biofilm Infections; Bjarnsholt, T., Jensen, P.Ø., Moser, C., Eds.; Springer: New York, NY, USA, 2011; pp. 251–266. [Google Scholar]
- Rumbaugh, K.P.; Carty, N.L. In vivo models of biofilm infection. In Biofilm Infections; Bjarnsholt, T., Jensen, P.Ø., Moser, C., Eds.; Springer: New York, NY, USA, 2011; pp. 267–290. [Google Scholar]
- Lebeaux, D.; Chauhan, A.; Rendueles, O.; Beloin, C. From in vitro to in vivo models of bacterial biofilm-related infections. Pathogens 2013, 2, 288–356. [Google Scholar] [CrossRef]
- Costerton, J.W. Overview of microbial biofilms. J. Ind. Microbiol. 1995, 15, 137–140. [Google Scholar] [CrossRef]
- Sandrin, T.R.; Goldstein, J.E.; Schumaker, S. MALDI TOF MS profiling of bacteria at the strain level: A review. Mass Spectrom. Rev. 2013, 32, 188–217. [Google Scholar] [CrossRef]
- Tanner, H.; Evans, J.T.; Gossain, S.; Hussain, A. Evaluation of three sample preparation methods for the direct identification of bacteria in positive blood cultures by MALDI-TOF. BMC Res. Notes 2017, 10, 48. [Google Scholar] [CrossRef]
- Fagerquist, C.K. Unlocking the proteomic information encoded in MALDI-TOF-MS data used for microbial identification and characterization. Expert Rev. Proteom. 2017, 14, 97–107. [Google Scholar] [CrossRef]
- Matuschek, E.; Brown, D.F.; Kahlmeter, G. Development of the EUCAST disk diffusion antimicrobial susceptibility testing method and its implementation in routine microbiology laboratories. Clin. Microbiol. Infect. 2014, 20, O255–O266. [Google Scholar] [CrossRef] [PubMed]
- Pannanusorn, S.; Fernandez, V.; Römling, U. Prevalence of biofilm formation in clinical isolates of Candida species causing bloodstream infection. Mycoses 2013, 56, 264–272. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.S.; Franklin, M.J. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 2008, 6, 199–210. [Google Scholar] [CrossRef]
- Stoodley, P.; Sauer, K.; Davies, D.G.; Costerton, J.W. Biofilms as complex differentiated communities. Ann. Rev. Microbiol. 2002, 56, 187–209. [Google Scholar] [CrossRef] [PubMed]
- Coraca-Huber, D.C.; Fille, M.; Hausdorfer, J.; Pfaller, K.; Nogler, M. Staphylococcus aureus biofilm formation and antibiotic susceptibility tests on polystyrene and metal surfaces. J. Appl. Microbiol. 2012, 112, 1235–1243. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Prakash, P.; Achra, A.; Singh, G.P.; Das, A.; Singh, R.K. Standardization and classification of in vitro biofilm formation by clinical isolates of Staphylococcus aureus. J. Global Infect. Dis. 2017, 9, 93–101. [Google Scholar]
- Seifi, K.; Kazemian, H.; Heidari, H.; Rezagholizadeh, F.; Saee, Y.; Shirvani, F.; Houri, H. Evaluation of biofilm formation among Klebsiella pneumoniae isolates and molecular characterization by ERIC-PCR. Jundishapur J. Microbiol. 2016, 9, e30682. [Google Scholar] [CrossRef]
- O’Toole, G.; Kaplan, H.B.; Kolter, R. Biofilm formation as microbial development. Annu. Rev. Microbiol. 2000, 54, 49–79. [Google Scholar] [CrossRef]
- Sauer, K.; Camper, A.K.; Ehrlich, G.D.; Costerton, J.W.; Davies, D.G. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 2002, 184, 1140–1154. [Google Scholar] [CrossRef]
- Southey-Pillig, C.J.; Davies, D.G.; Sauer, K. Characterization of temporal protein production in Pseudomonas aeruginosa biofilms. J. Bacteriol. 2005, 187, 8114–8126. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcal biofilms. Curr. Top. Microbiol. Immunol. 2008, 322, 207–228. [Google Scholar] [PubMed]
- Monds, R.D.; O’Toole, G.A. The developmental model of microbial biofilms: Ten years of a paradigm up for review. Trends Microbiol. 2009, 17, 73–87. [Google Scholar] [CrossRef] [PubMed]
- Lopez, D.; Vlamakis, H.; Kolter, R. Biofilms. Cold Spring Harbor Perspectives Biol. 2010, 2, a000398. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Flores-Mireles, A.L.; Cusumano, Z.T.; Takagi, E.; Hultgren, S.J.; Caparon, M.G. Host and bacterial proteases influence biofilm formation and virulence in a murine model of enterococcal catheter-associated urinary tract infection. NPJ Biofilms Microbiomes 2017, 3, 1–12. [Google Scholar] [CrossRef]
- Fraiha, R.O.; Pereira, A.P.R.; Brito, E.D.C.A.; Borges, C.L.; Parente, A.F.A.; Perdomo, R.T.; Macedo, M.L.R.; Weber, S.S. Stress conditions in the host induce persister cells and influence biofilm formation by Staphylococcus epidermidis RP62A. Rev. Soc. Bras. Med. Trop. 2019, 52. [Google Scholar] [CrossRef]
- Coraca-Huber, D.C.; Dichtl, S.; Steixner, S.; Nogler, M.; Weiss, G. Iron chelation destabilizes bacterial biofilms and potentiates the antimicrobial activity of antibiotics against coagulase-negative Staphylococci. Pathog. Dis. 2018, 76. [Google Scholar] [CrossRef]
- Ledala, N.; Zhang, B.; Seravalli, J.; Powers, R.; Somerville, G.A. Influence of iron and aeration on Staphylococcus aureus growth, metabolism, and transcription. J. Bacteriol. 2014, 196, 2178–2189. [Google Scholar] [CrossRef]
- Oliveira, F.; Franca, A.; Cerca, N. Staphylococcus epidermidis is largely dependent on iron availability to form biofilms. Int. J. Med. Microbiol. 2017, 307, 552–563. [Google Scholar] [CrossRef]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [Google Scholar] [CrossRef]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Høiby, N.; Bjarnsholt, T.; Givskov, M.; Molin, S.; Ciofu, O. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 2010, 35, 322–332. [Google Scholar] [CrossRef] [PubMed]
- Branda, S.S.; Vik, S.; Friedman, L.; Kolter, R. Biofilms: The matrix revisited. Trends Microbiol. 2005, 13, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W.; Lewandowski, Z.; Caldwell, D.E.; Korber, D.R.; Lappin-Scott, H.M. Microbial biofilms. Annu. Rev. Microbiol. 1995, 49, 711–745. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef]
- Coraca-Huber, D.C.; Fille, M.; Hausdorfer, J.; Pfaller, K.; Nogler, M. Evaluation of MBEC-HTP biofilm model for studies of implant associated infections. J. Orthop. Res. 2012, 30, 1176–1180. [Google Scholar] [CrossRef]
- Al-Ahmad, A.; Ameen, H.; Pelz, K.; Karygianni, L.; Wittmer, A.; Anderson, A.C.; Spitzmüller, B.; Hellwig, E. Antibiotic resistance and capacity for biofilm formation of different bacteria isolated from endodontic infections associated with root-filled teeth. J. Endod. 2014, 40, 223–230. [Google Scholar] [CrossRef]
- Qi, L.; Li, H.; Zhang, C.; Liang, B.; Li, J.; Wang, L.; Du, X.; Liu, X.; Qiu, S.; Song, H. Relationship between antibiotic resistance, biofilm formation, and biofilm-specific resistance in Acinetobacter baumannii. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef]
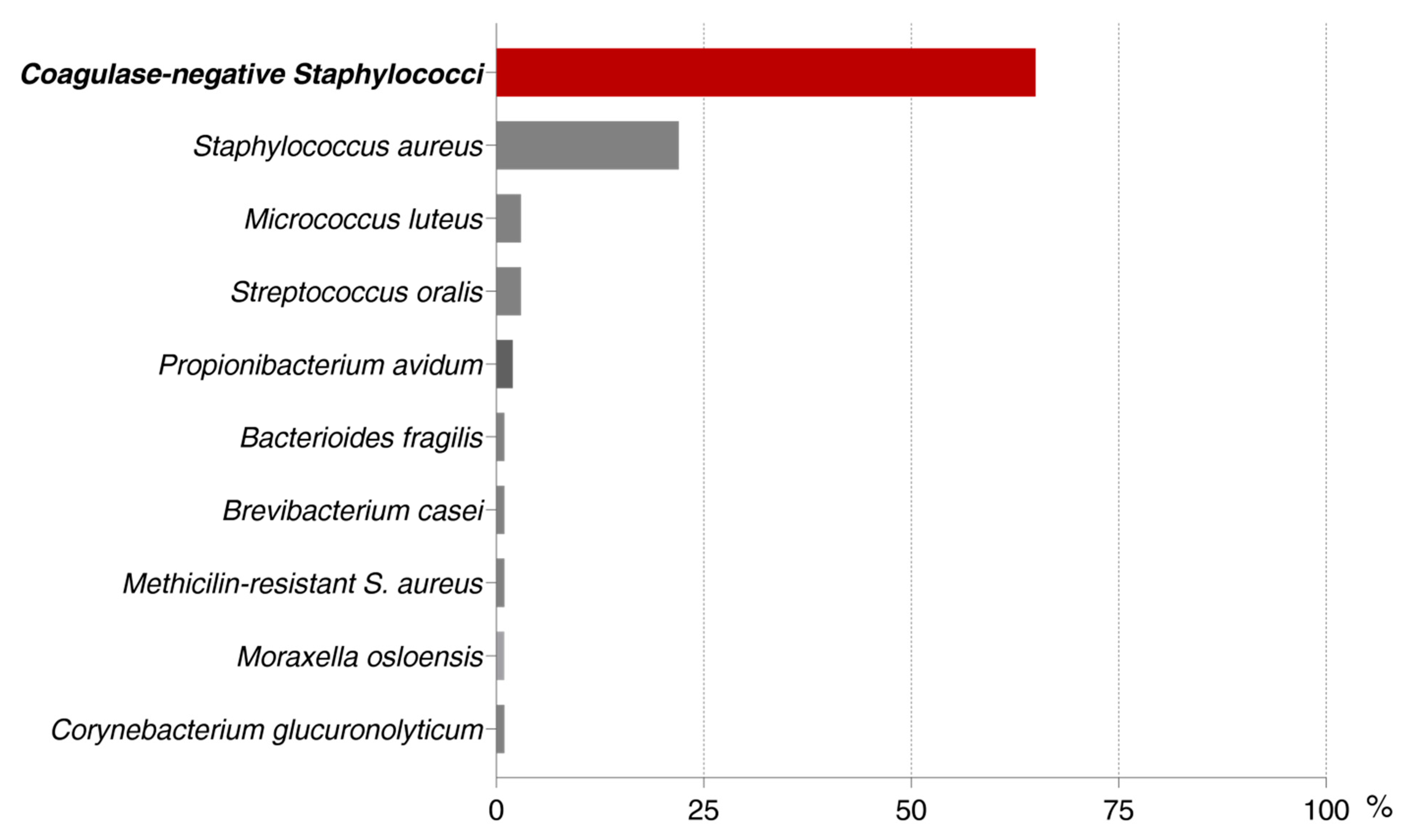


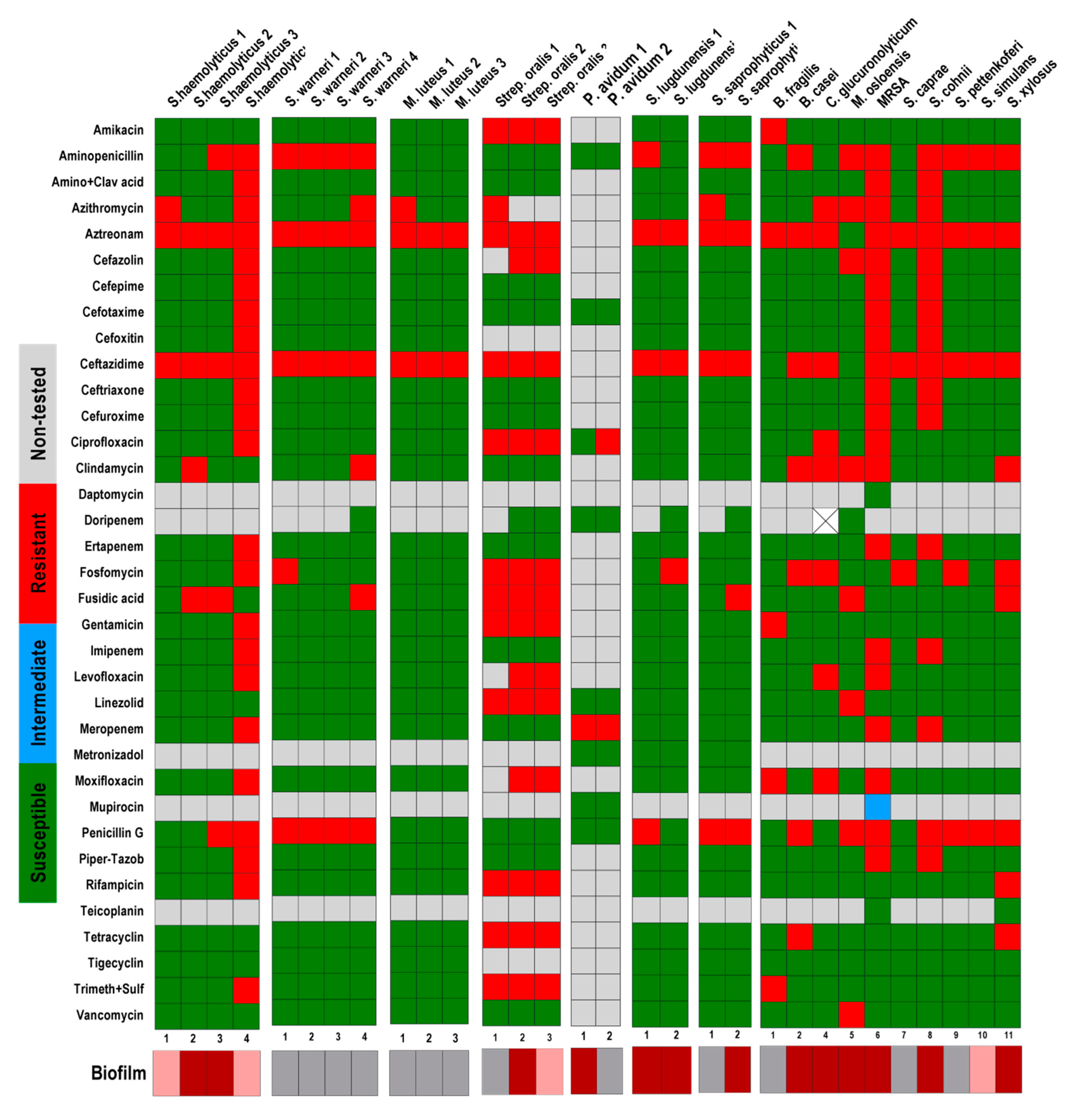
| INTENSITY OF BIOFILM FORMATION | ||||
|---|---|---|---|---|
| ORGANISMS | (HIGH) | (LOW) | (NO) | Total |
| n° Strains | n° Strains | n° Strains | n° Strains | |
| S. epidermidis | 34 | 16 | 12 | 62 |
| S. aureus | 13 | 5 | 4 | 22 |
| S. capitis | 6 | 4 | 4 | 14 |
| S. hominis | 6 | 5 | 1 | 12 |
| S. haemolyticus | 2 | 2 | - | 4 |
| S. warneri | - | - | 4 | 4 |
| M. luteus | - | - | 3 | 3 |
| Strep. oralis | 1 | 1 | 1 | 3 |
| P. avidum | 1 | - | 1 | 2 |
| S. lugdunensis | 2 | - | - | 2 |
| S. saprophyticus | 1 | - | 1 | 2 |
| B. fragilis | - | - | 1 | 1 |
| B. casei | 1 | - | - | 1 |
| C. glucuronolyticum | 1 | - | - | 1 |
| M. osloensis | 1 | - | - | 1 |
| MRSA | 1 | - | - | 1 |
| S. caprae | - | - | 1 | 1 |
| S. cohnii | 1 | - | - | 1 |
| S. pettenkoferi | - | - | 1 | 1 |
| S. simulans | - | 1 | - | 1 |
| S. xylosus | 1 | - | - | 1 |
| TOTAL | 72 | 34 | 34 | 140 |
| CFU | Total | ||||
|---|---|---|---|---|---|
| No | Low | High | |||
| MDR | No | 0 | 2 | 3 | 5 |
| MDR | 1 | 27 | 29 | 57 | |
| Total | 1 | 29 | 32 | 62 | |
| CFU | Total | ||||
|---|---|---|---|---|---|
| No | Low | High | |||
| XTT | No | 0 | 5 | 3 | 8 |
| Low | 0 | 4 | 10 | 14 | |
| High | 1 | 20 | 19 | 40 | |
| Total | 1 | 29 | 32 | 62 | |
| XTT | Total | ||||
|---|---|---|---|---|---|
| No | Low | High | |||
| MDR | No | 0 | 2 | 3 | 5 |
| MDR | 8 | 12 | 37 | 57 | |
| Total | 8 | 14 | 40 | 62 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coraça-Huber, D.C.; Kreidl, L.; Steixner, S.; Hinz, M.; Dammerer, D.; Fille, M. Identification and Morphological Characterization of Biofilms Formed by Strains Causing Infection in Orthopedic Implants. Pathogens 2020, 9, 649. https://doi.org/10.3390/pathogens9080649
Coraça-Huber DC, Kreidl L, Steixner S, Hinz M, Dammerer D, Fille M. Identification and Morphological Characterization of Biofilms Formed by Strains Causing Infection in Orthopedic Implants. Pathogens. 2020; 9(8):649. https://doi.org/10.3390/pathogens9080649
Chicago/Turabian StyleCoraça-Huber, Débora C., Lisa Kreidl, Stephan Steixner, Maximilian Hinz, Dietmar Dammerer, and Manfred Fille. 2020. "Identification and Morphological Characterization of Biofilms Formed by Strains Causing Infection in Orthopedic Implants" Pathogens 9, no. 8: 649. https://doi.org/10.3390/pathogens9080649
APA StyleCoraça-Huber, D. C., Kreidl, L., Steixner, S., Hinz, M., Dammerer, D., & Fille, M. (2020). Identification and Morphological Characterization of Biofilms Formed by Strains Causing Infection in Orthopedic Implants. Pathogens, 9(8), 649. https://doi.org/10.3390/pathogens9080649







