First Molecular Detection of Babesia ovis, Theileria spp., Anaplasma spp., and Ehrlichia ruminantium in Goats from Western Uganda
Abstract
1. Introduction
2. Results
2.1. Sampled Animal and Farm Demographics
2.2. Molecular Detection of Tick-borne Pathogens (TBPs) in Goats
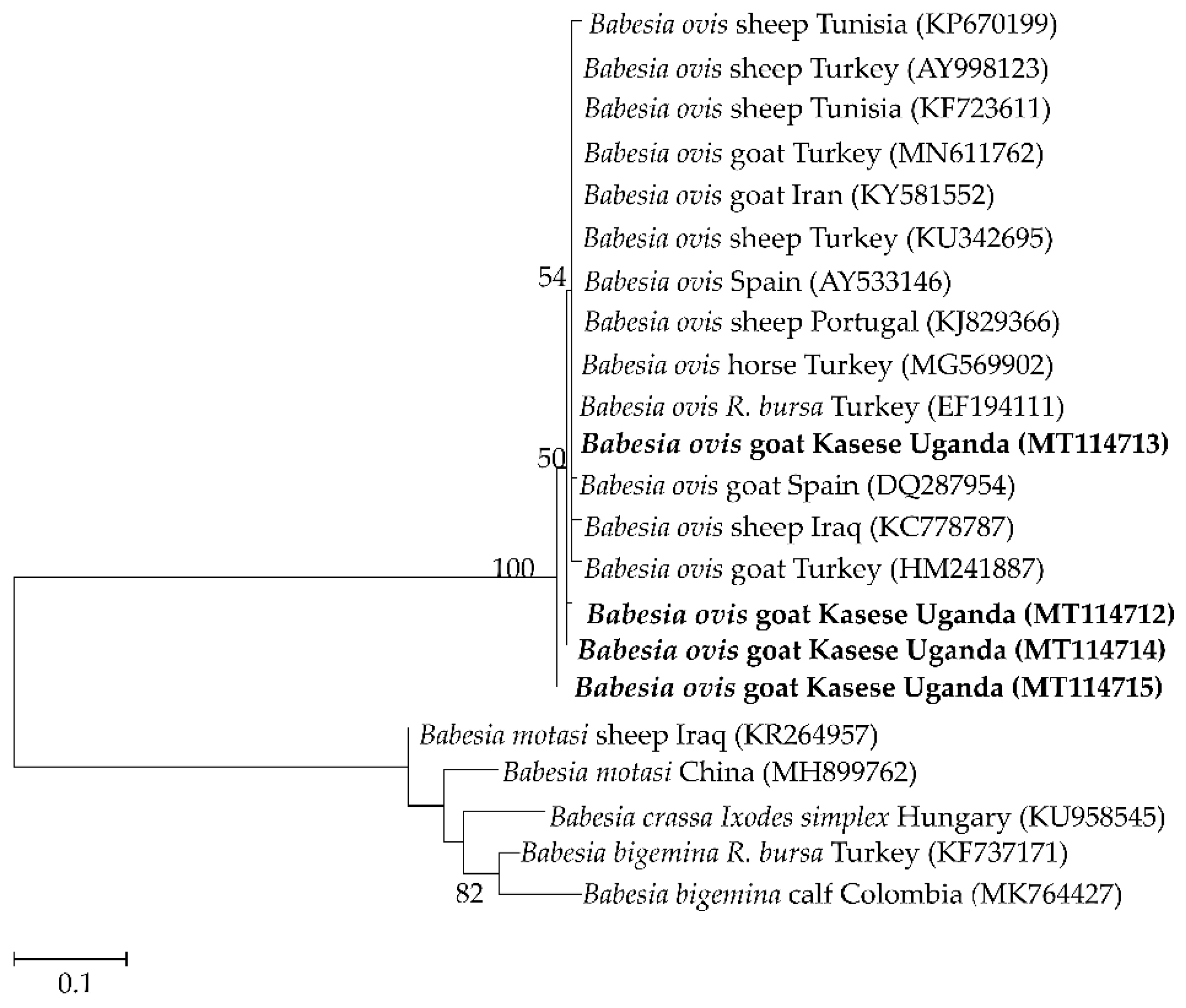
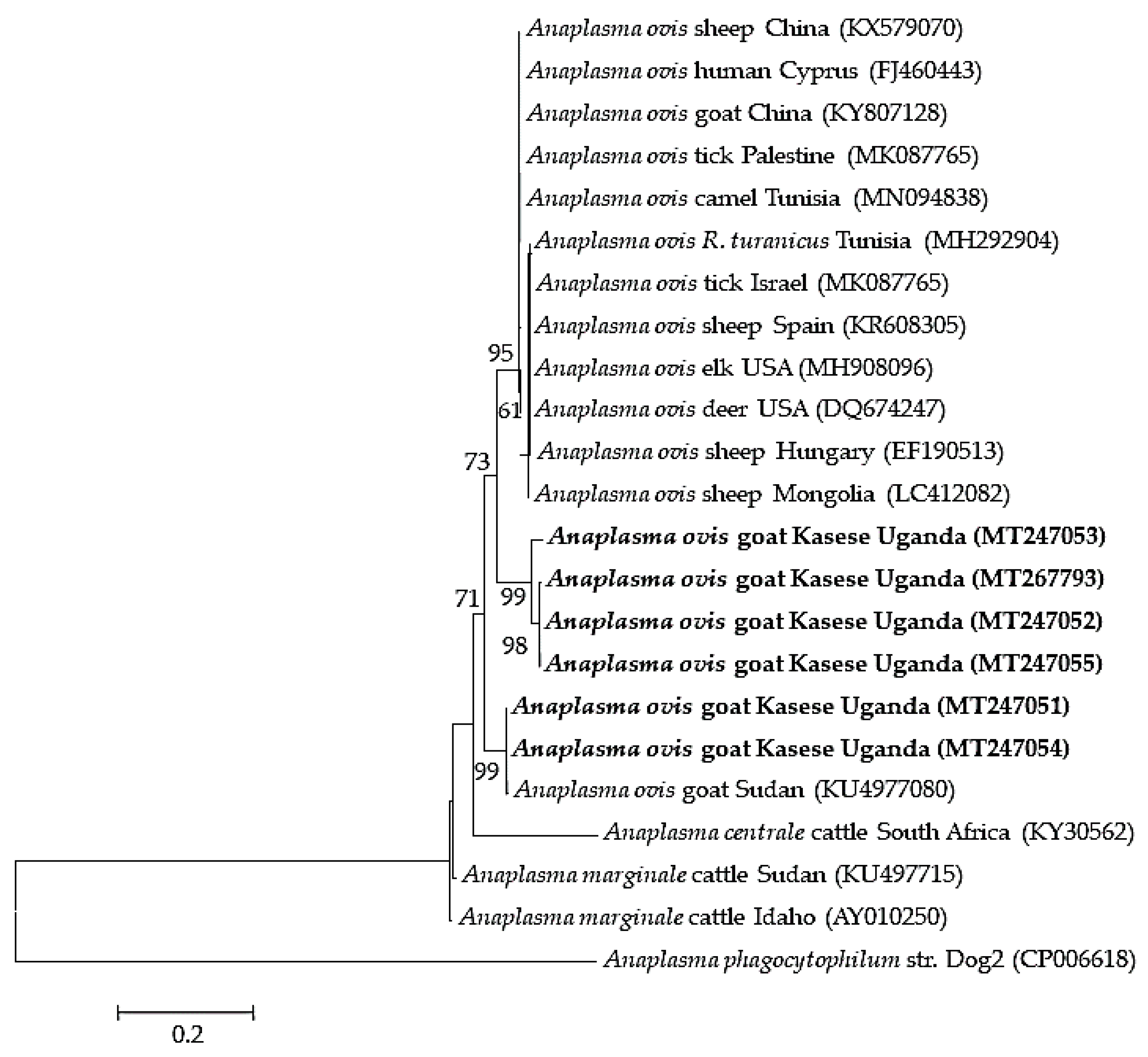
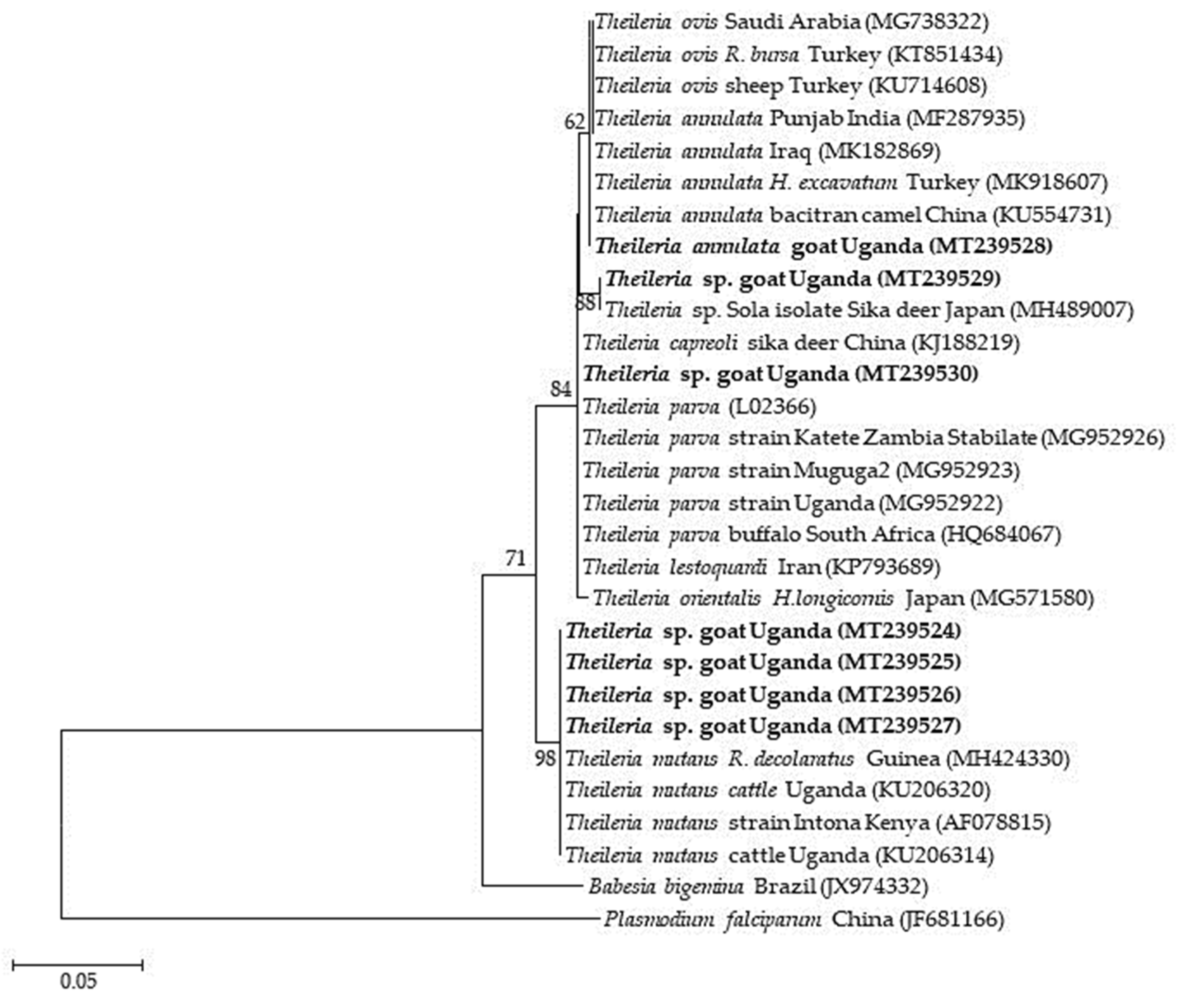
2.3. Sequence Similarities and Phylogenetic Analysis
2.4. Risk Factors Associated with the Detected TBPs
3. Discussion
4. Materials and Methods
4.1. Ethical Statements
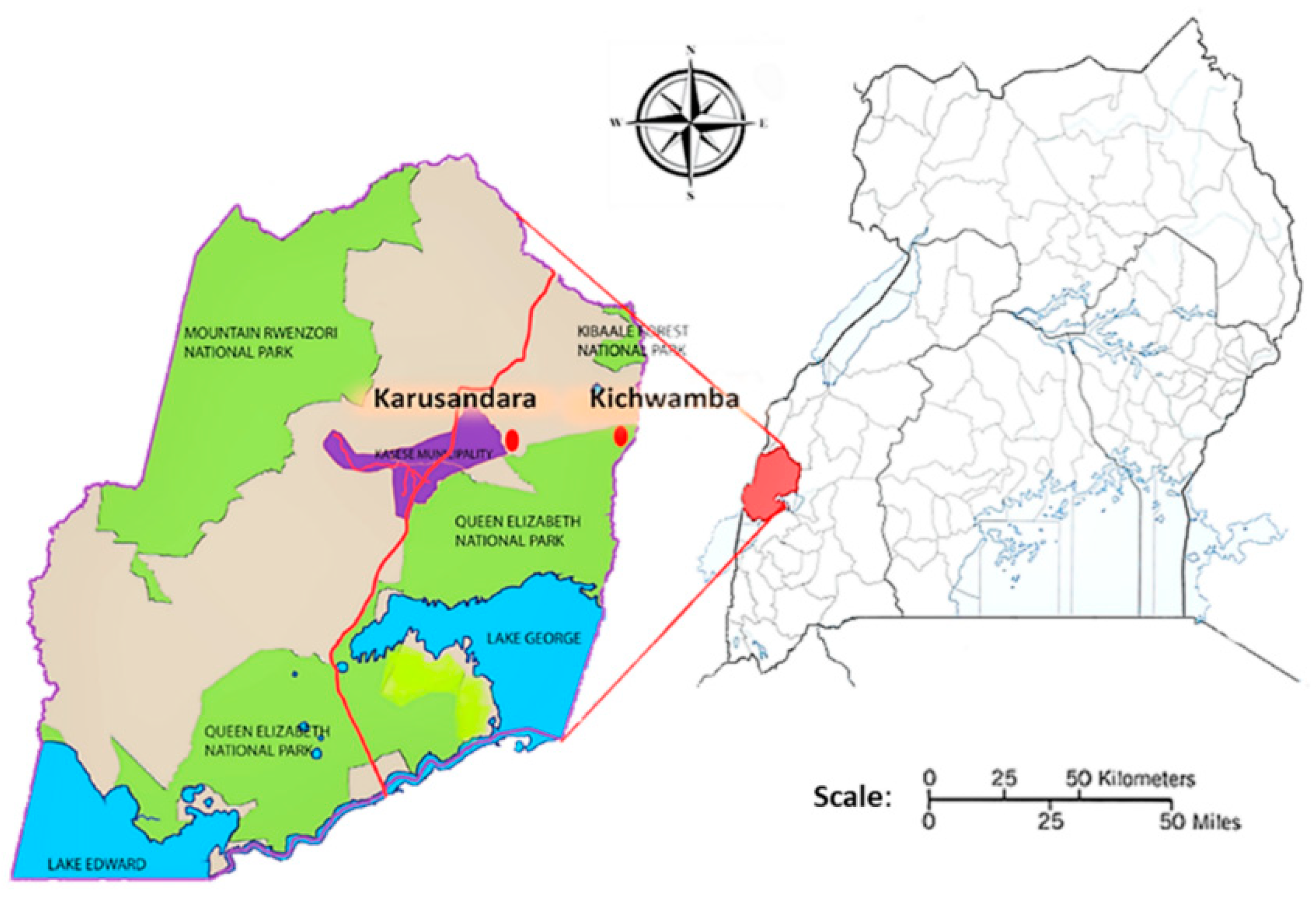
4.2. Sampling Area and Study Design
4.3. Sample Collection and DNA Extraction
4.4. Polymerase Chain Reaction Assays and DNA Sequencing
4.5. Phylogenetic Analysis
4.6. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Minjauw, B.; McLeod, A. Tick-Borne Diseases and Poverty: The Impact of Ticks and Tick-Borne Diseases on the Livelihoods of Small-Scale and Marginal Livestock Owners in India and Eastern and Southern Africa; DFID Animal Health Programme, Center for Tropical Veterinary Medicine: Edinburgh, UK, 2003; p. 124. [Google Scholar]
- Jongejan, F.; Uilenberg, G. Ticks and control methods. Rev. Sci. Tech. (Int. Off. Epizoot.) 1994, 13, 1201–1226. [Google Scholar] [CrossRef] [PubMed]
- El Hussein, A.M.; Majid, A.M.; Hassan, S.M. The present status of tick-borne diseases in the Sudan. Arch. Inst. Pasteur Tunis 2004, 81, 31–34. [Google Scholar] [PubMed]
- Ocaido, M.; Muwazi, R.T.; Opuda, J.A. Economic impact of ticks and tick-borne diseases on cattle production systems around Lake Mburo National Park in South Western Uganda. Trop. Anim. Health Prod. 2008, 41, 731–739. [Google Scholar] [CrossRef] [PubMed]
- Schnittger, L.; Yin, H.; Gubbels, M.-J.; Beyer, D.; Niemann, S.; Jongejan, F.; Ahmed, J.S. Phylogeny of sheep and goat Theileria and Babesia parasites. Parasitol. Res. 2003, 91, 398–406. [Google Scholar] [CrossRef] [PubMed]
- Yeruham, I.; Hadani, A.; Galker, F.; Rosen, S. Notes on the biology of the tick Rhipicephalus bursa (Canestrini and Fanzago, 1877) in Israel. Rev. Élev. Méd. Vét. Pays Trop. 1989, 42, 233–235. [Google Scholar] [PubMed]
- Ijaz, M.; Rehman, A.; Ali, M.M.; Umair, M.; Khalid, S.; Mehmood, K.; Hanif, A. Clinico-epidemiology and therapeutical trials on babesiosis in sheep and goats in Lahore, Pakistan. J. Anim. Plant Sci. 2013, 23, 666–669. [Google Scholar]
- Hasherni-Fesharki, R.; Uilenberg, G. Babesia crassa n.sp. (Sporozoa, Babesiidae) of domestic sheep in Iran. Vet. Q. 1981, 3, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Zaeemi, M.; Haddadzadeh, H.; Khazraiinia, P.; Kazemi, B.; Bandehpour, M. Identification of different Theileria species (Theileria lestoquardi, Theileria ovis, and Theileria annulata) in naturally infected sheep using nested PCR–RFLP. Parasitol. Res. 2010, 108, 837–843. [Google Scholar] [CrossRef] [PubMed]
- Khezri, M.; Habibi, G.; Esmaeil-Nia, K.; Afshari, A. The first genetic identification of Theileria ovis subtype KP019206 in sheep in Iran. Arch. Razi Inst. 2016, 71, 145–152. [Google Scholar]
- Bilgic, H.B.; Bakırcı, S.; Kose, O.; Unlu, A.H.; Hacılarlıoglu, S.; Eren, H.; Weir, W.; Karagenc, T. Prevalence of tick-borne haemoparasites in small ruminants in Turkey and diagnostic sensitivity of single-PCR and RLB. Parasites Vectors 2017, 10, 1–13. [Google Scholar] [CrossRef]
- Naz, S.; Maqbool, A.; Ahmed, S.; Ashraf, K.; Ahmed, N.; Saeed, K.; Latif, M.; Iqbal, J.; Ali, Z.; Shafi, K.; et al. Prevalence of theileriosis in small ruminants in Lahore-Pakistan. J. Vet. Anim. Sci. 2012, 2, 16–20. [Google Scholar]
- Kuttler, K.L. Anaplasma infections in wild and domestic ruminants: A review. J. Wildl. Dis. 1984, 20, 12–20. [Google Scholar] [CrossRef]
- Stuen, S.; Longbottom, D. Treatment and Control of Chlamydial and Rickettsial Infections in Sheep and Goats. Vet. Clin. N. Am. Food Anim. Pract. 2011, 27, 213–233. [Google Scholar] [CrossRef]
- Hornok, S.; De La Fuente, J.; Biró, N.; De Mera, I.G.F.; Meli, M.L.; Elek, V.; Gönczi, E.; Meili, T.; Tánczos, B.; Farkas, R.; et al. First Molecular Evidence of Anaplasma ovis and Rickettsia spp. in Keds (Diptera: Hippoboscidae) of Sheep and Wild Ruminants. Vector-Borne Zoonotic Dis. 2011, 11, 1319–1321. [Google Scholar] [CrossRef]
- Woldehiwet, Z. The natural history of Anaplasma phagocytophilum. Vet. Parasitol. 2010, 167, 108–122. [Google Scholar] [CrossRef]
- E Yunker, C. Heartwater in sheep and goats: A review. Onderstepoort J. Vet. Res. 1996, 63, 159–170. [Google Scholar]
- FAO Animal Production and Health Position Paper. Supporting Livelihoods and Building Resilience Through Peste des Petits Ruminants (PPR) and Small Ruminant Disease Control. Animal Production and Health Position Paper. 2013. Available online: http://www.fao.org/3/a-aq236e.pdf (accessed on 26 October 2020).
- FAO ASL2050. African Sustainable Livestock 2050–Livestock productions systems spotlight, Uganda, Chicken, Meat and Beef. Available online: www.fao.org/ag/againfo/programmes/en/ASL2050.html (accessed on 26 October 2020).
- Uilenberg, G.; Service, M.W. Babesiosis. In Encyclopedia of Arthropod-Transmitted Infections of Man and Domesticated Animals; Service, M.W., Ed.; CABI Publishing: Wallingford, UK, 2001; pp. 53–60. [Google Scholar]
- Sevinc, F.; Sevinc, M.; Ekici, O.D.; Yildiz, R.; Isik, N.; Aydoğdu, U. Babesia ovis infections: Detailed clinical and laboratory observations in the pre- and post-treatment periods of 97 field cases. Vet. Parasitol. 2013, 191, 35–43. [Google Scholar] [CrossRef]
- Rjeibi, M.R.; Gharbi, M.; Mhadhbi, M.; Mabrouk, W.; Ayari, B.; Nasfi, I.; Jedidi, M.; Sassi, L.; Rekik, M.; Darghouth, M.A. Prevalence of piroplasms in small ruminants in North-West Tunisia and the first genetic characterization of Babesia ovis in Africa. Parasite 2014, 21, 23. [Google Scholar] [CrossRef]
- Aouadi, A.; Leulmi, H.; Boucheikhchoukh, M.; Benakhla, A.; Raoult, D.; Parola, P. Molecular evidence of tick-borne hemoprotozoan-parasites (Theileria ovis and Babesia ovis) and bacteria in ticks and blood from small ruminants in Northern Algeria. Comp. Immunol. Microbiol. Infect. Dis. 2017, 50, 34–39. [Google Scholar] [CrossRef]
- Yeruham, I.; Hadani, A.; Galker, F. Some epizootiological and clinical aspects of ovine babesiosis caused by Babesia ovis—A review. Vet. Parasitol. 1998, 74, 153–163. [Google Scholar] [CrossRef]
- Altay, K.; Dumanli, N.; Aktas, M. Molecular identification, genetic diversity and distribution of Theileria and Babesia species infecting small ruminants. Vet. Parasitol. 2007, 147, 161–165. [Google Scholar] [CrossRef]
- Ranjbar-Bahadori, S.; Eckert, B.; Omidian, Z.; Shirazi, N.S.; Shayan, P. Babesia ovis as the main causative agent of sheep babesiosis in Iran. Parasitol. Res. 2011, 110, 1531–1536. [Google Scholar] [CrossRef]
- Esmaeilnejad, B.; Tavassoli, M.; Asri-Rezaei, S.; Dalir-Naghadeh, B.; Mardani, K.; Jalilzadeh-Amin, G.; Golabi, M.; Arjmand, J. PCR-Based Detection ofBabesia ovisinRhipicephalus bursaand Small Ruminants. J. Parasitol. Res. 2014, 2014, 1–6. [Google Scholar] [CrossRef]
- Aktaş, M.; Altay, K.; Dumanli, N. Determination of prevalence and risk factors for infection with Babesia ovis in small ruminants from Turkey by polymerase chain reaction. Parasitol. Res. 2006, 100, 797–802. [Google Scholar] [CrossRef]
- Stuen, S.; Kjølleberg, K. An investigation of lamb deaths on tick pastures in Norway. In Proceedings of the Third International Conference on Ticks and Tick-Borne Pathogens: Into the 21st Century; Kazimìrovà, M., Labuda, M., Nuttall, P.A., Eds.; Slovak Academy of Sciences: Bratislava, Slovakia, 2000; pp. 111–115. [Google Scholar]
- M’Ghirbi, Y.; Yaïch, H.; Ghorbel, A.; Bouattour, A. Anaplasma phagocytophilum in horses and ticks in Tunisia. Parasites Vectors 2012, 5, 180. [Google Scholar] [CrossRef]
- Belkahia, H.; Ben Said, M.; El Mabrouk, N.; Saidani, M.; Cherni, C.; Ben Hassen, M.; Bouattour, A.; Messadi, L. Seasonal dynamics, spatial distribution and genetic analysis of Anaplasma species infecting small ruminants from Northern Tunisia. Infect. Genet. Evol. 2017, 54, 66–73. [Google Scholar] [CrossRef]
- Yang, J.; Liu, Z.; Niu, Q.; Liu, J.; Han, R.; Guan, G.; Li, Y.; Liu, G.; Luo, J.; Yin, H. Anaplasma phagocytophilum in sheep and goats in central and southeastern China. Parasites Vectors 2016, 9, 1–7. [Google Scholar] [CrossRef][Green Version]
- Yoshimoto, K.; Matsuyama, Y.; Matsuda, H.; Sakamoto, L.; Matsumoto, K.; Yokoyama, N.; Inokuma, H. Detection of Anaplasma bovis and Anaplasma phagocytophilum DNA from Haemaphysalis megaspinosa in Hokkaido, Japan. Vet. Parasitol. 2010, 168, 170–172. [Google Scholar] [CrossRef]
- Oura, C.; Tait, A.; Asiimwe, B.; Lubega, G.W.; Weir, W. Theileria parva genetic diversity and haemoparasite prevalence in cattle and wildlife in and around Lake Mburo National Park in Uganda. Parasitol. Res. 2010, 108, 1365–1374. [Google Scholar] [CrossRef]
- Jahfari, S.; Coipan, E.C.; Fonville, M.; Van Leeuwen, A.D.; Hengeveld, P.; Heylen, D.; Heyman, P.; Van Maanen, C.; Butler, C.M.; Földvári, G.; et al. Circulation of four Anaplasma phagocytophilum ecotypes in Europe. Parasites Vectors 2014, 7, 1–11. [Google Scholar] [CrossRef]
- Magona, J.W.; Walubengo, J.; Olaho-Mukani, W.; Jonsson, N.N.; Welburn, S.C.; Eisler, M.C. Spatial variation of tick abundance and seroconversion rates of indigenous cattle to Anaplasma marginale, Babesia bigemina and Theileria parva infections in Uganda. Exp. Appl. Acarol. 2011, 55, 203–213. [Google Scholar] [CrossRef]
- Madeira, N.; Amarante, A.; Padovani, C.R. Diversity of Ectoparasites in Sheep Flocks in Sao Paulo, Brazil. Trop. Anim. Health Prod. 2000, 32, 225–232. [Google Scholar] [CrossRef]
- Ahmed, J.S.; Yin, H.; Bakheit, M.; Liu, Z.; Mehlhorn, H.; Seitzer, U. Small Ruminant Theileriosis. In Parasitology; Springer: Berlin/Heidelberg, Germany, 2011; Volume 2, pp. 135–153. [Google Scholar] [CrossRef]
- Gebrekidan, H.; Hailu, A.; Kassahun, A.; Rohoušová, I.; Maia, C.; Talmi-Frank, D.; Warburg, A.; Baneth, G. Theileria infection in domestic ruminants in northern Ethiopia. Vet. Parasitol. 2014, 200, 31–38. [Google Scholar] [CrossRef]
- Weny, G.; Okwee-Acai, J.; Okech, S.G.; Tumwine, G.; Ndyanabo, S.; Abigaba, S.; Goldberg, T.L. Prevalence and Risk Factors Associated with Hemoparasites in Cattle and Goats at the Edge of Kibale National Park, Western Uganda. J. Parasitol. 2017, 103, 69–74. [Google Scholar] [CrossRef]
- Ghai, R.R.; Mutinda, M.; Ezenwa, V.O. Limited sharing of tick-borne hemoparasites between sympatric wild and domestic ungulates. Vet. Parasitol. 2016, 226, 167–173. [Google Scholar] [CrossRef]
- Brown, C.G.D.; Ilhan, T.; Kirvar, E.; Thomas, M.; Wilkie, G.; Leemans, I.; Hooshmand-Rad, P. Theileria lestoquardi and T. annulata in Cattle, Sheep, and Goats: In Vitro and in Vivo Studies a. Ann. N. Y. Acad. Sci. 1998, 849, 44–51. [Google Scholar] [CrossRef]
- Taha, K.; Salih, D.; Ali, A.; Omer, R.; El Hussein, A. Naturally occurring infections of cattle with Theileria lestoquardi and sheep with Theileria annulata in the Sudan. Vet. Parasitol. 2013, 191, 143–145. [Google Scholar] [CrossRef]
- Macfie, J.S. Babesiosis and trypanosomiasis at Accra, Gold Coast, West Africa. Ann. Trop. Med. Parasitol. 1915, 9, 457–494. [Google Scholar] [CrossRef]
- Lee, S.-H.; Moumouni, P.F.A.; Galon, E.M.S.; Vudriko, P.; Liu, M.; Benedicto, B.; Tumwebaze, M.A.; Boldbaatar, D.; Umemiya-Shirafuji, R.; Fukumoto, S.; et al. Differential diagnosis and molecular characterization of Theileria spp. in sika deer (Cervus nippon) in Hokkaido, Japan. Parasitol. Int. 2019, 70, 23–26. [Google Scholar] [CrossRef]
- El Imam, A.H.; Hassan, S.M.; A Gameel, A.; El Hussein, A.M.; Taha, K.M.; Oosthuizen, M.C. Molecular identification of different Theileria and Babesia species infecting sheep in Sudan. Ann. Parasitol. 2016, 62, 47–54. [Google Scholar]
- Kahn, C.M.; Scott, L.; Aiello, S.E. The Merck Veterinary Manual, 9th ed.; National Publishing Co.: Philadelphia, PA, USA, 2005; pp. 146–148. [Google Scholar]
- Dolan, T.T. Theileriosis: A comprehensive review. Rev. Sci. Tech. Off. Int. Epizoot. 1989, 8, 11–36. [Google Scholar] [CrossRef] [PubMed]
- El Hussein, A.M.; ElGhali, A.A.; Mohammed, S.A. Efficacy of buparvaquone in the treatment of malignant theileriosis of sheep in Ed-Damer Province, N. State, Sudan: A field trial. Sud. J. Vet. Res. 1993, 12, 51–57. [Google Scholar]
- Ringo, A.E.; Moumouni, P.F.A.; Lee, S.-H.; Liu, M.; Khamis, Y.H.; Gao, Y.; Guo, H.; Zheng, W.; Efstratiou, A.; Galon, E.M.S.; et al. Molecular detection and characterization of tick-borne protozoan and rickettsial pathogens isolated from cattle on Pemba Island, Tanzania. Ticks Tick-Borne Dis. 2018. [Google Scholar] [CrossRef]
- Lee, S.-H.; Mossaad, E.; Ibrahim, A.M.; Ismail, A.A.; Moumouni, P.F.A.; Liu, M.; Ringo, A.E.; Gao, Y.; Guo, H.; Li, J.; et al. Detection and molecular characterization of tick-borne pathogens infecting sheep and goats in Blue Nile and West Kordofan states in Sudan. Ticks Tick-Borne Dis. 2018, 9, 598–604. [Google Scholar] [CrossRef]
- Altay, K.; Dumanli, N.; Holman, P.J.; Aktaş, M. Detection of Theileria ovis in naturally infected sheep by nested PCR. Vet. Parasitol. 2005, 127, 99–104. [Google Scholar] [CrossRef] [PubMed]
- The Ministry of Agriculture, Animal Industry and Fisheries of the Republic of Uganda. Goat Production Manual; National Agricultural Advisory Services (NAADs): Kampala, Uganda, 2005.
- Abebe, R.; Tatek, M.; Megersa, B.; Sheferaw, D. Prevalence of small ruminant ectoparasites and associated risk factors in selected districts of Tigray Region, Ethiopia. Glob. Vet. 2011, 7, 433–437. [Google Scholar]
- Ringo, A.E.; Moumouni, P.F.A.; Taioe, M.; Jirapattharasate, C.; Liu, M.; Wang, G.; Gao, Y.; Guo, H.; Lee, S.-H.; Zheng, W.; et al. Molecular analysis of tick-borne protozoan and rickettsial pathogens in small ruminants from two South African provinces. Parasitol. Int. 2018, 67, 144–149. [Google Scholar] [CrossRef]
- Ben Said, M.; Belkahia, H.; Alberti, A.; Zobba, R.; Bousrih, M.; Yahiaoui, M.; Daaloul-Jedidi, M.; Mamlouk, A.; Gharbi, M.; Messadi, L. Molecular Survey ofAnaplasmaSpecies in Small Ruminants Reveals the Presence of Novel Strains Closely Related toA. phagocytophilumin Tunisia. Vector-Borne Zoonotic Dis. 2015, 15, 580–590. [Google Scholar] [CrossRef] [PubMed]
- De La Fuente, J.; Ruybal, P.; Mtshali, M.S.; Naranjo, V.; Shuqing, L.; Mangold, A.J.; Rodríguez, S.D.; Jiménez, R.; Vicente, J.; Moretta, R.; et al. Analysis of world strains of Anaplasma marginale using major surface protein 1a repeat sequences. Vet. Microbiol. 2007, 119, 382–390. [Google Scholar] [CrossRef]
- Han, R.; Yang, J.; Liu, Z.; Gao, S.; Niu, Q.; Hassan, M.A.; Luo, J.; Yin, H. Characterization of Anaplasma ovis strains using the major surface protein 1a repeat sequences. Parasites Vectors 2017, 10, 447. [Google Scholar] [CrossRef]
- Vudriko, P.; Okwee-Acai, J.; Tayebwa, D.S.; Byaruhanga, J.; Kakooza, S.; Wampande, E.; Omara, R.; Muhindo, J.B.; Tweyongyere, R.; Owiny, D.O.; et al. Emergence of multi-acaricide resistant Rhipicephalus ticks and its implication on chemical tick control in Uganda. Parasites Vectors 2016, 9, 1–13. [Google Scholar] [CrossRef]
- Benedicto, B.; Ceylan, O.; Moumouni, P.F.A.; Lee, S.-H.; Tumwebaze, M.A.; Li, J.; Galon, E.M.; Liu, M.; Li, Y.; Ji, S.; et al. Molecular Detection and Assessment of Risk Factors for Tick-Borne Diseases in Sheep and Goats from Turkey. Acta Parasitol. 2020, 65, 723–732. [Google Scholar] [CrossRef] [PubMed]
- Aktaş, M.; Altay, K.; Dumanlı, N. Development of a polymerase chain reaction method for diagnosis of Babesia ovis infection in sheep and goats. Vet. Parasitol. 2005, 133, 277–281. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Zhang, S.; Jia, L.; Xue, S.; Yu, L.; Kamyingkird, K.; Moumouni, P.F.A.; Moussa, A.A.E.M.; Zhou, M.; Zhang, Y.; et al. Molecular Detection of Theileria Species in Sheep from Northern China. J. Vet. Med. Sci. 2013, 75, 1227–1230. [Google Scholar] [CrossRef] [PubMed]
- Torina, A.; Agnone, A.; Blanda, V.; Alongi, A.; D’Agostino, R.; Caracappa, S.; Marino, A.M.; Di Marco, V.; De La Fuente, J. Development and validation of two PCR tests for the detection of and differentiation between Anaplasma ovis and Anaplasma marginale. Ticks Tick-Borne Dis. 2012, 3, 283–287. [Google Scholar] [CrossRef] [PubMed]
- E Barlough, J.; E Madigan, J.; DeRock, E.; Dumler, J.S.; Bakken, J.S. Protection against Ehrlichia equi is conferred by prior infection with the human granulocytotropic Ehrlichia (HGE agent). J. Clin. Microbiol. 1995, 33, 3333–3334. [Google Scholar] [CrossRef] [PubMed]
- Farougou, S.; Adakal, H.; Biguezoton, A.S.; Boko, C. Prevalence of Amblyomma variegatum infection by Ehrlichia ruminantium in cattle extensive herds in Benin. Rev. Med. Vet. 2012, 163, 261–266. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
| Parameter | Number (n = 201) | Percentage (%) |
|---|---|---|
| Source | ||
| Karusandara | 103 | 51.2 |
| Kichwamba | 98 | 48.8 |
| Breed | ||
| Local | 162 | 80.6 |
| Cross | 39 | 19.4 |
| Sex | ||
| Female | 182 | 90.5 |
| Male | 19 | 9.5 |
| Age | ||
| Young | 112 | 55.7 |
| Adult | 89 | 44.3 |
| Body condition score (BCS) | ||
| ≤3 | 160 | 79.6 |
| >3 | 41 | 20.4 |
| Tick infestation | ||
| Infested | 78 | 38.8 |
| Not infested | 123 | 61.2 |
| Parameter | No. of Farms | Percentage (%) |
|---|---|---|
| Farming system | (n = 19) | |
| Free range | 10 | 52.6 |
| Paddocking | 4 | 21.1 |
| Communal | 3 | 15.8 |
| Tethering | 2 | 10.5 |
| Herd size | ||
| ≥30 | 14 | 73.7 |
| <30 | 5 | 26.3 |
| Acaricide application | ||
| Application | 12 | 63.2 |
| None | 7 | 36.8 |
| Interaction with wildlife | ||
| Interaction | 9 | 47.4 |
| None | 10 | 52.6 |
| Disease challenges | ||
| Observed | 8 | 42.1 |
| None | 11 | 57.9 |
| Wildlife Encountered | (n = 9) | |
| Antelopes | 3 | 33.3 |
| Elephants | 3 | 33.3 |
| Warthogs | 2 | 22.2 |
| Buffaloes | 2 | 22.2 |
| Hares | 2 | 22.2 |
| Primary Disease challenges | (n = 8) | |
| Fever | 4 | 50.0 |
| Helminths | 2 | 25.0 |
| Abortion | 1 | 12.5 |
| Abscesses | 1 | 12.5 |
| Overall (%) | Babesia ovis (%) | Theileria spp. (%) | Anaplasma ovis (%) | Anaplasma phagocytophilum (%) | Ehrlichia ruminantium (%) | |
|---|---|---|---|---|---|---|
| Karusandara (n = 103) | 52 (50.5) | 4 (3.9) | 25 (24.3) | 8 (7.8) | 15 (4.9) | 0 (0.0) |
| Kichwamba (n = 98) | 20 (20.4) | 7 (7.1) | 2 (2.0) | 3 (3.1) | 7 (7.1) | 1 (1.0) |
| Total (n = 201) | 72 (35.8) | 11 (5.5) | 27 (13.4) | 11 (5.5) | 22 (10.9) | 1 (0.5) |
| p-value | 0.00001 * | 0.30985 | 0.00001 * | 0.14260 | 0.09213 | N/A 1 |
| Pathogen Species | No. of Positives (n = 201) | Detection Rate % |
|---|---|---|
| Single infections | ||
| Babesia ovis | 7 | 3.5 |
| Theileria spp. | 17 | 8.5 |
| Anaplasma ovis | 3 | 1.5 |
| Anaplasma phagocytophilum | 16 | 7.9 |
| Ehrlichia ruminantium | 1 | 0.49 |
| Coinfections | ||
| Double | ||
| Babesia ovis + Anaplasma ovis | 1 | 0.49 |
| Babesia ovis + Theileria spp. | 1 | 0.49 |
| Babesia ovis + Anaplasma phagocytophilum | 3 | 1.49 |
| Anaplasma ovis + Theileria spp. | 4 | 1.99 |
| Anaplasma ovis + Anaplasma phagocytophilum | 2 | 0.99 |
| Theileria spp. + Anaplasma phagocytophilum | 5 | 2.49 |
| Triple | ||
| Theileria spp. + Anaplasma ovis + Anaplasma phagocytophilum | 1 | 0.49 |
| Pathogen | Parameter | p-Value | OR | Confidence Interval |
|---|---|---|---|---|
| B. ovis | Wildlife interaction | 0.020 | 10.0 | 1.25–76.99 |
| Theileria spp. | Subcounty | 0.003 | 19.9 | 3.84–103.46 |
| BCS | 0.011 | 3.90 | 1.35–11.42 | |
| Tick infestation | 0.04 | 0.22 | 0.05–0.94 | |
| A. phagocytophilum | Breed | 0.005 | 0.18 | 0.05–0.60 |
| Tick infestation | 0.030 | 0.30 | 0.09–0.93 | |
| Herd size | 0.023 | 0.16 | 0.03–0.78 | |
| Wildlife interaction | 0.001 | 7.97 | 2.15–29.54 |
| Pathogen | Target Gene | Primer Sequence (5′->3′) | Annealing Temp (°C) | Amplicon Size (bp) | Reference |
|---|---|---|---|---|---|
| Babesia ovis | ssu rRNA | TGGGCAGGACCTTGGTTCTTCT CCGCGTAGCGCCGGCTAAATA | 62 | 549 | [61] |
| Theileria spp. | 18S rRNA | GAAACGGCTACCACATCT AGTTTCCCCGTGTTGAGT TTAAACCTCTTCCAGAGT TCAGCCTTGCGACCATAC | 55 | 778 581 | [62] |
| Theileria ovis | ssu rRNA | TCGAGACCTTCGGGT TCCGGACATTGTAAAACAAA | 53 | 520 | [52] |
| Theileria lestoquardi | 18S rRNA | GTGCCGCAAGTGAGTCA GGACTGATGAGAAGACGATGAG | 52 | 730 | [43] |
| Anaplasma ovis | msp4 | TGAAGGGAGCGGGGTCATGGG GAGTAATTGCAGCCAGGCACTCT | 62 | 347 | [63] |
| Anaplasma phagocytophilum | 16S rRNA | TCCTGGCTCAGAACGAACGCTGGCGGC AGTCACTGACCCAACCTTAAATGGCTG GTCGAACGGATTATTCTTTATAGCTTGC CCCTTCCGTTAAGAAGGATCTAATCTCC | 50 | 1433 926 | [64] |
| Ehrlichia ruminantium | pCS20 | ACTAGTAGAAATTGCACAATCYAT RCTDGCWGCTTTYTGTTCAGCTAK ACTAGTAGAAATTGCACAATCYAT TGATAACTTGGWGCRRGDARTCCTT | 61 | 400 278 | [65] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tumwebaze, M.A.; Byamukama, B.; Tayebwa, D.S.; Byaruhanga, J.; Angwe, M.K.; Galon, E.M.; Liu, M.; Lee, S.-H.; Ringo, A.E.; Adjou Moumouni, P.F.; et al. First Molecular Detection of Babesia ovis, Theileria spp., Anaplasma spp., and Ehrlichia ruminantium in Goats from Western Uganda. Pathogens 2020, 9, 895. https://doi.org/10.3390/pathogens9110895
Tumwebaze MA, Byamukama B, Tayebwa DS, Byaruhanga J, Angwe MK, Galon EM, Liu M, Lee S-H, Ringo AE, Adjou Moumouni PF, et al. First Molecular Detection of Babesia ovis, Theileria spp., Anaplasma spp., and Ehrlichia ruminantium in Goats from Western Uganda. Pathogens. 2020; 9(11):895. https://doi.org/10.3390/pathogens9110895
Chicago/Turabian StyleTumwebaze, Maria Agnes, Benedicto Byamukama, Dickson Stuart Tayebwa, Joseph Byaruhanga, Martin Kamilo Angwe, Eloiza May Galon, Mingming Liu, Seung-Hun Lee, Aaron Edmond Ringo, Paul Franck Adjou Moumouni, and et al. 2020. "First Molecular Detection of Babesia ovis, Theileria spp., Anaplasma spp., and Ehrlichia ruminantium in Goats from Western Uganda" Pathogens 9, no. 11: 895. https://doi.org/10.3390/pathogens9110895
APA StyleTumwebaze, M. A., Byamukama, B., Tayebwa, D. S., Byaruhanga, J., Angwe, M. K., Galon, E. M., Liu, M., Lee, S.-H., Ringo, A. E., Adjou Moumouni, P. F., Li, J., Li, Y., Ji, S., Vudriko, P., & Xuan, X. (2020). First Molecular Detection of Babesia ovis, Theileria spp., Anaplasma spp., and Ehrlichia ruminantium in Goats from Western Uganda. Pathogens, 9(11), 895. https://doi.org/10.3390/pathogens9110895







