Molecular Characterization of the Multidrug Resistant Escherichia coli ST131 Clone
Abstract
:1. Introduction
2. Global Epidemiology of ST131
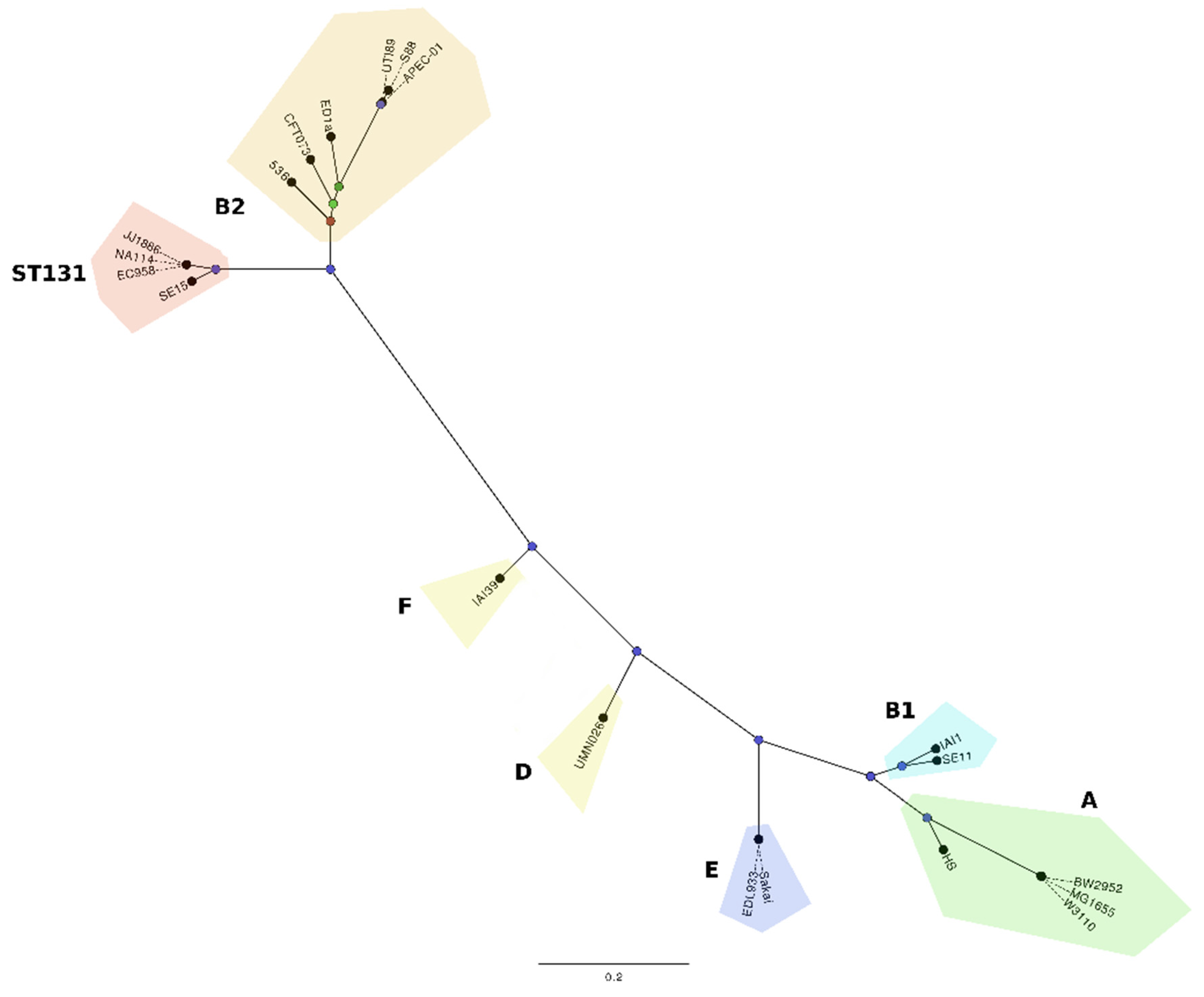
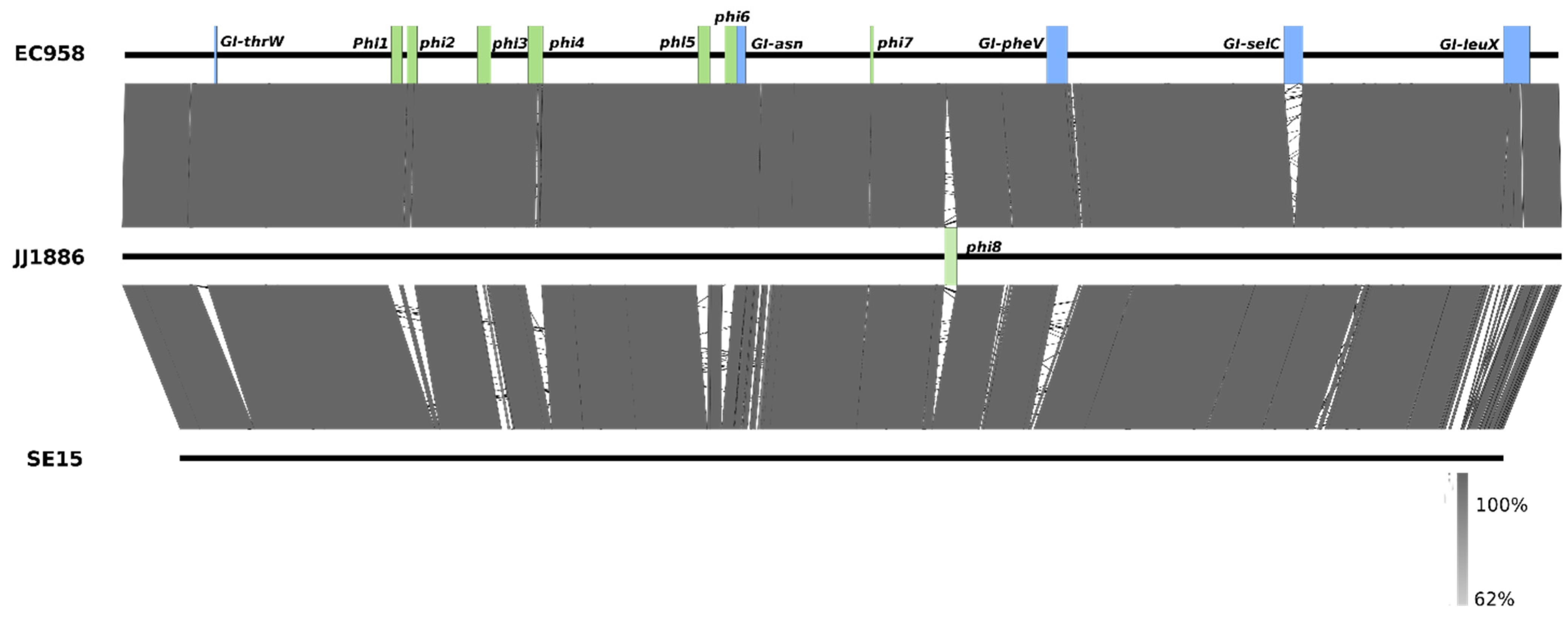
3. Molecular Characterisation of the ST131 Reference Strain EC958
4. Virulence of E. coli ST131
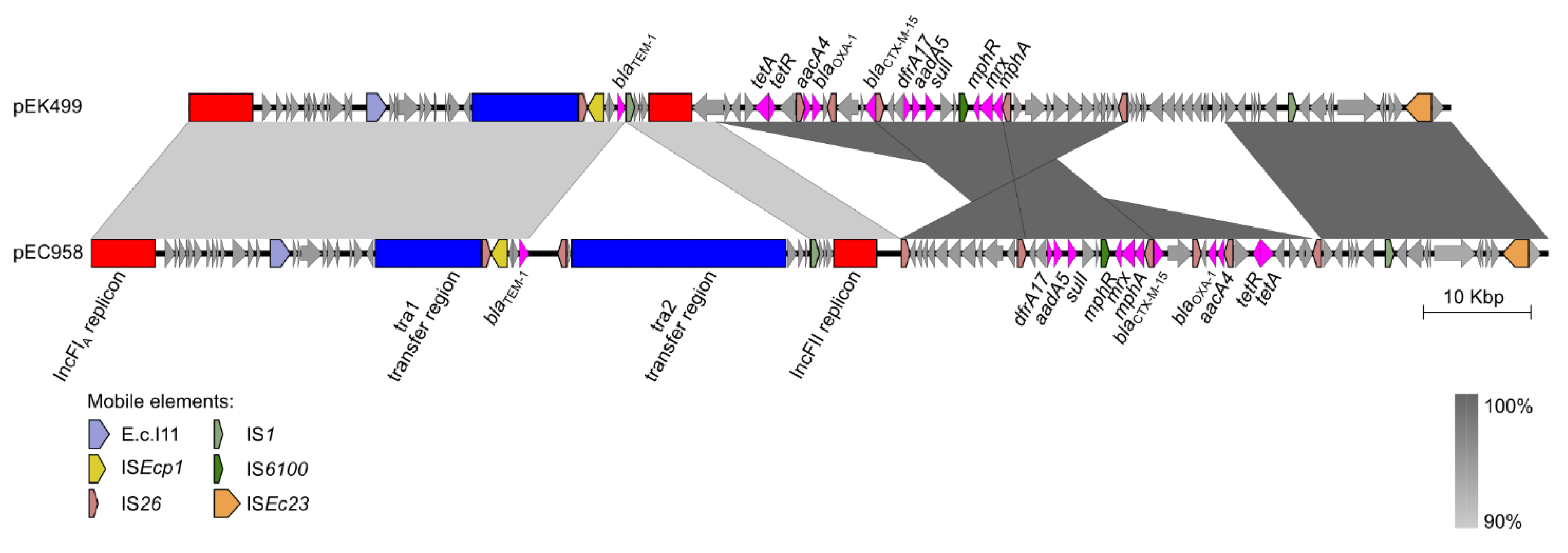
5. Plasmids of ST131
6. Conclusions
Acknowledgments
Conflicts of Interest
References
- Flores-Mireles, A.L.; Walker, J.N.; Caparon, M.; Hultgren, S.J. Urinary tract infections: Epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 2015, 13, 269–284. [Google Scholar] [CrossRef] [PubMed]
- Riley, L.W. Pandemic lineages of extraintestinal pathogenic Escherichia coli. Clin. Microbiol. Infect. 2014, 20, 380–390. [Google Scholar] [CrossRef] [PubMed]
- Mathers, A.J.; Peirano, G.; Pitout, J.D. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant enterobacteriaceae. Clin. Microbiol. Rev. 2015, 28, 565–591. [Google Scholar] [CrossRef] [PubMed]
- Coque, T.M.; Novais, A.; Carattoli, A.; Poirel, L.; Pitout, J.; Peixe, L.; Baquero, F.; Canton, R.; Nordmann, P. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum beta-lactamase ctx-m-15. Emerg. Infect. Dis. 2008, 14, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.H.; Reddy, S.; Cheesbrough, J.; Bolton, F.J.; Willshaw, G.; Cheasty, T.; Fox, A.J.; Upton, M. Major uropathogenic Escherichia coli strain isolated in the northwest of England identified by multilocus sequence typing. J. Clin. Microbiol. 2008, 46, 1076–1080. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nicolas-Chanoine, M.H.; Blanco, J.; Leflon-Guibout, V.; Demarty, R.; Alonso, M.P.; Canica, M.M.; Park, Y.J.; Lavigne, J.P.; Pitout, J.; Johnson, J.R. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 2008, 61, 273–281. [Google Scholar] [CrossRef] [PubMed]
- D’Andrea, M.M.; Arena, F.; Pallecchi, L.; Rossolini, G.M. CTX-M-type β-lactamases: A successful story of antibiotic resistance. Int. J. Med. Microbiol. 2013, 303, 305–317. [Google Scholar] [CrossRef] [PubMed]
- Canton, R.; Coque, T.M. The CTX-M β-lactamase pandemic. Curr. Opin. Microbiol. 2006, 9, 466–475. [Google Scholar] [CrossRef] [PubMed]
- Nicolas-Chanoine, M.H.; Bertrand, X.; Madec, J.Y. Escherichia coli ST131, an intriguing clonal group. Clin. Microbiol. Rev. 2014, 27, 543–574. [Google Scholar] [CrossRef] [PubMed]
- Rogers, B.A.; Sidjabat, H.E.; Paterson, D.L. Escherichia coli O25b-ST131: A pandemic, multiresistant, community-associated strain. J. Antimicrob. Chemother. 2011, 66, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, R.; Johnson, J.R. A new clone sweeps clean: The enigmatic emergence of Escherichia coli sequence type 131. Antimicrob. Agents Chemother. 2014, 58, 4997–5004. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, Z.A.; Doi, Y. Escherichia coli sequence type 131: Epidemiology and challenges in treatment. Expert Rev. Anti Infect. Ther. 2014, 12, 597–609. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.R.; Tchesnokova, V.; Johnston, B.; Clabots, C.; Roberts, P.L.; Billig, M.; Riddell, K.; Rogers, P.; Qin, X.; Butler-Wu, S.; et al. Abrupt emergence of a single dominant multidrug-resistant strain of Escherichia coli. J. Infect. Dis. 2013, 207, 919–928. [Google Scholar] [CrossRef] [PubMed]
- Toh, H.; Oshima, K.; Toyoda, A.; Ogura, Y.; Ooka, T.; Sasamoto, H.; Park, S.H.; Iyoda, S.; Kurokawa, K.; Morita, H.; et al. Complete genome sequence of the wild-type commensal Escherichia coli strain SE15, belonging to phylogenetic group B2. J. Bacteriol. 2010, 192, 1165–1166. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Forde, B.M.; Ben Zakour, N.L.; Stanton-Cook, M.; Phan, M.D.; Totsika, M.; Peters, K.M.; Chan, K.G.; Schembri, M.A.; Upton, M.; Beatson, S.A. The complete genome sequence of Escherichia coli EC958: A high quality reference sequence for the globally disseminated multidrug resistant E. coli O25b:H4-ST131 clone. PLoS ONE 2014, 9, e104400. [Google Scholar] [CrossRef] [PubMed]
- Andersen, P.S.; Stegger, M.; Aziz, M.; Contente-Cuomo, T.; Gibbons, H.S.; Keim, P.; Sokurenko, E.V.; Johnson, J.R.; Price, L.B. Complete genome sequence of the epidemic and highly virulent CTX-M-15-producing H30-RX subclone of escherichia coli st131. Genome Announc. 2013, 1. [Google Scholar] [CrossRef] [PubMed]
- Avasthi, T.S.; Kumar, N.; Baddam, R.; Hussain, A.; Nandanwar, N.; Jadhav, S.; Ahmed, N. Genome of multidrug-resistant uropathogenic Escherichia coli strain NA114 from India. J. Bacteriol. 2011, 193, 4272–4273. [Google Scholar] [CrossRef] [PubMed]
- Price, L.B.; Johnson, J.R.; Aziz, M.; Clabots, C.; Johnston, B.; Tchesnokova, V.; Nordstrom, L.; Billig, M.; Chattopadhyay, S.; Stegger, M.; et al. The epidemic of extended-spectrum-β-lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone, H30-RX. MBio 2013, 4, e00377–00313. [Google Scholar] [CrossRef] [PubMed]
- Petty, N.K.; Ben Zakour, N.L.; Stanton-Cook, M.; Skippington, E.; Totsika, M.; Forde, B.M.; Phan, M.D.; Gomes Moriel, D.; Peters, K.M.; Davies, M.; et al. Global dissemination of a multidrug resistant Escherichia coli clone. Proc. Natl. Acad. Sci. USA 2014, 111, 5694–5699. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Linardopoulou, E.V.; Billig, M.; Tchesnokova, V.; Price, L.B.; Johnson, J.R.; Chattopadhyay, S.; Sokurenko, E.V. Role of homologous recombination in adaptive diversification of extraintestinal Escherichia coli. J. Bacteriol. 2013, 195, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Totsika, M.; Beatson, S.A.; Sarkar, S.; Phan, M.D.; Petty, N.K.; Bachmann, N.; Szubert, M.; Sidjabat, H.E.; Paterson, D.L.; Upton, M.; et al. Insights into a multidrug resistant Escherichia coli pathogen of the globally disseminated ST131 lineage: Genome analysis and virulence mechanisms. PLoS ONE 2011, 6, e26578. [Google Scholar] [CrossRef] [PubMed]
- Totsika, M.; Kostakioti, M.; Hannan, T.J.; Upton, M.; Beatson, S.A.; Janetka, J.W.; Hultgren, S.J.; Schembri, M.A. A FimH inhibitor prevents acute bladder infection and treats chronic cystitis caused by multidrug-resistant uropathogenic Escherichia coli ST131. J. Infect. Dis. 2013, 208, 921–928. [Google Scholar] [CrossRef] [PubMed]
- Floyd, R.V.; Upton, M.; Hultgren, S.J.; Wray, S.; Burdyga, T.V.; Winstanley, C. Escherichia coli-mediated impairment of ureteric contractility is uropathogenic E. coli specific. J. Infect. Dis. 2012, 206, 1589–1596. [Google Scholar] [CrossRef] [PubMed]
- Phan, M.D.; Peters, K.M.; Sarkar, S.; Lukowski, S.W.; Allsopp, L.P.; Gomes Moriel, D.; Achard, M.E.; Totsika, M.; Marshall, V.M.; Upton, M.; et al. The serum resistome of a globally disseminated multidrug resistant uropathogenic Escherichia coli clone. PLoS Genet. 2013, 9, e1003834. [Google Scholar] [CrossRef] [PubMed]
- Langridge, G.C.; Phan, M.D.; Turner, D.J.; Perkins, T.T.; Parts, L.; Haase, J.; Charles, I.; Maskell, D.J.; Peters, S.E.; Dougan, G.; et al. Simultaneous assay of every Salmonella typhi gene using one million transposon mutants. Genome Res. 2009, 19, 2308–2316. [Google Scholar] [CrossRef] [PubMed]
- Bateman, S.L.; Seed, P.C. Epigenetic regulation of the nitrosative stress response and intracellular macrophage survival by extraintestinal pathogenic Escherichia coli. Mol. Microbiol. 2012, 83, 908–925. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef] [PubMed]
- Lanza, V.F.; de Toro, M.; Garcillan-Barcia, M.P.; Mora, A.; Blanco, J.; Coque, T.M.; de la Cruz, F. Plasmid flux in Escherichia coli ST131 sublineages, analyzed by plasmid constellation network (placnet), a new method for plasmid reconstruction from whole genome sequences. PLoS Genet. 2014, 10, e1004766. [Google Scholar] [CrossRef] [PubMed]
- Phan, M.D.; Forde, B.M.; Peters, K.M.; Sarkar, S.; Hancock, S.; Stanton-Cook, M.; Ben Zakour, N.L.; Upton, M.; Beatson, S.A.; Schembri, M.A. Molecular characterization of a multidrug resistance IncF plasmid from the globally disseminated Escherichia coli ST131 clone. PLoS ONE 2015, 10, e0122369. [Google Scholar] [CrossRef] [PubMed]
- Woodford, N.; Carattoli, A.; Karisik, E.; Underwood, A.; Ellington, M.J.; Livermore, D.M. Complete nucleotide sequences of plasmids pEK204, pEK499, and pEK516, encoding CTX-M enzymes in three major Escherichia coli lineages from the united kingdom, all belonging to the international O25:H4-ST131 clone. Antimicrob. Agents Chemother. 2009, 53, 4472–4482. [Google Scholar] [CrossRef] [PubMed]
- Phan, M.D.; Peters, K.M.; Sarkar, S.; Forde, B.M.; Lo, A.W.; Stanton-Cook, M.; Roberts, L.W.; Upton, M.; Beatson, S.A.; Schembri, M.A. Third-generation cephalosporin resistance conferred by a chromosomally encoded blaCMY-23 gene in the Escherichia coli ST131 reference strain EC958. J. Antimicrob. Chemother. 2015, 70, 1969–1972. [Google Scholar] [CrossRef] [PubMed]
- Johnson, T.J.; Hargreaves, M.; Shaw, K.; Snippes, P.; Lynfield, R.; Aziz, M.; Price, L.B. Complete genome sequence of a carbapenem-resistant extraintestinal pathogenic Escherichia coli strain belonging to the sequence type 131 H30R subclade. Genome Announc. 2015, 3. [Google Scholar] [CrossRef] [PubMed]
- Accogli, M.; Giani, T.; Monaco, M.; Giufre, M.; Garcia-Fernandez, A.; Conte, V.; D’Ancona, F.; Pantosti, A.; Rossolini, G.M.; Cerquetti, M. Emergence of Escherichia coli ST131 sub-clone H30 producing VIM-1 and KPC-3 carbapenemases, Italy. J. Antimicrob. Chemother. 2014, 69, 2293–2296. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cai, J.C.; Zhang, R.; Hu, Y.Y.; Zhou, H.W.; Chen, G.X. Emergence of Escherichia coli sequence type 131 isolates producing KPC-2 carbapenemase in China. Antimicrob. Agents Chemother. 2014, 58, 1146–1152. [Google Scholar] [CrossRef] [PubMed]
- Naas, T.; Cuzon, G.; Gaillot, O.; Courcol, R.; Nordmann, P. When carbapenem-hydrolyzing β-lactamase KPC meets Escherichia coli ST131 in France. Antimicrob. Agents Chemother. 2011, 55, 4933–4934. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schembri, M.A.; Zakour, N.L.B.; Phan, M.-D.; Forde, B.M.; Stanton-Cook, M.; Beatson, S.A. Molecular Characterization of the Multidrug Resistant Escherichia coli ST131 Clone. Pathogens 2015, 4, 422-430. https://doi.org/10.3390/pathogens4030422
Schembri MA, Zakour NLB, Phan M-D, Forde BM, Stanton-Cook M, Beatson SA. Molecular Characterization of the Multidrug Resistant Escherichia coli ST131 Clone. Pathogens. 2015; 4(3):422-430. https://doi.org/10.3390/pathogens4030422
Chicago/Turabian StyleSchembri, Mark A., Nouri L. Ben Zakour, Minh-Duy Phan, Brian M. Forde, Mitchell Stanton-Cook, and Scott A. Beatson. 2015. "Molecular Characterization of the Multidrug Resistant Escherichia coli ST131 Clone" Pathogens 4, no. 3: 422-430. https://doi.org/10.3390/pathogens4030422
APA StyleSchembri, M. A., Zakour, N. L. B., Phan, M.-D., Forde, B. M., Stanton-Cook, M., & Beatson, S. A. (2015). Molecular Characterization of the Multidrug Resistant Escherichia coli ST131 Clone. Pathogens, 4(3), 422-430. https://doi.org/10.3390/pathogens4030422




