The Role of Artificial Intelligence in Herpesvirus Detection, Transmission, and Predictive Modeling: With a Special Focus on Marek’s Disease Virus
Abstract
1. Introduction
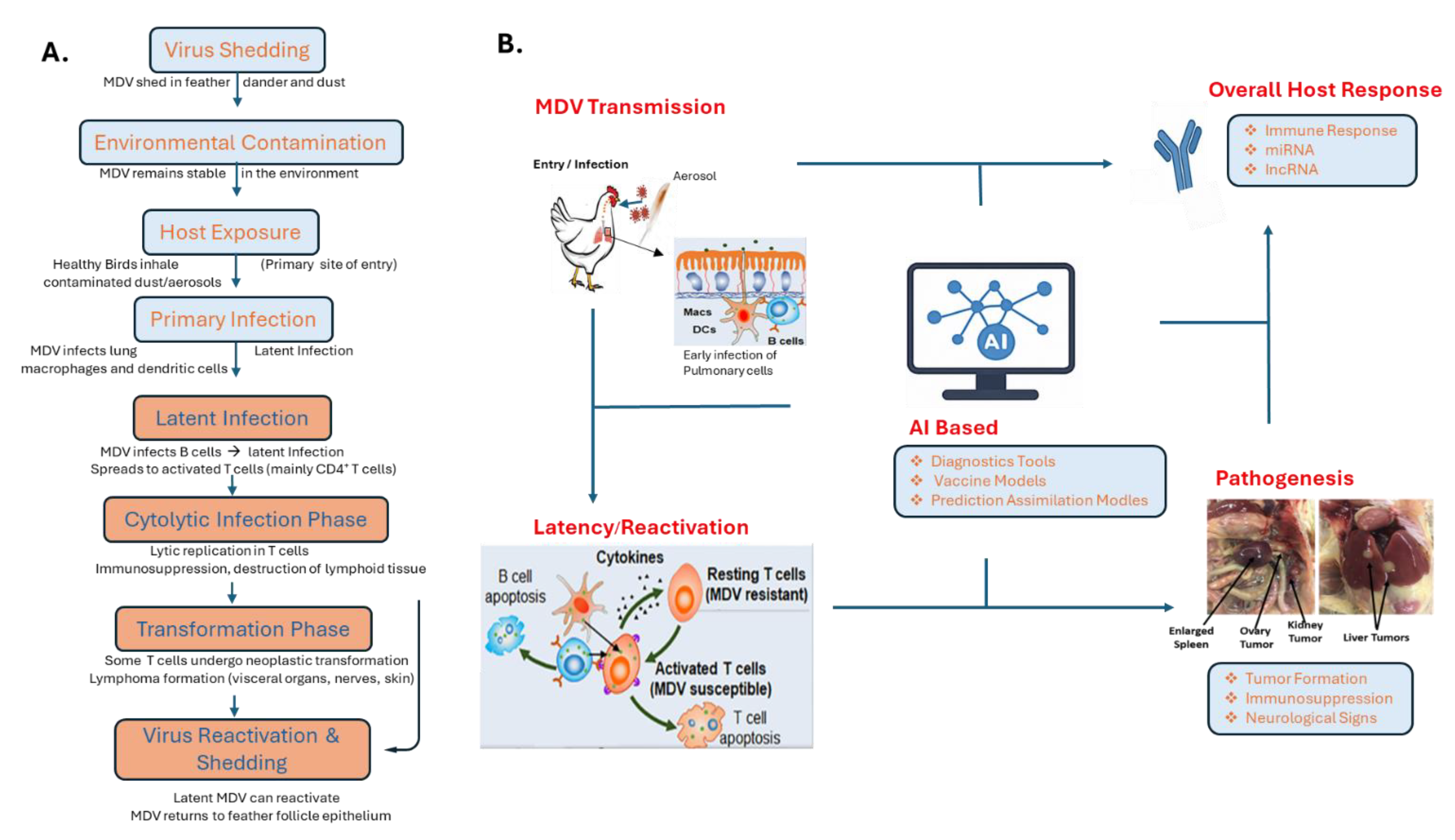
2. Overview of Marek’s Disease Virus (MDV)
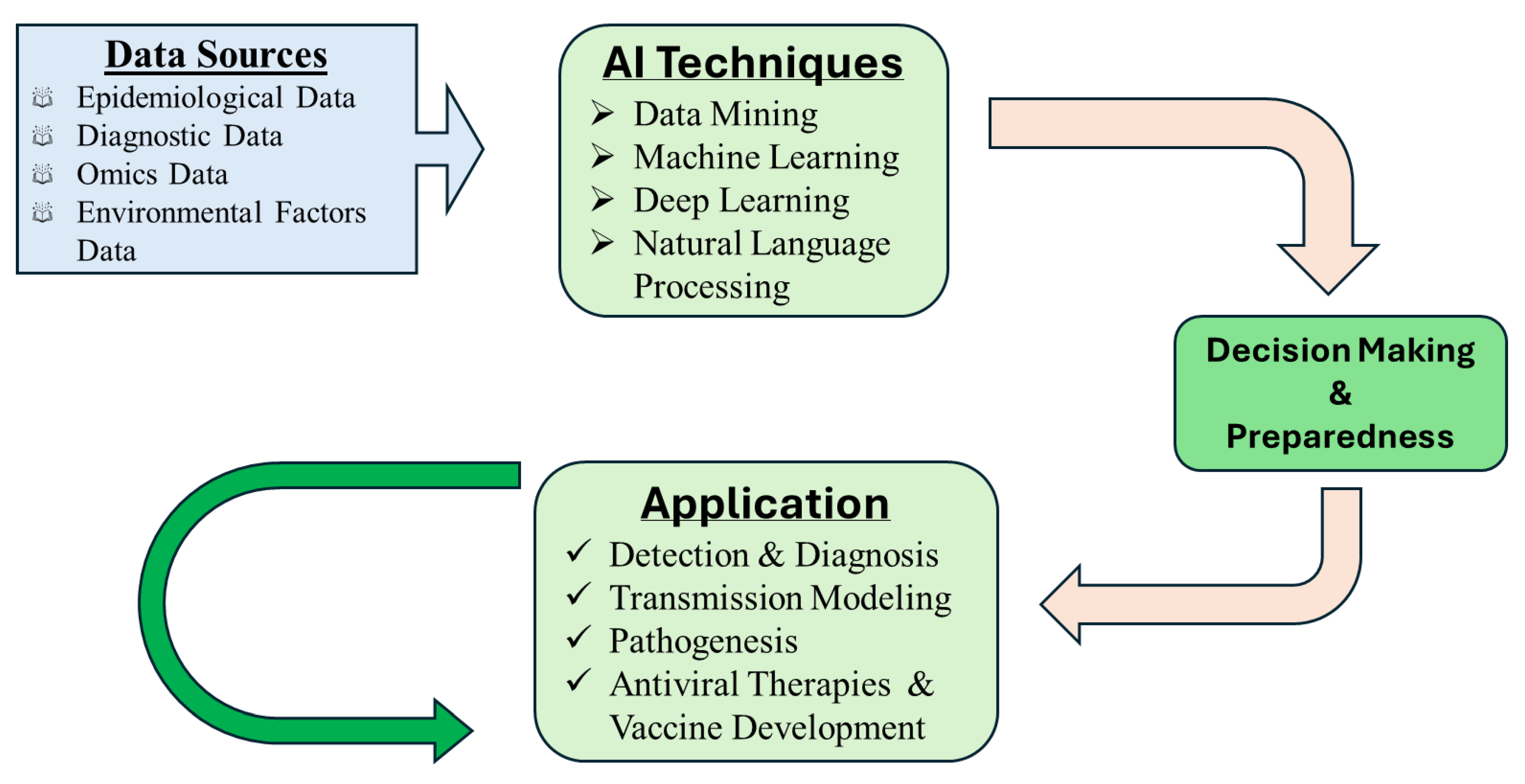
3. Role of AI in Infectious Disease Research
3.1. Disease Surveillance and Outbreak Prediction
3.2. AI for Diagnostic Imaging, Molecular Analysis and Pattern Recognition
3.3. Genomic and Pathogen Evolution Studies
4. AI Applications Specific to Marek’s Disease Virus
4.1. Predictive Modeling of Virulence and Vaccine Breaks
4.2. Early Detection Using Behavioral Data
4.3. AI in Understanding MDV Pathogenesis
4.4. AI in Immune Evasion and Viral Persistence
4.5. Enhancing Breeding Programs
5. Ethical and Regulatory Considerations
6. Challenges and Limitation in Applying AI to MDV Management
7. Strategic Directions for Advancing AI in MDV Research and Control
8. Conclusions
Funding
Conflicts of Interest
References
- Mcelwain, T.F.; Thumbi, S.M. Animal pathogens and their impact on animal health, the economy, food security, food safety and public health. Rev. Sci. Tech. 2017, 36, 423–433. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Šudomová, M.; Hassan, S.T.S. Herpesvirus Diseases in Humans and Animals: Recent Developments, Challenges, and Charting Future Paths. Pathogens 2023, 12, 1422. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Branch-Elliman, W.; Sundermann, A.J.; Wiens, J.; Shenoy, E.S. The future of automated infection detection: Innovation to transform practice (Part III/III). Antimicrob. Steward. Healthc. Epidemiol. 2023, 3, e26. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pavlin, J.A.; Mostashari, F.; Kortepeter, M.G.; Hynes, N.A.; Chotani, R.A.; Mikol, Y.B.; Ryan, M.A.; Neville, J.S.; Gantz, D.T.; Writer, J.V.; et al. Innovative surveillance methods for rapid detection of disease outbreaks and bioterrorism: Results of an interagency workshop on health indicator surveillance. Am. J. Public Health 2003, 93, 1230–1235. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Scott, G.Y.; Aborode, A.T.; Adesola, R.O.; Elebesunu, E.E.; Agyapong, J.; Ibrahim, A.M.; Andigema, A.N.Y.B.S.; Kwarteng, S.; Onifade, I.A.; Adeoye, A.F.; et al. Transforming early microbial detection: Investigating innovative biosensors for emerging infectious diseases. Adv. Biomark. Sci. Technol. 2024, 6, 59–71. [Google Scholar] [CrossRef]
- Gatherer, D.; Depledge, D.P.; Hartley, C.A.; Szpara, M.L.; Vaz, P.K.; Benko, M.; Brandt, C.R.; Bryant, N.A.; Dastjerdi, A.; Doszpoly, A.; et al. ICTV Virus Taxonomy Profile: Herpesviridae 2021. J. Gen. Virol. 2021, 102, 001673. [Google Scholar] [CrossRef]
- Marek, J. Multiple Nervenentzündung (Polyneuritis) bei Hühnern. Dtsch. Tierärztliche Wochenschr. 1907, 15, 417–421. [Google Scholar]
- Osterrieder, N.; Kamil, J.P.; Schumacher, D.; Tischer, B.K.; Trapp, S. Marek’s disease virus: From miasma to model, Nature reviews. Microbiology 2006, 4, 283–294. [Google Scholar] [PubMed]
- Morrow, C.; Fehler, F. Marek’s Disease; Davison, F., Nair, V., Eds.; Institute for Animal Health, Compton Laboratory: Berkshire, UK, 2004; pp. 49–61. [Google Scholar]
- Akbar, H.; Fasick, J.J.; Ponnuraj, N.; Jarosinski, K.W. Purinergic signaling during Marek’s disease in chickens. Sci. Rep. 2023, 13, 2044. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Witter, R.L. The changing landscape of Marek’s disease. Avian Pathol. 1998, 27 (Suppl. S1), S46–S53. [Google Scholar]
- Bull, J.J.; Antia, R. Which ‘imperfect vaccines’ encourage the evolution of higher virulence? Evol. Med. Public Health 2022, 10, 202–213. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Shao, Z.; Zhao, R.; Yuan, S.; Ding, M.; Wang, Y. Tracing the evolution of AI in the past decade and forecasting the emerging trends. Expert Syst. Appl. 2022, 209, 118221. [Google Scholar] [CrossRef]
- Bhat, A.K. The Evolution of AI: From Foundations to Future Prospects. IEEE Computer Society. 11 March 2025. Available online: https://www.computer.org/publications/tech-news/research/evolution-of-ai (accessed on 1 August 2025).
- Shao, Z.; Shen, Z.; Yuan, S.; Tang, J.; Wang, Y.; Wu, L.; Zheng, W. AI 2000: A Decade of Artificial Intelligence. TechVision Reports, 2020. In WebSci’20: Proceedings of the 12th ACM Conference on Web Science, Southampton, UK, 7–10 July 2020; Association for Computing Machinery: New York, NY, USA, 2020; pp. 345–354. [Google Scholar] [CrossRef]
- Faiyazuddin, M.; Rahman, S.J.Q.; Anand, G.; Siddiqui, R.K.; Mehta, R.; Khatib, M.N.; Gaidhane, S.; Zahiruddin, Q.S.; Hussain, A.; Sah, R. The Impact of Artificial Intelligence on Healthcare: A Comprehensive Review of Advancements in Diagnostics, Treatment, and Operational Efficiency. Health Sci. Rep. 2025, 8, e70312. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Khalifa, M.; Albadawy, M. AI in diagnostic imaging: Revolutionising accuracy and efficiency. Comput. Methods Programs Biomed. Update 2024, 5, 100146. [Google Scholar] [CrossRef]
- Pinto-Coelho, L. How Artificial Intelligence Is Shaping Medical Imaging Technology: A Survey of Innovations and Applications. Bioengineering 2023, 10, 1435. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Maniaci, A.; Lavalle, S.; Gagliano, C.; Lentini, M.; Masiello, E.; Parisi, F.; Iannella, G.; Cilia, N.D.; Salerno, V.; Cusumano, G.; et al. The Integration of Radiomics and Artificial Intelligence in Modern Medicine. Life 2024, 14, 1248. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kinsulie, O.C.; Idris, I.; Aliyu, V.A.; Shahzad, S.; Banwo, O.G.; Ogunleye, S.C.; Olorunshola, M.; Okedoyin, D.O.; Ugwu, C.; Oladapo, I.P.; et al. The potential application of artificial intelligence in veterinary clinical practice and biomedical research. Front. Vet. Sci. 2024, 11, 1347550. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jarosinski, K.W.; Tischer, B.K.; Trapp, S.; Osterrieder, N. Marek’s disease virus: Lytic replication, oncogenesis and control. Exp. Rev. Vaccines 2006, 5, 761–772. [Google Scholar] [CrossRef]
- Jarosinski, K.W. Interindividual spread of herpesviruses. Adv. Anat. Embryol. Cell Biol. 2017, 223, 195–224. [Google Scholar] [CrossRef]
- Boodhoo, N.; Gurung, A.; Sharif, S.; Behboudi, S. Marek’s disease in chickens: A review with focus on immunology. Vet. Res. 2016, 47, 119. [Google Scholar] [CrossRef]
- Wannaratana, S.; Prakairungnamthip, D.; Tunterak, W.; Charoenvisal, N.; Limpavithayakul, K.; Areeraksakul, P.; Sasipreeyajan, J.; Thontiravong, A. Pathogenicity and transmissibility of Marek’s disease virus isolated from chickens in Thailand. Poult. Sci. 2025, 104, 105519. [Google Scholar] [CrossRef]
- Nair, V. Evolution of Marek’s disease—A paradigm for incessant race between the pathogen and the host. Vet. J. 2005, 170, 175–183. [Google Scholar] [CrossRef]
- Defending Against Marek’s Disease with Genetics and Epigenetics. Available online: https://tellus.ars.usda.gov/stories/articles/defending-against-mareks-disease-genetics-and-epigenetics (accessed on 29 August 2025).
- Schat, K.A.; Nair, V. Marek’s disease. In Diseases of Poultry, 13th ed.; Swayne, D.E., Ed.; Wiley-Blackwell: Indianapolis, IN, USA, 2013; pp. 515–552. [Google Scholar]
- Read, A.F.; Baigent, S.J.; Powers, C.; Kgosana, L.B.; Blackwell, L.; Smith, L.P.; Kennedy, D.A.; Walkden-Brown, S.W.; Nair, V.K. Imperfect Vaccination Can Enhance the Transmission of Highly Virulent Pathogens. PLoS Biol. 2015, 13, e1002198. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- van Hulten, M.C.W.; Cruz-Coy, J.; Gergen, L.; Pouwels, H.; Ten Dam, G.B.; Verstegen, I.; de Groof, A.; Morsey, M.; Tarpey, I. Efficacy of a turkey herpesvirus double construct vaccine (HVT-ND-IBD) against challenge with different strains of Newcastle disease, infectious bursal disease and Marek’s disease viruses. Avian Pathol. 2021, 50, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, D.A.; Cairns, C.; Jones, M.J.; Bell, A.S.; Salathé, R.M.; Baigent, S.J.; Nair, V.K.; Dunn, P.A.; Read, A.F. Industry-Wide Surveillance of Marek’s Disease Virus on Commercial Poultry Farms. Avian Dis. 2017, 61, 53–164. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bertzbach, L.D.; Conradie, A.M.; You, Y.; Kaufer, B.B. Latest Insights into Marek’s Disease Virus Pathogenesis and Tumorigenesis. Cancers 2020, 12, 647. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wajid, S.J.; Katz, M.E.; Renz, K.G.; Walkden-Brown, S.W. Prevalence of Marek’s Disease Virus in Different Chicken Populations in Iraq and Indicative Virulence Based on Sequence Variation in the EcoRI-Q (meq) Gene. Avian Dis. 2013, 57, 562–568. [Google Scholar] [CrossRef] [PubMed]
- World Organization for Animal Health (WOAH). “Marek’s Disease.” Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. 2023. Chapter 3.3.13. Available online: https://www.woah.org/fileadmin/Home/fr/Health_standards/tahm/3.03.13_MAREK_DIS.pdf (accessed on 5 September 2025).
- Schat, K.A.; Skinner, M.A. Avian Immunosuppressive Diseases and Immunoevasion. In Avian Immunology, 2nd ed.; Schat, K.A., Kaspers, B., Kaiser, P., Eds.; Academic Press: Cambridge, MA, USA, 2014; pp. 275–297. [Google Scholar] [CrossRef]
- The Poultry Site. (n.d.). Marek’s Disease Control in Broiler Breeds. Available online: https://www.thepoultrysite.com/articles/mareks-disease-control-in-broiler-breeds (accessed on 11 May 2025).
- Real-Time PCR for the Detection of Marek’s Disease Virus. Iowa State University Digital Repository, Iowa State University. Available online: https://dr.lib.iastate.edu/server/api/core/bitstreams/9f5fb6ec-afa4-432d-88e1-d7a02eac7d29/content (accessed on 20 June 2025).
- Kalita, A.J.; Subba, M.; Adil, S.; Wani, M.A.; Beigh, Y.A.; Shafi, M. Application of artificial intelligence and machine learning in poultry disease detection and diagnosis: A review: AI and Machine learning in poultry disease diagnosis. Lett. Anim. Biol. 2025, 5, 1–6. [Google Scholar] [CrossRef]
- Ojo, R.O.; Ajayi, A.O.; Owolabi, H.A.; Oyedele, L.O.; Akanbi, L.A. Internet of Things and Machine Learning Techniques in Poultry Health and Welfare Management: A Systematic Literature Review. Comput. Electron. Agric. 2022, 200, 107266. [Google Scholar] [CrossRef]
- Dhankani, V.; Kutz, J.N.; Schiffer, J.T. Herpes Simplex Virus-2 Genital Tract Shedding Is Not Predictable over Months or Years in Infected Persons. PLoS Comput. Biol. 2014, 10, e1003922. [Google Scholar] [CrossRef]
- Ye, Y.; Pandey, A.; Bawden, C.; Sumsuzzman, D.M.; Rajput, R.; Shoukat, A.; Singer, B.H.; Moghadas, S.M.; Galvani, A.P. Integrating artificial intelligence with mechanistic epidemiological modeling: A scoping review of opportunities and challenges. Nat. Commun. 2025, 16, 581. [Google Scholar] [CrossRef] [PubMed]
- Kraemer, M.U.G.; Tsui, J.L.H.; Chang, S.Y.; Lytras, S.; Khurana, M.P.; Vanderslott, S.; Bajaj, S.; Scheidwasser, N.; Curran-Sebastian, J.L.; Semenova, E.; et al. Artificial intelligence for modelling infectious disease epidemics. Nature 2025, 638, 623–635. [Google Scholar] [CrossRef]
- Spicknall, I.H.; Looker, K.J.; Gottlieb, S.L.; Chesson, H.W.; Schiffer, J.T.; Elmes, J.; Boily, M.-C. Review of mathematical models of HSV-2 vaccination: Implications for vaccine development. Vaccine 2019, 37, 7007–7014. [Google Scholar] [CrossRef]
- Vargas-Santiago, M.; León-Velasco, D.A.; Maldonado-Sifuentes, C.E.; Chanona-Hernandez, L. A State-of-the-Art Review of Artificial Intelligence (AI) Applications in Healthcare: Advances in Diabetes, Cancer, Epidemiology, and Mortality Prediction. Computers 2025, 14, 143. [Google Scholar] [CrossRef]
- Walsh, D.P.; Ma, T.F.; Ip, H.S.; Zhu, J. Artificial intelligence and avian influenza: Using machine learning to enhance active surveillance for avian influenza viruses. Transbound. Emerg. Dis. 2019, 66, 2537–2545. [Google Scholar] [CrossRef]
- El Morr, C.; Ozdemir, D.; Asdaah, Y.; Saab, A.; El-Lahib, Y.; Sokhn, E.S. AI-based epidemic and pandemic early warning systems: A systematic scoping review. Health Inform. J. 2024, 30, 14604582241275844. [Google Scholar] [CrossRef] [PubMed]
- Herrick, K.A.; Huettmann, F.; Lindgren, M.A. A global model of avian influenza prediction in wild birds: The importance of northern regions. Vet. Res. 2013, 44, 42. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Musa, E.; Nia, Z.M.; Bragazzi, N.L.; Leung, D.; Lee, N.; Kong, J.D. Avian Influenza: Lessons from Past Outbreaks and an Inventory of Data Sources, Mathematical and AI Models, and Early Warning Systems for Forecasting and Hotspot Detection to Tackle Ongoing Outbreaks. Healthcare 2024, 12, 1959. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yajie, L.; Johar, M.G.M.; Hajamydeen, A.I. Poultry Disease Early Detection Methods Using Deep Learning Technology. Indones. J. Electr. Eng. Comput. Sci. 2023, 32, 1712–1723. [Google Scholar] [CrossRef]
- Taleb, H.M.; Mahrose, K.; Abdel-Halim, A.A.; Kasem, H.; Ramadan, G.S.; Fouad, A.M.; Khafaga, A.F.; Khalifa, N.E.; Kamal, M.; Salem, H.M.; et al. Using Artificial Intelligence to Improve Poultry Productivity—A Review. Ann. Anim. Sci. 2024, 25, 23–33. [Google Scholar] [CrossRef]
- Cuan, K.; Zhang, T.; Li, Z.; Huang, J.; Ding, Y.; Fang, C. Automatic Newcastle disease detection using sound technology and deep learning method. Comput. Electron. Agric. 2022, 194, 106740. [Google Scholar] [CrossRef]
- Karras, A.; Karras, C.; Sioutas, S.; Makris, C.; Katselis, G.; Hatzilygeroudis, I.; Theodorou, J.A.; Tsolis, D. An Integrated GIS-Based Reinforcement Learning Approach for Efficient Prediction of Disease Transmission in Aquaculture. Information 2023, 14, 583. [Google Scholar] [CrossRef]
- Ardui, S.; Ameur, A.; Vermeesch, J.R.; Hestand, M.S. Single molecule real-time (SMRT) sequencing comes of age: Applications and utilities for medical diagnostics. Nucleic Acids Res. 2018, 46, 2159–2168. [Google Scholar] [CrossRef]
- de Souza, A.I.; da Silva, A.C.; Ramos, R.T.J. Artificial intelligence and machine learning in viral genomics and precision medicine. Brief. Bioinform. 2021, 22, bbaa162. [Google Scholar] [CrossRef]
- Esteva, A.; Robicquet, A.; Ramsundar, B.; Kuleshov, V.; DePristo, M.; Chou, K.; Dean, J. A guide to deep learning in healthcare. Nat. Med. 2019, 25, 24–29. [Google Scholar] [CrossRef]
- Topol, E.J. High-performance medicine: The convergence of human and artificial intelligence. Nat. Med. 2019, 25, 44–56. [Google Scholar] [CrossRef]
- Whitley, R.J.; Roizman, B. Herpes simplex viruses. In Fields Virology, 4th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Hagerstown, MD, USA, 2001; pp. 2461–2509. [Google Scholar]
- Liu, Y.; Chen, P.H.C.; Krause, J.; Peng, L. How to read articles that use machine learning: Users’ guides to the medical literature. JAMA 2020, 322, 1806–1816. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Luo, W.; Phung, D.; Tran, T.; Gupta, S.; Rana, S.; Karmakar, C.; Venkatesh, S. Guidelines for developing and reporting machine learning predictive models in biomedical research: A multidisciplinary view. NPJ Digit. Med. 2022, 5, e323. [Google Scholar] [CrossRef] [PubMed]
- Madani, A.; Arnaout, R.; Mofrad, M.; Arnaout, R. Fast and accurate view classification of echocardiograms using deep learning. NPJ Digit. Med. 2018, 1, 6. [Google Scholar] [CrossRef]
- Liu, H.; Ma, K.; Liu, M.; Yang, C.; Huang, X.; Zhao, Y.; Qi, K. Histologic findings and viral antigen distribution in natural coinfection of layer hens with subgroup J avian leukosis virus, Marek’s disease virus, and reticuloendotheliosis virus. J. Vet. Diagn. Investig. 2019, 31, 761–765. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ehsan, Z.; Mohtavipour, S. Broiler-Net: A Deep Convolutional Framework for Broiler Behavior Recognition in Cage-Free Poultry Houses. arXiv 2024, arXiv:2401.12176. [Google Scholar]
- Min, S.; Lee, B.; Yoon, S. Deep learning in bioinformatics. Brief. Bioinform. 2017, 18, 851–869. [Google Scholar] [CrossRef] [PubMed]
- Ergun, H.; Alkan, C.; Bilgen, T. Unsupervised deep learning approaches for clustering and visualizing single-cell transcriptomic data. Brief. Bioinform. 2021, 22, bbaa318. [Google Scholar] [CrossRef]
- Torkamaneh, D.; Boyle, B.; Belzile, F. Efficient genome-wide genotyping strategies and data integration pipelines for crop research. Brief. Bioinform. 2021, 22, bbab060. [Google Scholar] [CrossRef]
- Poplin, R.; Chang, P.C.; Alexander, D.; Schwartz, S.; Colthurst, T.; Ku, A.; DePristo, M.A. A universal SNP and small-indel variant caller using deep neural networks. Nat. Biotechnol. 2018, 36, 983–987. [Google Scholar] [CrossRef]
- Huang, P.J.; Chang, J.H.; Lin, H.H.; Li, Y.X.; Lee, C.C.; Su, C.T.; Li, Y.L.; Chang, M.T.; Weng, S.; Cheng, W.H.; et al. DeepVariant-on-Spark: Small-Scale Genome Analysis Using a Cloud-Based Computing Framework. Comput. Math Methods Med. 2020, 1, 7231205. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Villanueva-Miranda, I.; Xiao, G.; Xie, Y. Artificial intelligence in early warning systems for infectious disease surveillance: A systematic review. Front. Public Health. 2025, 13, 1609615. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lee, J.; Yoon, W.; Kim, S.; Kim, D.; Kim, S.; So, C.H.; Kang, J. BioBERT: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2020, 36, 1234–1240. [Google Scholar] [CrossRef] [PubMed]
- Neumann, M.; King, D.; Beltagy, I.; Ammar, W. ScispaCy: Fast and robust models for biomedical natural language processing. arXiv 2019. [Google Scholar] [CrossRef]
- Walkden-Brown, S.W.; Islam, A.F.M.F.; Reddy, S.M.; Renz, K.G. Marek’s disease: Still a significant threat to the poultry industry. Poult. Sci. 2019, 98, 5286–5295. [Google Scholar] [CrossRef]
- Kim, T.; Hearn, C.J.; Mays, J.; Velez-Irizarry, D.; Reddy, S.M.; Spatz, S.J.; Cheng, H.H.; Dunn, J.R. Phenotypic Characterization of Recombinant Marek’s Disease Virus in Live Birds Validates Polymorphisms Associated with Virulence. Viruses 2023, 15, 2263. [Google Scholar] [CrossRef] [PubMed]
- Trimpert, J.; Groenke, N.; Jenckel, M.; He, S.; Kunec, D.; Szpara, M.L.; Spatz, S.J.; Osterrieder, N.; McMahon, D.P. A phylogenomic analysis of Marek’s disease virus reveals independent paths to virulence in Eurasia and North America. Evol. Appl. 2017, 10, 1091–1101. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, Y.; Lan, X.; Wang, Y.; Lin, Y.; Yu, Z.; Guo, R.; Li, K.; Cui, H.; Qi, X.; Wang, Y.; et al. Emerging natural recombinant Marek’s disease virus between vaccine and virulence strains and their pathogenicity. Transbound. Emerg. Dis. 2022, 69, e1702–e1709. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Cui, Z.; Wu, C.; Yu, Y.; Tian, R.; Xie, H.; Jin, Z.; Fan, W.; Xie, W.; Huang, Z.; et al. DeepEBV: A deep learning model to predict Epstein-Barr virus (EBV) integration sites. Bioinformatics 2021, 37, 3405–3411. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Hong, H.Y.; Dong, X.Y.; Xu, L.P.; Zhang, X.H.; Wang, Y.; Yan, C.H.; Chen, H.; Chen, Y.H.; Han, W.; et al. Machine learning algorithm as a prognostic tool for Epstein-Barr virus reactivation after haploidentical hematopoietic stem cell transplantation. Blood Sci. 2022, 5, 51–59. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Franzo, G.; Fusaro, A.; Snoeck, C.J.; Dodovski, A.; Van Borm, S.; Steensels, M.; Christodoulou, V.; Onita, I.; Burlacu, R.; Sánchez, A.S.; et al. Evaluation of Different Machine Learning Approaches to Predict Antigenic Distance Among Newcastle Disease Virus (NDV) Strains. Viruses 2025, 17, 567. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zheng, L.P.; Teng, M.; Li, G.X.; Zhang, W.K.; Wang, W.D.; Liu, J.L.; Li, L.Y.; Yao, Y.; Nair, V.; Luo, J. Current Epidemiology and Co-Infections of Avian Immunosuppressive and Neoplastic Diseases in Chicken Flocks in Central China. Viruses 2022, 14, 2599. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Li, M.; Wang, P.; Li, Q.; Deng, Q.; Shi, M.; Mo, M.; Wei, T.; Huang, T.; Wei, P. Reemergence of reticuloendotheliosis virus and Marek’s disease virus co-infection in Yellow-Chickens in Southern China. Poult. Sci. 2021, 100, 101099. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chen, T.; Guestrin, C. XGBoost: A scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar] [CrossRef]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Liu, T.Y. LightGBM: A highly efficient gradient boosting decision tree. Adv. Neural Inf. Process. Syst. 2017, 30, 3146–3154. [Google Scholar]
- Yoo, D.S.; Song, Y.H.; Choi, D.W.; Lim, J.S.; Lee, K.; Kang, T. Machine learning-driven dynamic risk prediction for highly pathogenic avian influenza at poultry farms in Republic of Korea: Daily risk estimation for individual premises. Transbound. Emerg. Dis. 2022, 69, 2667–2681. [Google Scholar] [CrossRef] [PubMed]
- Lipkin, E.; Smith, J.; Soller, M.; Burt, D.W.; Fulton, J.E. Mapping quantitative trait loci regions associated with Marek’s disease on chicken autosomes by means of selective DNA pooling. Sci. Rep. 2024, 14, 31896. [Google Scholar] [CrossRef]
- Gul, H.; Habib, G.; Khan, I.M.; Rahman, S.U.; Khan, N.M.; Wang, H.; Khan, N.U.; Liu, Y. Genetic resilience in chickens against bacterial, viral and protozoal pathogens. Front. Vet. Sci. 2022, 9, 1032983. [Google Scholar] [CrossRef]
- Nasirahmadi, A.; Gonzalez, J.; Sturm, B.; Hensel, O. AI applications for behavior-based monitoring in animal production systems: A review. Comput. Electron. Agric. 2022, 196, 106889. [Google Scholar] [CrossRef]
- Lee, H.; Kim, M.J.; Choi, H.; Cho, K.H. Early detection of Marek’s Disease in poultry using deep learning-based gait analysis. Poult. Sci. 2022, 101, 101940. [Google Scholar] [CrossRef]
- Talebi, R.; Zulkifli, I.; Alimon, A.R. Welfare assessment in poultry through automated behavior monitoring: Recent advances and future perspectives. Animals 2023, 13, 435. [Google Scholar] [CrossRef]
- Madavaraju, K.; Koganti, R.; Volety, I.; Yadavalli, T.; Shukla, D. Herpes Simplex Virus Cell Entry Mechanisms: An Update. Front. Cell Infect. Microbiol. 2021, 10, 617578. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Connolly, S.; Jackson, J.; Jardetzky, T.S.; Longnecker, R. Fusing structure and function: A structural view of the herpesvirus entry machinery. Nat. Rev. Microbiol. 2011, 9, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Akkutay-Yoldar, Z.; Yoldar, M.T.; Akkaş, Y.B.; Şurak, S.; Garip, F.; Turan, O.; Ekizoğlu, B.; Yüca, O.C.; Özkul, A.; Ünver, B. A web-based artificial intelligence system for label-free virus classification and detection of cytopathic effects. Sci. Rep. 2025, 15, 5904. [Google Scholar] [CrossRef] [PubMed]
- Schang, L.M.; Hu, M.; Cortes, E.F.; Sun, K. Chromatin-mediated epigenetic regulation of HSV-1 transcription as a potential target in antiviral therapy. Antivir. Res. 2021, 192, 105103. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kristie, T.M. Dynamic modulation of HSV chromatin drives initiation of infection and provides targets for epigenetic therapies. Virology 2015, 479–480, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Tahir, M.; Norouzi, M.; Khan, S.S.; Davie, J.R.; Yamanaka, S.; Ashraf, A. Artificial intelligence and deep learning algorithms for epigenetic sequence analysis: A review for epigeneticists and AI experts. Comput. Biol. Med. 2024, 183, 109302. [Google Scholar] [CrossRef]
- Vider-Shalit, T.; Fishbain, V.; Raffaeli, S.; Louzoun, Y. Phase-dependent immune evasion of herpesviruses. J. Virol. 2007, 81, 9536–9545. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sorel, O.; Dewals, B.G. The Critical Role of Genome Maintenance Proteins in Immune Evasion During Gammaherpesvirus Latency. Front. Microbiol. 2019, 9, 3315. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Suzich, J.B.; Cliffe, A.R. Strength in diversity: Understanding the pathways to herpes simplex virus reactivation. Virology 2018, 522, 81–91. [Google Scholar] [CrossRef]
- Canova, P.N.; Charron, A.J.; Leib, D.A. Models of Herpes Simplex Virus Latency. Viruses 2024, 16, 747. [Google Scholar] [CrossRef]
- Gilyazova, I.; Korytina, G.; Kochetova, O.; Savelieva, O.; Mikhaylova, E.; Vershinina, Z.; Chumakova, A.; Markelov, V.; Abdeeva, G.; Karunas, A.; et al. Advances in Genomics and Postgenomics in Poultry Science: Current Achievements and Future Directions. Int. J. Mol. Sci. 2025, 26, 8285. [Google Scholar] [CrossRef]
- Floridi, L.; Cowls, J.; Beltrametti, M.; Chiarello, F.; Chatila, R.; Dignum, V.; Vayena, E. AI4People—An ethical framework for a good AI society: Opportunities, risks, principles, and recommendations. Minds Mach. 2018, 28, 689–707. [Google Scholar] [CrossRef]
- Mittelstadt, B.D.; Allo, P.; Taddeo, M.; Wachter, S.; Floridi, L. The ethics of algorithms: Mapping the debate. Big Data Soc. 2016, 3, 2053951716679679. [Google Scholar] [CrossRef]
- Cambon-Thomsen, A.; Rial-Sebbag, E.; Knoppers, B.M. Trends in ethical and legal frameworks for the use of human biobanks. Eur. Respir. J. 2007, 30, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Mehrabi, N.; Morstatter, F.; Saxena, N.; Lerman, K.; Galstyan, A. A survey on bias and fairness in machine learning. ACM Comput. Surv. (CSUR) 2021, 54, 1–35. [Google Scholar] [CrossRef]
- Obermeyer, Z.; Powers, B.; Vogeli, C.; Mullainathan, S. Dissecting racial bias in an algorithm used to manage the health of populations. Science 2019, 366, 447–453. [Google Scholar] [CrossRef]
- Richards, N.M.; King, J.H. Big data ethics. Wake Forest L. Rev. 2014, 49, 393–432. [Google Scholar]
- Cakic, S.; Popovic, T.; Krco, S.; Nedic, D.; Babic, D.; Jovovic, I. Developing Edge AI Computer Vision for Smart Poultry Farms Using Deep Learning and HPC. Sensors 2023, 23, 3002. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- World Health Organization. Ethics and Governance of Artificial Intelligence for Health: WHO Guidance. 2021. Available online: https://www.who.int/publications/i/item/9789240029200 (accessed on 18 July 2025).
- Vokinger, K.N.; Feuerriegel, S.; Kesselheim, A.S. Mitigating bias in machine learning for medicine. Commun. Med. 2021, 1, 25. [Google Scholar] [CrossRef]
- Xiao, S.; Dhand, N.K.; Wang, Z.; Hu, K.; Thomson, P.C.; House, J.K.; Khatkar, M.S. Review of applications of deep learning in veterinary diagnostics and animal health. Front. Vet. Sci. 2025, 12, 1511522. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Oduoye, M.O.; Fatima, E.; Muzammil, M.A.; Dave, T.; Irfan, H.; Fariha, F.N.U.; Marbell, A.; Ubechu, S.C.; Scott, G.Y.; Elebesunu, E.E. Impacts of the advancement in artificial intelligence on laboratory medicine in low- and middle-income countries: Challenges and recommendations—A literature review. Health Sci. Rep. 2024, 7, e1794. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Coghlan, S.; Quinn, T. Ethics of using artificial intelligence (AI) in veterinary medicine. AI Soc. 2024, 39, 2337–2348. [Google Scholar] [CrossRef]
- Mao, A.; Huang, E.; Gan, H.; Liu, K. FedAAR: A Novel Federated Learning Framework for Animal Activity Recognition with Wearable Sensors. Animals 2022, 12, 2142. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Teo, Z.L.; Jin, L.; Li, S.; Miao, D.; Zhang, X.; Ng, W.Y.; Tan, T.F.; Lee, D.M.; Chua, K.J.; Heng, J.; et al. Federated machine learning in healthcare: A systematic review on clinical applications and technical architecture. Cell Rep. Med. 2024, 5, 101419. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Eden, R.; Chukwudi, I.; Bain, C.; Barbieri, S.; Callaway, L.; de Jersey, S.; George, Y.; Gorse, A.-D.; Lawley, M.; Marendy, P.; et al. A scoping review of the governance of federated learning in healthcare. npj Digit. Med. 2025, 8, 427. [Google Scholar] [CrossRef]
- Boguslav, M.R.; Kiehl, A.; Kott, D.; Strecker, G.J.; Webb, T.; Saklou, N.; Ward, T.; Kirby, M. Fine-tuning foundational models to code diagnoses from veterinary health records. arXiv 2024, arXiv:2410.15186. [Google Scholar] [CrossRef]
- Tong, Q.; Wang, J.; Yang, W.; Wu, S.; Zhang, W.; Sun, C.; Xu, K. Edge AI-enabled chicken health detection based on enhanced FCOS-Lite and knowledge distillation. arXiv 2024, arXiv:2407.09562. [Google Scholar] [CrossRef]
- Szlosek, D.; Coyne, M.; Riggot, J.; Knight, K.; McCrann, D.J.; Kincaid, D. Development and validation of a machine learning algorithm for clinical wellness visit classification in cats and dogs. arXiv 2024, arXiv:2406.10314. [Google Scholar] [CrossRef]
- Longo, L.; Brcic, M.; Cabitza, F.; Choi, J.; Confalonieri, R.; Del Ser, J.; Guidotti, R.; Hayashi, Y.; Herrera, F.; Holzinger, A.; et al. Explainable Artificial Intelligence (XAI) 2.0: A manifesto of open challenges and interdisciplinary research directions. Inf. Fusion 2024, 106, 102301. [Google Scholar] [CrossRef]
- Antoniadi, A.M.; Du, Y.; Guendouz, Y.; Wei, L.; Mazo, C.; Becker, B.A.; Mooney, C. Current Challenges and Future Opportunities for XAI in Machine Learning-Based Clinical Decision Support Systems: A Systematic Review. Appl. Sci. 2021, 11, 5088. [Google Scholar] [CrossRef]
- Ball, J.C. This AI Helps Detect Wildlife Health Issues in Real Time. WIRED. 16 August 2021. Available online: https://www.wired.com/story/this-ai-helps-detect-wildlife-health-issues-in-real-time/ (accessed on 20 July 2025).
- Joslyn, S.K.; Faulkner, J.; Ma, D.; Appleby, R. Commentary: Comparison of radiological interpretation made by veterinary radiologists and state-of-the-art commercial AI software for canine and feline radiographic studies. Front. Vet. Sci. 2025, 12, 1615947. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Shukla, R. AI in Veterinary Digital Pathology & Laboratory Diagnostics. VOSD. 24 July 2025. Available online: https://www.vosd.in/ai-in-digital-pathology-human-vs-veterinary/?utm_source=chatgpt.com (accessed on 5 September 2025).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akbar, H. The Role of Artificial Intelligence in Herpesvirus Detection, Transmission, and Predictive Modeling: With a Special Focus on Marek’s Disease Virus. Pathogens 2025, 14, 937. https://doi.org/10.3390/pathogens14090937
Akbar H. The Role of Artificial Intelligence in Herpesvirus Detection, Transmission, and Predictive Modeling: With a Special Focus on Marek’s Disease Virus. Pathogens. 2025; 14(9):937. https://doi.org/10.3390/pathogens14090937
Chicago/Turabian StyleAkbar, Haji. 2025. "The Role of Artificial Intelligence in Herpesvirus Detection, Transmission, and Predictive Modeling: With a Special Focus on Marek’s Disease Virus" Pathogens 14, no. 9: 937. https://doi.org/10.3390/pathogens14090937
APA StyleAkbar, H. (2025). The Role of Artificial Intelligence in Herpesvirus Detection, Transmission, and Predictive Modeling: With a Special Focus on Marek’s Disease Virus. Pathogens, 14(9), 937. https://doi.org/10.3390/pathogens14090937





