Bacteria-Based Roles in Solid Tumors: Potential for Prevention and Treatment
Abstract
1. Introduction
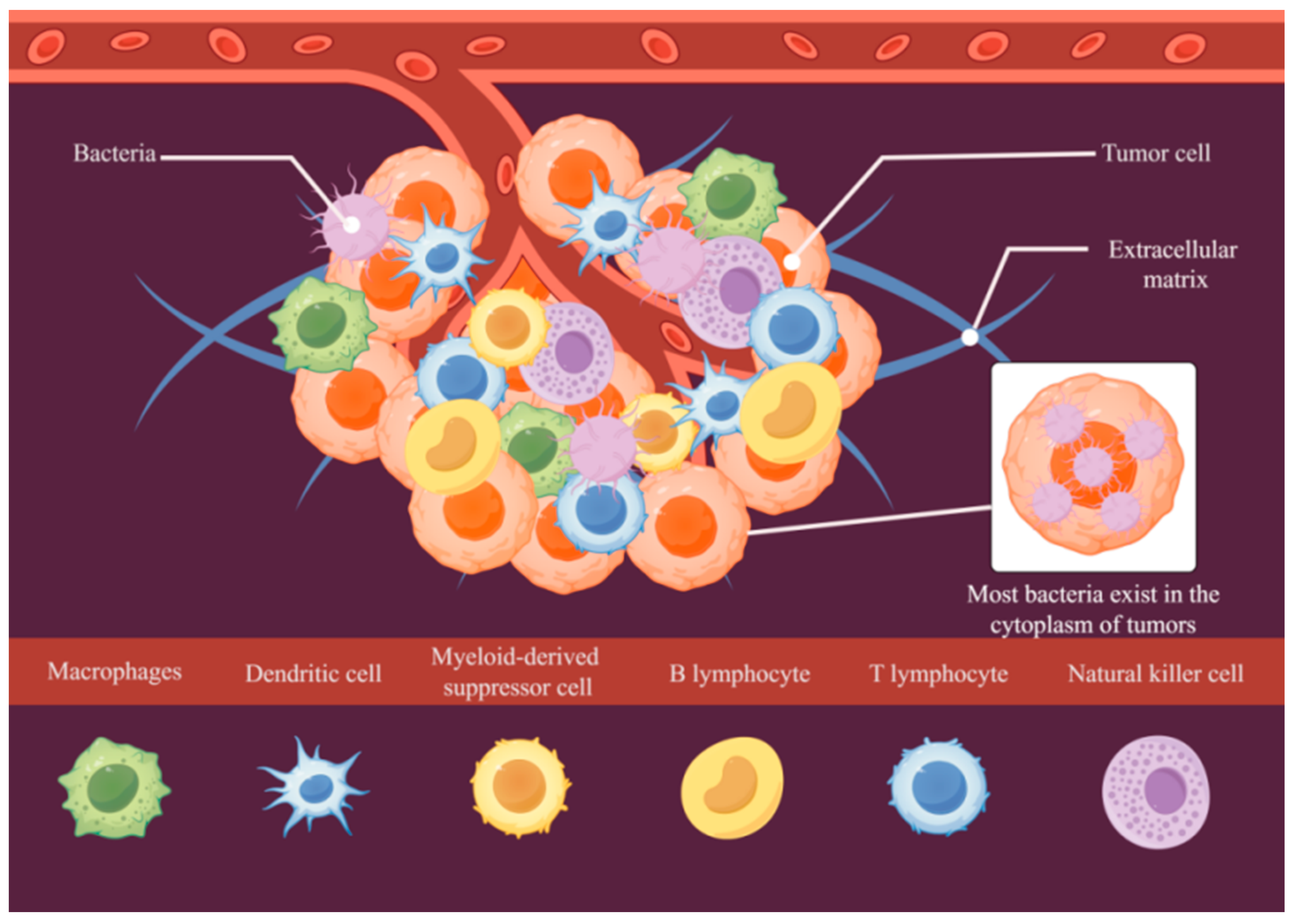
2. Tumor Microenvironment
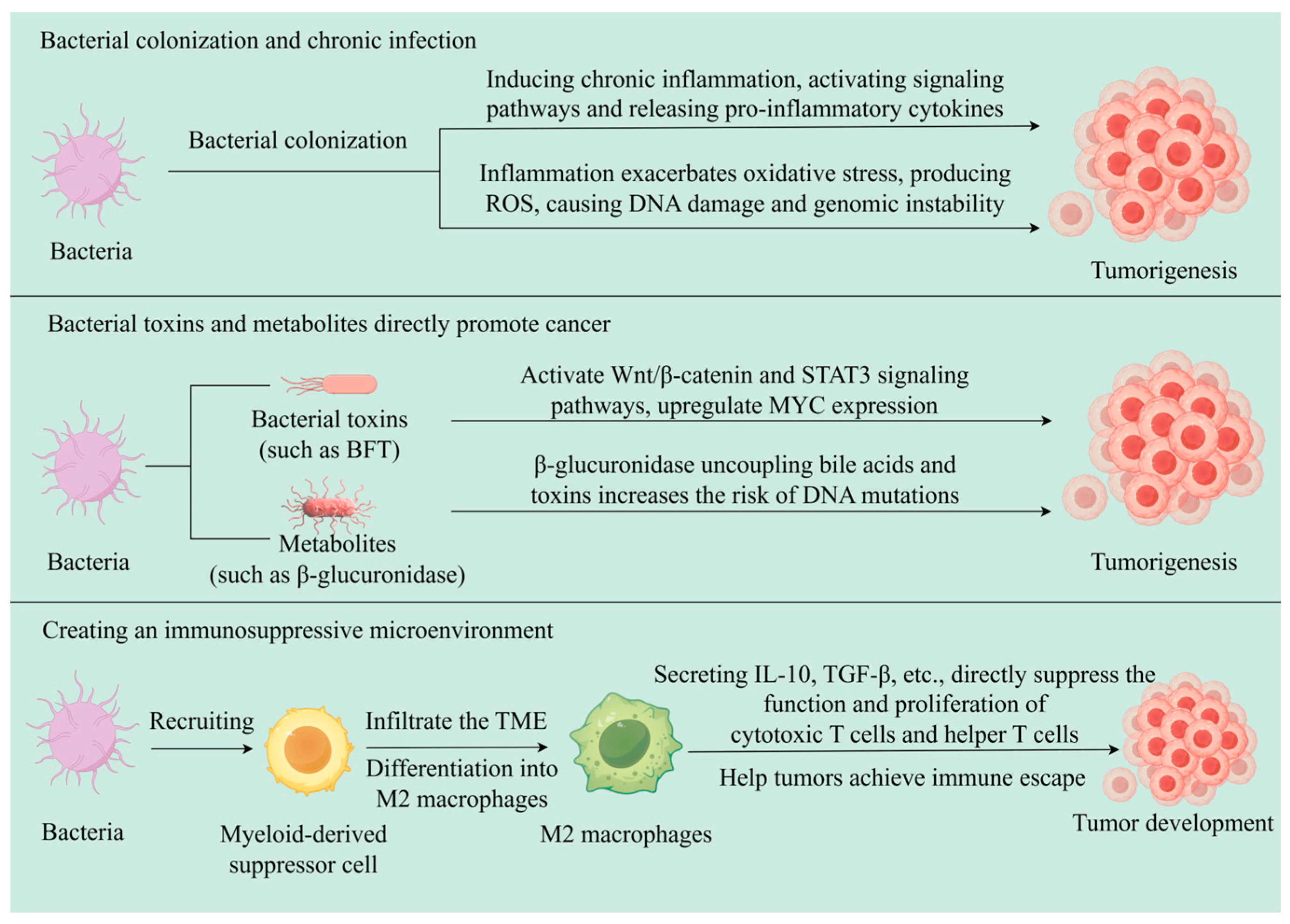
3. Relationship Between Bacteria and Tumors
3.1. Colorectal Cancer and Bacteria
3.2. Breast Cancer and Bacteria
3.3. Pancreatic Cancer and Bacteria
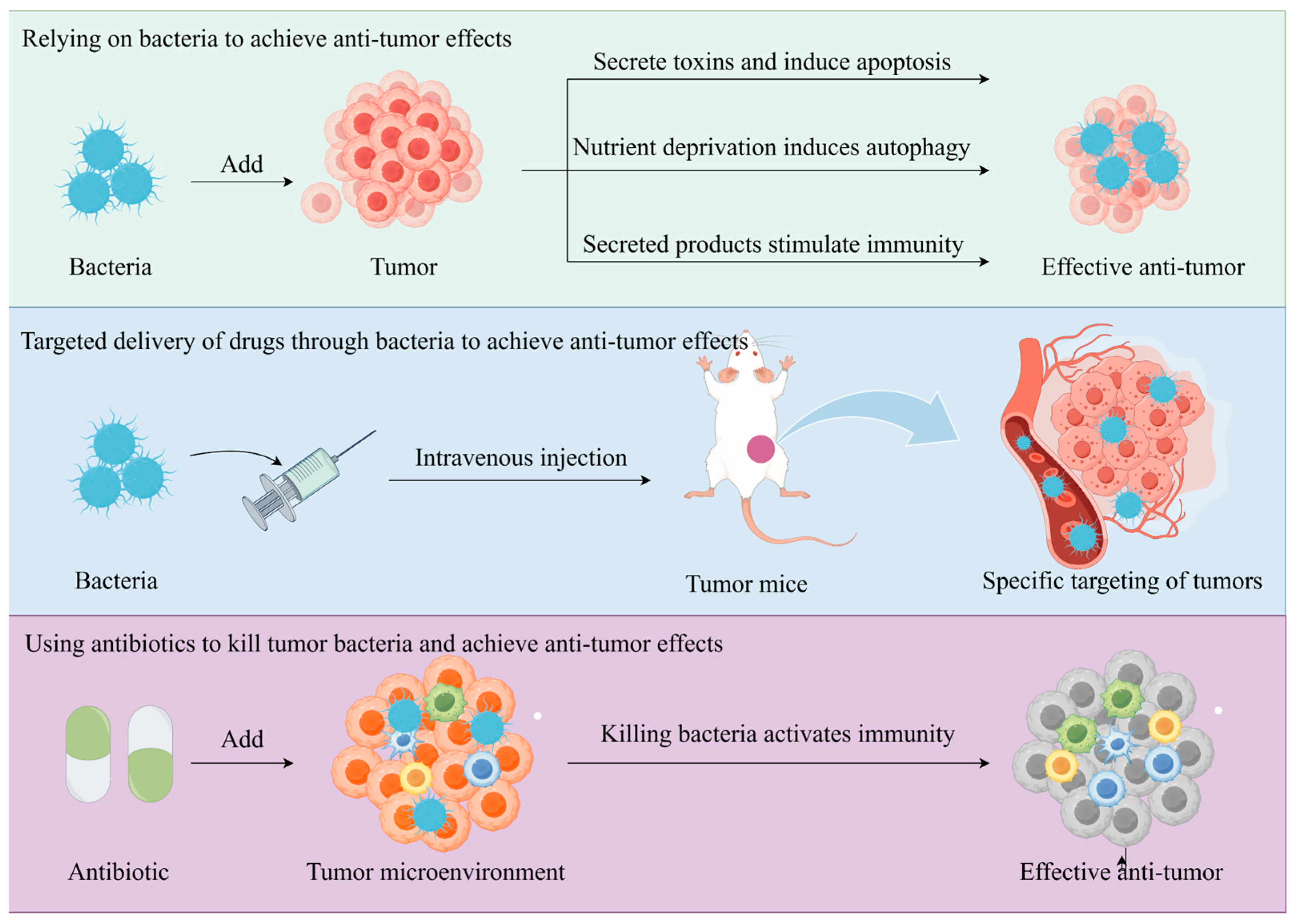
4. Bacteria for Tumor Prevention and Treatment
4.1. Antitumor Effects of Bacterial Components or Products
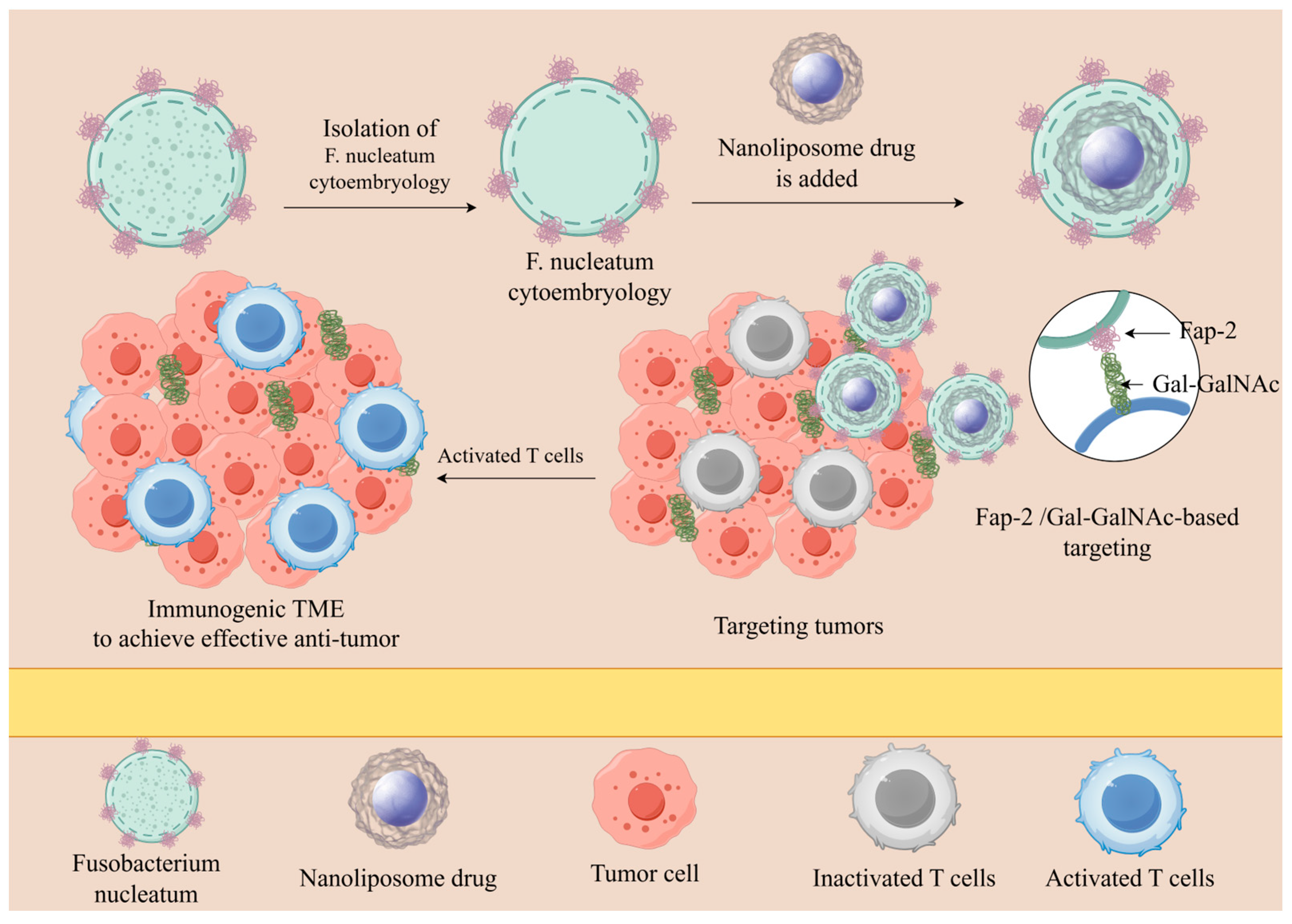
4.2. Natural Targeting of Bacteria to Tumors
4.3. Treatment of Tumors by Destroying Bacteria
5. Bacteria as Tumor Markers
6. Bacterial Therapy-Related Clinical Trials
7. Limitations of Bacterial Therapy
8. Conclusions and Future Prospects of Bacterial Therapy
Author Contributions
Funding
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Sung, H.; Me, J.F.; Siegel, R.L.; Soerjomataram, I.; Dvm, A.J. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Waldman, A.D.; Fritz, J.M.; Lenardo, M.J. A guide to cancer immunotherapy: From T cell basic science to clinical practice. Nat. Rev. Immunol. 2020, 20, 651–668. [Google Scholar] [CrossRef]
- Nejman, D.; Livyatan, I.; Fuks, G.; Gavert, N.; Zwang, Y.; Geller, L.T.; Rotter-Maskowitz, A.; Weiser, R.; Mallel, G.; Gigi, E.; et al. The human tumor microbiome is composed of tumor type–specific intracellular bacteria. Science 2020, 368, 973–980. [Google Scholar] [CrossRef]
- Pushalkar, S.; Hundeyin, M.; Daley, D.; Zambirinis, C.P.; Kurz, E.; Mishra, A.; Mohan, N.; Aykut, B.; Usyk, M.; Torres, L.E. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov. 2018, 8, 403–416. [Google Scholar] [CrossRef]
- Winkler, J.; Abisoye-Ogunniyan, A.; Metcalf, K.J.; Werb, Z. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat. Commun. 2020, 11, 5120. [Google Scholar] [CrossRef]
- Bahuguna, A.; Dubey, S.K. Relevance of tumor microbiome in cancer incidence, prognosis, and its clinical implications in therapeutics. Biochim. Biophys. Acta BBA-Rev. Cancer 2023, 1878, 188956. [Google Scholar] [CrossRef]
- Gruenbacher, G.; Thurnher, M. Mevalonate metabolism in cancer. Cancer Lett. 2015, 356, 192–196. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Cheng, P.; Huang, Q.; Hu, J.; Huang, L.; Hu, G. Immunocytes interact directly with cancer cells in the tumor microenvironment: One coin with two sides and future perspectives. Front. Immunol. 2024, 15, 1388176. [Google Scholar] [CrossRef] [PubMed]
- Wein, T.; Sorek, R. Bacterial origins of human cell-autonomous innate immune mechanisms. Nat. Rev. Immunol. 2022, 22, 629–638. [Google Scholar] [CrossRef]
- Fu, A.; Yao, B.; Dong, T.; Chen, Y.; Yao, J.; Liu, Y.; Li, H.; Bai, H.; Liu, X.; Zhang, Y. Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell 2022, 185, 1356–1372.e26. [Google Scholar] [CrossRef] [PubMed]
- Galeano Niño, J.L.; Wu, H.; LaCourse, K.D.; Kempchinsky, A.G.; Baryiames, A.; Barber, B.; Futran, N.; Houlton, J.; Sather, C.; Sicinska, E. Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature 2022, 611, 810–817. [Google Scholar] [CrossRef]
- Jiang, H.; Li, L.; Bao, Y.; Cao, X.; Ma, L. Microbiota in tumors: New factor influencing cancer development. Cancer Gene Ther. 2024, 31, 1773–1785. [Google Scholar] [CrossRef]
- Kalaora, S.; Nagler, A.; Nejman, D.; Alon, M.; Barbolin, C.; Barnea, E.; Ketelaars, S.L.C.; Cheng, K.; Vervier, K.; Shental, N.; et al. Identification of bacteria-derived HLA-bound peptides in melanoma. Nature 2021, 592, 138–143. [Google Scholar] [CrossRef]
- Fu, A.; Yao, B.; Dong, T.; Cai, S. Emerging roles of intratumor microbiota in cancer metastasis. Trends Cell Biol. 2023, 33, 583–593. [Google Scholar] [CrossRef]
- Kim, J.; Lee, H.K. Potential Role of the Gut Microbiome in Colorectal Cancer Progression. Front. Immunol. 2022, 12, 807648. [Google Scholar] [CrossRef]
- Polk, D.B.; Peek, R.M., Jr. Helicobacter pylori: Gastric cancer and beyond. Nat. Rev. Cancer 2010, 10, 403–414. [Google Scholar] [CrossRef]
- Cullin, N.; Antunes, C.A.; Straussman, R.; Stein-Thoeringer, C.K.; Elinav, E. Microbiome and cancer. Cancer Cell 2021, 39, 1317–1341. [Google Scholar] [CrossRef]
- Kostic, A.D.; Gevers, D.; Pedamallu, C.S.; Michaud, M.; Duke, F.; Earl, A.M.; Ojesina, A.I.; Jung, J.; Bass, A.J.; Tabernero, J. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012, 22, 292–298. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-K. A Comparison Study of Age and Colorectal Cancer-Related Gut Bacteria. Front. Cell. Infect. Microbiol. 2021, 11, 606490. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, Q.; Xu, R.; Li, X.; Hong, Z. Research progress on the correlation between intestinal flora and colorectal cancer. Front. Oncol. 2024, 14, 1416806. [Google Scholar] [CrossRef] [PubMed]
- Bartchewsky, W., Jr.; Martini, M.R.; Masiero, M.; Squassoni, A.C.; Alvarez, M.C.; Ladeira, M.S.; Salvatore, D.; Trevisan, M.; Pedrazzoli, J., Jr.; Ribeiro, M.L. Effect of Helicobacter pylori infection on IL-8, IL-1β and COX-2 expression in patients with chronic gastritis and gastric cancer. Scand. J. Gastroenterol. 2009, 44, 153–161. [Google Scholar] [CrossRef]
- Rokkas, T.; Rokka, A.; Portincasa, P. A systematic review and meta-analysis of the role of Helicobacter pylori eradication in preventing gastric cancer. Ann. Gastroenterol. 2017, 30, 414. [Google Scholar] [CrossRef]
- Kuo, S.-H.; Yeh, K.-H.; Wu, M.-S.; Lin, C.-W.; Hsu, P.-N.; Wang, H.-P.; Chen, L.-T.; Cheng, A.-L. Helicobacter pylori eradication therapy is effective in the treatment of early-stage H pylori–positive gastric diffuse large B-cell lymphomas. Blood J. Am. Soc. Hematol. 2012, 119, 4838–4844. [Google Scholar] [CrossRef]
- Kuo, S.-H.; Chen, L.-T.; Lin, C.-W.; Yeh, K.-H.; Shun, C.-T.; Tzeng, Y.-S.; Liou, J.-M.; Wu, M.-S.; Hsu, P.-N.; Cheng, A.-L. Expressions of the CagA protein and CagA-signaling molecules predict Helicobacter pylori dependence of early-stage gastric DLBCL. Blood J. Am. Soc. Hematol. 2017, 129, 188–198. [Google Scholar] [CrossRef]
- Bhavsar, A.P.; Guttman, J.A.; Finlay, B.B. Manipulation of host-cell pathways by bacterial pathogens. Nature 2007, 449, 827–834. [Google Scholar] [CrossRef] [PubMed]
- Tekle, G.E.; Garrett, W.S. Bacteria in cancer initiation, promotion and progression. Nat. Rev. Cancer 2023, 23, 600–618. [Google Scholar] [CrossRef]
- Cao, Y.; Xia, H.; Tan, X.; Shi, C.; Ma, Y.; Meng, D.; Zhou, M.; Lv, Z.; Wang, S.; Jin, Y. Intratumoural microbiota: A new frontier in cancer development and therapy. Signal Transduct. Target. Ther. 2024, 9, 15. [Google Scholar] [CrossRef] [PubMed]
- Cuevas-Ramos, G.; Petit, C.R.; Marcq, I.; Boury, M.; Oswald, E.; Nougayrède, J.-P. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc. Natl. Acad. Sci. USA 2010, 107, 11537–11542. [Google Scholar] [CrossRef]
- Hong, Y.; Boiti, A.; Vallone, D.; Foulkes, N.S. Reactive Oxygen Species Signaling and Oxidative Stress: Transcriptional Regulation and Evolution. Antioxidants 2024, 13, 312. [Google Scholar] [CrossRef] [PubMed]
- An, X.; Yu, W.; Liu, J.; Tang, D.; Yang, L.; Chen, X. Oxidative cell death in cancer: Mechanisms and therapeutic opportunities. Cell Death Dis. 2024, 15, 556. [Google Scholar] [CrossRef]
- Herrera, L.A.; Benítez-Bribiesca, L.; Mohar, A.; Ostrosky-Wegman, P. Role of infectious diseases in human carcinogenesis. Environ. Mol. Mutagen. 2005, 45, 284–303. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, L.; Xu, C.; Wang, Y.; Wang, Z.; Chen, M.; Jiang, Z.; Pan, J.; Yang, C.; Li, X.; et al. Cross-talk between the gut microbiota and monocyte-like macrophages mediates an inflammatory response to promote colitis-associated tumourigenesis. Gut 2020, 70, 1495–1506. [Google Scholar] [CrossRef]
- Hill, M.J. Chronic bacterial infection and subsequent human carcinogenesis. Eur. J. Cancer Prev. 1995, 4, 127–128. [Google Scholar]
- Little, M.S.; Pellock, S.J.; Walton, W.G.; Tripathy, A.; Redinbo, M.R. Structural basis for the regulation of β-glucuronidase expression by human gut Enterobacteriaceae. Proc. Natl. Acad. Sci. USA 2018, 115, E152–E161. [Google Scholar] [CrossRef]
- Chang, A.H.; Parsonnet, J. Role of bacteria in oncogenesis. Clin. Microbiol. Rev. 2010, 23, 837–857. [Google Scholar] [CrossRef]
- Peterson, J.; Garges, S.; Giovanni, M.; McInnes, P.; Wang, L.; Schloss, J.A.; Bonazzi, V.; McEwen, J.E.; Wetterstrand, K.A.; Deal, C. The NIH human microbiome project. Genome Res. 2009, 19, 2317–2323. [Google Scholar] [CrossRef]
- Round, J.L.; Mazmanian, S.K. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl Acad. Sci. USA 2010, 107, 12204–12209. [Google Scholar] [CrossRef]
- Wu, S.; Morin, P.J.; Maouyo, D.; Sears, C.L. Bacteroides fragilis enterotoxin induces c-Myc expression and cellular proliferation. Gastroenterology 2003, 124, 392–400. [Google Scholar] [CrossRef]
- Wu, S.; Rhee, K.-J.; Zhang, M.; Franco, A.; Sears, C.L. Bacteroides fragilis toxin stimulates intestinal epithelial cell shedding and γ-secretase-dependent E-cadherin cleavage. J. Cell Sci. 2007, 120, 1944–1952. [Google Scholar] [CrossRef]
- Wu, S.; Powell, J.; Mathioudakis, N.; Kane, S.; Fernandez, E.; Sears, C.L. Bacteroides fragilis enterotoxin induces intestinal epithelial cell secretion of interleukin-8 through mitogen-activated protein kinases and a tyrosine kinase-regulated nuclear factor-κB pathway. Infect. Immun. 2004, 72, 5832–5839. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Rhee, K.-J.; Albesiano, E.; Rabizadeh, S.; Wu, X.; Yen, H.-R.; Huso, D.L.; Brancati, F.L.; Wick, E.; McAllister, F.; et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 2009, 15, 1016–1022. [Google Scholar] [CrossRef] [PubMed]
- Chiu, D.K.-C.; Tse, A.P.-W.; Xu, I.M.-J.; Di Cui, J.; Lai, R.K.-H.; Li, L.L.; Koh, H.-Y.; Tsang, F.H.-C.; Wei, L.L.; Wong, C.-M.; et al. Hypoxia inducible factor HIF-1 promotes myeloid-derived suppressor cells accumulation through ENTPD2/CD39L1 in hepatocellular carcinoma. Nat. Commun. 2017, 8, 517. [Google Scholar] [CrossRef]
- Kostic, A.D.; Chun, E.; Robertson, L.; Glickman, J.N.; Gallini, C.A.; Michaud, M.; Clancy, T.E.; Chung, D.C.; Lochhead, P.; Hold, G.L.; et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013, 14, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Canny, G.O.; McCormick, B.A. Bacteria in the intestine, helpful residents or enemies from within? Infect. Immun. 2008, 76, 3360–3373. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Kang, M.; Martin, A. Microbiome and colorectal cancer: Unraveling host-microbiota interactions in colitis-associated colorectal cancer development. Semin. Immunol. 2017, 32, 3–13. [Google Scholar] [CrossRef]
- Genua, F.; Raghunathan, V.; Jenab, M.; Gallagher, W.M.; Hughes, D.J. The role of gut barrier dysfunction and microbiome dysbiosis in colorectal cancer development. Front. Oncol. 2021, 11, 626349. [Google Scholar] [CrossRef]
- Ye, P.; Xi, Y.; Huang, Z.; Xu, P. Linking obesity with colorectal cancer: Epidemiology and mechanistic insights. Cancers 2020, 12, 1408. [Google Scholar] [CrossRef]
- Wilson, M.R.; Jiang, Y.; Villalta, P.W.; Stornetta, A.; Boudreau, P.D.; Carrá, A.; Brennan, C.A.; Chun, E.; Ngo, L.; Samson, L.D. The human gut bacterial genotoxin colibactin alkylates DNA. Science 2019, 363, eaar7785. [Google Scholar] [CrossRef]
- Dejea, C.M.; Fathi, P.; Craig, J.M.; Boleij, A.; Taddese, R.; Geis, A.L.; Wu, X.; DeStefano Shields, C.E.; Hechenbleikner, E.M.; Huso, D.L. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 2018, 359, 592–597. [Google Scholar] [CrossRef]
- Chen, T.; Li, Q.; Wu, J.; Wu, Y.; Peng, W.; Li, H.; Wang, J.; Tang, X.; Peng, Y.; Fu, X. Fusobacterium nucleatum promotes M2 polarization of macrophages in the microenvironment of colorectal tumours via a TLR4-dependent mechanism. Cancer Immunol. Immunother. 2018, 67, 1635–1646. [Google Scholar] [CrossRef]
- Yu, T.; Guo, F.; Yu, Y.; Sun, T.; Ma, D.; Han, J.; Qian, Y.; Kryczek, I.; Sun, D.; Nagarsheth, N. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 2017, 170, 548–563.e16. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Tang, L.; Feng, Y.; Zhao, S.; Han, M.; Zhang, C.; Yuan, G.; Zhu, J.; Cao, S.; Wu, Q. A purified membrane protein from Akkermansia muciniphila or the pasteurised bacterium blunts colitis associated tumourigenesis by modulation of CD8+ T cells in mice. Gut 2020, 69, 1988–1997. [Google Scholar] [CrossRef]
- Álvarez-Mercado, A.I.; del Valle Cano, A.; Fernández, M.F.; Fontana, L. Gut microbiota and breast cancer: The dual role of microbes. Cancers 2023, 15, 443. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Wei, Z.; Tian, T.; Bose, D.; Shih, N.N.; Feldman, M.D.; Khoury, T.; De Michele, A.; Robertson, E.S. Prognostic correlations with the microbiome of breast cancer subtypes. Cell Death Dis. 2021, 12, 831. [Google Scholar] [CrossRef]
- Zhu, J.; Liao, M.; Yao, Z.; Liang, W.; Li, Q.; Liu, J.; Yang, H.; Ji, Y.; Wei, W.; Tan, A.; et al. Breast cancer in postmenopausal women is associated with an altered gut metagenome. Microbiome 2018, 6, 136. [Google Scholar] [CrossRef] [PubMed]
- Kovács, P.; Csonka, T.; Kovács, T.; Sári, Z.; Ujlaki, G.; Sipos, A.; Karányi, Z.; Szeőcs, D.; Hegedűs, C.; Uray, K. Lithocholic acid, a metabolite of the microbiome, increases oxidative stress in breast cancer. Cancers 2019, 11, 1255. [Google Scholar] [CrossRef]
- Parhi, L.; Alon-Maimon, T.; Sol, A.; Nejman, D.; Shhadeh, A.; Fainsod-Levi, T.; Yajuk, O.; Isaacson, B.; Abed, J.; Maalouf, N. Breast cancer colonization by Fusobacterium nucleatum accelerates tumor growth and metastatic progression. Nat. Commun. 2020, 11, 3259. [Google Scholar] [CrossRef]
- Geller, L.T.; Barzily-Rokni, M.; Danino, T.; Jonas, O.H.; Shental, N.; Nejman, D.; Gavert, N.; Zwang, Y.; Cooper, Z.A.; Shee, K. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 2017, 357, 1156–1160. [Google Scholar] [CrossRef]
- Riquelme, E.; Zhang, Y.; Zhang, L.; Montiel, M.; Zoltan, M.; Dong, W.; Quesada, P.; Sahin, I.; Chandra, V.; San Lucas, A. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell 2019, 178, 795–806.e12. [Google Scholar] [CrossRef]
- Brennan, C.A.; Garrett, W.S. Fusobacterium nucleatum–symbiont, opportunist and oncobacterium. Nat. Rev. Microbiol. 2019, 17, 156–166. [Google Scholar] [CrossRef]
- Castellarin, M.; Warren, R.L.; Freeman, J.D.; Dreolini, L.; Krzywinski, M.; Strauss, J.; Barnes, R.; Watson, P.; Allen-Vercoe, E.; Moore, R.A. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012, 22, 299–306. [Google Scholar] [CrossRef]
- Bullman, S.; Pedamallu, C.S.; Sicinska, E.; Clancy, T.E.; Zhang, X.; Cai, D.; Neuberg, D.; Huang, K.; Guevara, F.; Nelson, T. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 2017, 358, 1443–1448. [Google Scholar] [CrossRef] [PubMed]
- Lopès, A.; Billard, E.; Casse, A.H.; Villéger, R.; Veziant, J.; Roche, G.; Carrier, G.; Sauvanet, P.; Briat, A.; Pagès, F.; et al. Colibactin-positive E.coli induce a procarcinogenic immune environment leading to immunotherapy resistance in colorectal cancer. Int. J. Cancer 2020, 146, 3147–3159. [Google Scholar] [CrossRef]
- Arthur, J.C.; Perez-Chanona, E.; Mühlbauer, M.; Tomkovich, S.; Uronis, J.M.; Fan, T.-J.; Campbell, B.J.; Abujamel, T.; Dogan, B.; Rogers, A.B. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 2012, 338, 120–123. [Google Scholar] [CrossRef] [PubMed]
- Raisch, J.; Rolhion, N.; Dubois, A.; Darfeuille-Michaud, A.; Bringer, M.-A. Intracellular colon cancer-associated Escherichia coli promote protumoral activities of human macrophages by inducing sustained COX-2 expression. Lab. Investig. 2015, 95, 296–307. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Guo, B.; Gao, R.; Zhu, Q.; Qin, H. Microbiota disbiosis is associated with colorectal cancer. Front. Microbiol. 2015, 6, 20. [Google Scholar] [CrossRef]
- Richard, M.L.; Liguori, G.; Lamas, B.; Brandi, G.; da Costa, G.; Hoffmann, T.W.; Di Simone, M.P.; Calabrese, C.; Poggioli, G.; Langella, P.; et al. Mucosa-associated microbiota dysbiosis in colitis associated cancer. Gut Microbes 2017, 9, 131–142. [Google Scholar] [CrossRef]
- Wang, X.; Jia, Y.; Wen, L.; Mu, W.; Wu, X.; Liu, T.; Liu, X.; Fang, J.; Luan, Y.; Chen, P.; et al. Porphyromonas gingivalis Promotes Colorectal Carcinoma by Activating the Hematopoietic NLRP3 Inflammasome. Cancer Res. 2021, 81, 2745–2759. [Google Scholar] [CrossRef]
- Lo, C.; Wu, D.; Jao, S.; Wu, C.; Lin, C.; Chuang, C.; Lin, Y.; Chen, C.; Chen, Y.; Chen, J.; et al. Enrichment of Prevotella intermedia in human colorectal cancer and its additive effects with Fusobacterium nucleatum on the malignant transformation of colorectal adenomas. J. Biomed. Sci. 2022, 29, 88. [Google Scholar] [CrossRef]
- Xuan, C.; Shamonki, J.M.; Chung, A.; DiNome, M.L.; Chung, M.; Sieling, P.A.; Lee, D.J. Microbial Dysbiosis Is Associated with Human Breast Cancer. PLoS ONE 2014, 9, e83744. [Google Scholar] [CrossRef] [PubMed]
- Parida, S.; Wu, S.; Siddharth, S.; Wang, G.; Muniraj, N.; Nagalingam, A.; Hum, C.; Mistriotis, P.; Hao, H.; Talbot, C.C., Jr. A procarcinogenic colon microbe promotes breast tumorigenesis and metastatic progression and concomitantly activates notch and β-catenin axes. Cancer Discov. 2021, 11, 1138–1157. [Google Scholar] [CrossRef]
- Knippel, R.J.; Drewes, J.L.; Sears, C.L. The cancer microbiome: Recent highlights and knowledge gaps. Cancer Discov. 2021, 11, 2378–2395. [Google Scholar] [CrossRef]
- Ghaddar, B.; Biswas, A.; Harris, C.; Omary, M.B.; Carpizo, D.R.; Blaser, M.J.; De, S. Tumor microbiome links cellular programs and immunity in pancreatic cancer. Cancer Cell 2022, 40, 1240–1253. [Google Scholar] [CrossRef]
- Fan, X.; Alekseyenko, A.V.; Wu, J.; Peters, B.A.; Jacobs, E.J.; Gapstur, S.M.; Purdue, M.P.; Abnet, C.C.; Stolzenberg-Solomon, R.; Miller, G.; et al. Human oral microbiome and prospective risk for pancreatic cancer: A population-based nested case-control study. Gut 2018, 67, 120–127. [Google Scholar] [CrossRef]
- Aykut, B.; Pushalkar, S.; Chen, R.; Li, Q.; Abengozar, R.; Kim, J.I.; Shadaloey, S.A.; Wu, D.; Preiss, P.; Verma, N.; et al. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature 2019, 574, 264–267. [Google Scholar] [CrossRef]
- Badger, J.L.; Stins, M.F.; Kim, K.S. Citrobacter freundii Invades and Replicates in Human Brain Microvascular Endothelial Cells. Infect Immun. 1999, 67, 4208–4215. [Google Scholar] [CrossRef] [PubMed]
- Felgner, S.; Kocijancic, D.; Frahm, M.; Weiss, S. Bacteria in Cancer Therapy: Renaissance of an Old Concept. Int. J. Microbiol. 2016, 2016, 8451728. [Google Scholar] [CrossRef] [PubMed]
- Gurbatri, C.R.; Arpaia, N.; Danino, T. Engineering bacteria as interactive cancer therapies. Science 2022, 378, 858–864. [Google Scholar] [CrossRef]
- Wang, M.; Rousseau, B.; Qiu, K.; Huang, G.; Zhang, Y.; Su, H.; Le Bihan-Benjamin, C.; Khati, I.; Artz, O.; Foote, M.B. Killing tumor-associated bacteria with a liposomal antibiotic generates neoantigens that induce anti-tumor immune responses. Nat. Biotechnol. 2023, 42, 1263–1274. [Google Scholar] [CrossRef]
- Kwon, S.-Y.; Thi-Thu Ngo, H.; Son, J.; Hong, Y.; Min, J.-J. Exploiting bacteria for cancer immunotherapy. Nat. Rev. Clin. Oncol. 2024, 21, 569–589. [Google Scholar] [CrossRef]
- Zhu, C.; Liu, C.; Wu, Q.; Sheng, T.; Zhou, R.; Ren, E.; Zhang, R.; Zhao, Z.; Shi, J.; Shen, X.; et al. Remolding the tumor microenvironment by bacteria augments adoptive T cell therapy in advanced-stage solid tumors. Signal Transduct. Target. Ther. 2024, 9, 307. [Google Scholar] [CrossRef] [PubMed]
- Leschner, S.; Westphal, K.; Dietrich, N.; Viegas, N.; Jablonska, J.; Lyszkiewicz, M.; Lienenklaus, S.; Falk, W.; Gekara, N.; Loessner, H. Tumor invasion of Salmonella enterica serovar Typhimurium is accompanied by strong hemorrhage promoted by TNF-α. PLoS ONE 2009, 4, e6692. [Google Scholar] [CrossRef]
- Forbes, N.S. Engineering the perfect (bacterial) cancer therapy. Nat. Rev. Cancer 2010, 10, 785–794. [Google Scholar] [CrossRef]
- Middlebrook, J.L.; Dorland, R.B. Bacterial toxins: Cellular mechanisms of action. Microbiol. Rev. 1984, 48, 199–221. [Google Scholar] [CrossRef] [PubMed]
- Staedtke, V.; Roberts, N.J.; Bai, R.-Y.; Zhou, S. Clostridium novyi-NT in cancer therapy. Genes Dis. 2016, 3, 144–152. [Google Scholar] [CrossRef]
- Flickinger, J.C., Jr.; Rodeck, U.; Snook, A.E. Listeria monocytogenes as a vector for cancer immunotherapy: Current understanding and progress. Vaccines 2018, 6, 48. [Google Scholar] [CrossRef]
- Ganai, S.; Arenas, R.B.; Sauer, J.P.; Bentley, B.; Forbes, N.S. In tumors Salmonella migrate away from vasculature toward the transition zone and induce apoptosis. Cancer Gene Ther. 2011, 18, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Uchugonova, A.; Zhang, Y.; Salz, R.; Liu, F.; Suetsugu, A.; Zhang, L.; Koenig, K.; Hoffman, R.M.; Zhao, M. Imaging the different mechanisms of prostate cancer cell-killing by tumor-targeting Salmonella typhimurium A1-R. Anticancer Res. 2015, 35, 5225–5229. [Google Scholar]
- Lee, C.H.; Lin, S.T.; Liu, J.J.; Chang, W.W.; Hsieh, J.L.; Wang, W.K. Salmonella induce autophagy in melanoma by the downregulation of AKT/mTOR pathway. Gene Ther. 2014, 21, 309–316. [Google Scholar] [CrossRef]
- Uchugonova, A.; Zhao, M.; Zhang, Y.; Weinigel, M.; Koenig, K.; Hoffman, R.M. Cancer-cell killing by engineered Salmonella imaged by multiphoton tomography in live mice. Anticancer Res. 2012, 32, 4331–4337. [Google Scholar]
- Liu, B.; Jiang, Y.; Dong, T.; Zhao, M.; Wu, J.; Li, L.; Chu, Y.; She, S.; Zhao, H.; Hoffman, R.M. Blockage of autophagy pathway enhances Salmonella tumor-targeting. Oncotarget 2016, 7, 22873. [Google Scholar] [CrossRef]
- Saccheri, F.; Pozzi, C.; Avogadri, F.; Barozzi, S.; Faretta, M.; Fusi, P.; Rescigno, M. Bacteria-induced gap junctions in tumors favor antigen cross-presentation and antitumor immunity. Sci. Transl. Med. 2010, 2, 44ra57. [Google Scholar] [CrossRef]
- Chang, W.-W.; Lai, C.-H.; Chen, M.-C.; Liu, C.-F.; Kuan, Y.-D.; Lin, S.-T.; Lee, C.-H. Salmonella enhance chemosensitivity in tumor through connexin 43 upregulation. Int. J. Cancer 2013, 133, 1926–1935. [Google Scholar] [CrossRef]
- Avogadri, F.; Martinoli, C.; Petrovska, L.; Chiodoni, C.; Transidico, P.; Bronte, V.; Longhi, R.; Colombo, M.P.; Dougan, G.; Rescigno, M. Cancer immunotherapy based on killing of Salmonella-infected tumor cells. Cancer Res. 2005, 65, 3920–3927. [Google Scholar] [CrossRef]
- Kim, S.H.; Castro, F.; Paterson, Y.; Gravekamp, C. High efficacy of a Listeria-based vaccine against metastatic breast cancer reveals a dual mode of action. Cancer Res. 2009, 69, 5860–5866. [Google Scholar] [CrossRef]
- Phan, T.X.; Nguyen, V.H.; Duong, M.T.-Q.; Hong, Y.; Choy, H.E.; Min, J.-J. Activation of inflammasome by attenuated Salmonella typhimurium in bacteria-mediated cancer therapy. Microbiol. Immunol. 2015, 59, 664–675. [Google Scholar] [CrossRef]
- Beutler, B.; Cerami, A. The biology of cachectin/TNF--a primary mediator of the host response. Annu. Rev. Immunol. 1989, 7, 625–655. [Google Scholar] [CrossRef] [PubMed]
- Kocijancic, D.; Leschner, S.; Felgner, S.; Komoll, R.-M.; Frahm, M.; Pawar, V.; Weiss, S. Therapeutic benefit of Salmonella attributed to LPS and TNF-α is exhaustible and dictated by tumor susceptibility. Oncotarget 2017, 8, 36492. [Google Scholar] [CrossRef] [PubMed]
- Dobrovolskaia, M.A.; Vogel, S.N. Toll receptors, CD14, and macrophage activation and deactivation by LPS. Microbes Infect. 2002, 4, 903–914. [Google Scholar] [CrossRef] [PubMed]
- Sfondrini, L.; Rossini, A.; Besusso, D.; Merlo, A.; Tagliabue, E.; Mènard, S.; Balsari, A. Antitumor activity of the TLR-5 ligand flagellin in mouse models of cancer. J. Immunol. 2006, 176, 6624–6630. [Google Scholar] [CrossRef]
- Agrawal, N.; Bettegowda, C.; Cheong, I.; Geschwind, J.-F.; Drake, C.G.; Hipkiss, E.L.; Tatsumi, M.; Dang, L.H.; Diaz, L.A., Jr.; Pomper, M. Bacteriolytic therapy can generate a potent immune response against experimental tumors. Proc. Natl. Acad. Sci. USA 2004, 101, 15172–15177. [Google Scholar] [CrossRef]
- Shinnoh, M.; Horinaka, M.; Yasuda, T.; Yoshikawa, S.; Morita, M.; Yamada, T.; Miki, T.; Sakai, T. Clostridium butyricum MIYAIRI 588 shows antitumor effects by enhancing the release of TRAIL from neutrophils through MMP-8. Int. J. Oncol. 2013, 42, 903–911. [Google Scholar] [CrossRef]
- Kim, K.; Jeong, J.H.; Lim, D.; Hong, Y.; Lim, H.-J.; Kim, G.-J.; Shin, S.; Lee, J.-J.; Yun, M.; Harris, R.A.; et al. L-Asparaginase delivered by Salmonella typhimurium suppresses solid tumors. Mol. Ther.-Oncolytics 2015, 2, 15007. [Google Scholar] [CrossRef]
- Bredon, M.; le Malicot, K.; Louvet, C.; Evesque, L.; Gonzalez, D.; Tougeron, D.; Sokol, H. Faecalibacteriumprausnitzii Is Associated With Clinical Response to Immune Checkpoint Inhibitors in Patients with Advanced Gastric Adenocarcinoma: Results of Microbiota Analysis of PRODIGE 59-FFCD 1707-DURIGAST Trial. Gastroenterology 2025, 168, 601–603. [Google Scholar] [CrossRef]
- Ganai, S.; Arenas, R.B.; Forbes, N.S. Tumour-targeted delivery of TRAIL using Salmonella typhimurium enhances breast cancer survival in mice. Br. J. Cancer 2009, 101, 1683–1691. [Google Scholar] [CrossRef]
- Zheng, J.H.; Nguyen, V.H.; Jiang, S.-N.; Park, S.-H.; Tan, W.; Hong, S.H.; Shin, M.G.; Chung, I.-J.; Hong, Y.; Bom, H.-S.; et al. Two-step enhanced cancer immunotherapy with engineered Salmonella typhimurium secreting heterologous flagellin. Sci. Transl. Med. 2017, 9, eaak9537. [Google Scholar] [CrossRef] [PubMed]
- Wilson, W.R.; Hay, M.P. Targeting hypoxia in cancer therapy. Nat. Rev. Cancer 2011, 11, 393–410. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Ma, S.; Wu, H.; Zheng, L.; Yi, Y.; Liu, G.; Li, B.; Sun, J.; Du, Y.; Wang, B.; et al. Zonated Copper-Driven Breast Cancer Progression Countered by a Copper-Depleting Nanoagent for Immune and Metabolic Reprogramming. Adv. Sci. 2025, 12, 2412434. [Google Scholar] [CrossRef] [PubMed]
- Manoochehri Khoshinani, H.; Afshar, S.; Najafi, R. Hypoxia: A double-edged sword in cancer therapy. Cancer Investig. 2016, 34, 536–545. [Google Scholar] [CrossRef]
- Fu, J.; Wu, Q.; Dang, Y.; Lei, X.; Feng, G.; Chen, M.; Yu, X.-Y. Synergistic therapy using doxorubicin-loading and nitric oxide-generating hollow Prussian blue nanoparticles with photoacoustic imaging potential against breast cancer. Int. J. Nanomed. 2021, 16, 6003–6016. [Google Scholar] [CrossRef] [PubMed]
- Muhamad, N.; Plengsuriyakarn, T.; Na-Bangchang, K. Application of active targeting nanoparticle delivery system for chemotherapeutic drugs and traditional/herbal medicines in cancer therapy: A systematic review. Int. J. Nanomed. 2018, 13, 3921–3935. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.; Fan, M.; Liu, H.; Zhang, Y.; Dai, X.; Li, H.; Zhou, X.; Hu, S.; Yang, X.; Jin, Y. Self-propelled and near-infrared-phototaxic photosynthetic bacteria as photothermal agents for hypoxia-targeted cancer therapy. ACS Nano 2020, 15, 1100–1110. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Zang, Z.; Chen, Z.; Cui, L.; Chang, Z.; Ma, A.; Yin, T.; Liang, R.; Han, Y.; Wu, Z. Nanophotosensitizer-engineered Salmonella bacteria with hypoxia targeting and photothermal-assisted mutual bioaccumulation for solid tumor therapy. Biomaterials 2019, 214, 119226. [Google Scholar] [CrossRef]
- Duong, M.T.-Q.; Qin, Y.; You, S.-H.; Min, J.-J. Bacteria-cancer interactions: Bacteria-based cancer therapy. Exp. Mol. Med. 2019, 51, 1–15. [Google Scholar] [CrossRef]
- Ijaz, M.; Hasan, I.; Chaudhry, T.H.; Huang, R.; Zhang, L.; Hu, Z.; Tan, Q.; Guo, B. Bacterial derivatives mediated drug delivery in cancer therapy: A new generation strategy. J. Nanobiotechnol. 2024, 22, 510. [Google Scholar] [CrossRef]
- Dadgar-Zankbar, L.; Elahi, Z.; Shariati, A.; Khaledi, A.; Razavi, S.; Khoshbayan, A. Exploring the role of Fusobacterium nucleatum in colorectal cancer: Implications for tumor proliferation and chemoresistance. Cell Commun. Signal. 2024, 22, 547. [Google Scholar] [CrossRef]
- Lee, J.B.; Kim, K.-A.; Cho, H.Y.; Kim, D.; Kim, W.K.; Yong, D.; Lee, H.; Yoon, S.S.; Han, D.H.; Han, Y.D.; et al. Association between Fusobacterium nucleatum and patient prognosis in metastatic colon cancer. Sci. Rep. 2021, 11, 20263. [Google Scholar] [CrossRef]
- Alon-Maimon, T.; Mandelboim, O.; Bachrach, G. Fusobacterium nucleatum and cancer. Periodontology 2000 2022, 89, 166–180. [Google Scholar] [CrossRef]
- Pawelek, J.M.; Low, K.B.; Bermudes, D. Tumor-targeted Salmonella as a novel anticancer vector. Cancer Res. 1997, 57, 4537–4544. [Google Scholar]
- Diaz, L.A., Jr.; Cheong, I.; Foss, C.A.; Zhang, X.; Peters, B.A.; Agrawal, N.; Bettegowda, C.; Karim, B.; Liu, G.; Khan, K. Pharmacologic and toxicologic evaluation of C. novyi-NT spores. Toxicol. Sci. 2005, 88, 562–575. [Google Scholar] [CrossRef] [PubMed]
- Kasinskas, R.W.; Forbes, N.S. Salmonella typhimurium lacking ribose chemoreceptors localize in tumor quiescence and induce apoptosis. Cancer Res. 2007, 67, 3201–3209. [Google Scholar] [CrossRef]
- Lee, C.-H.; Wu, C.-L.; Shiau, A.-L. Systemic administration of attenuated Salmonella choleraesuis carrying thrombospondin-1 gene leads to tumor-specific transgene expression, delayed tumor growth and prolonged survival in the murine melanoma model. Cancer Gene Ther. 2005, 12, 175–184. [Google Scholar] [CrossRef]
- Westphal, K.; Leschner, S.; Jablonska, J.; Loessner, H.; Weiss, S. Containment of Tumor-Colonizing Bacteria by Host Neutrophils. Cancer Res. 2008, 68, 2952–2960. [Google Scholar] [CrossRef]
- Branzk, N.; Lubojemska, A.; Hardison, S.E.; Wang, Q.; Gutierrez, M.G.; Brown, G.D.; Papayannopoulos, V. Neutrophils sense microbe size and selectively release neutrophil extracellular traps in response to large pathogens. Nat. Immunol. 2014, 15, 1017–1025. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.A.; Zhang, Q.; Szalay, A.A. Establishment and characterization of conditions required for tumor colonization by intravenously delivered bacteria. Biotechnol. Bioeng. 2008, 100, 567–578. [Google Scholar] [CrossRef]
- Zhou, S.; Gravekamp, C.; Bermudes, D.; Liu, K. Tumour-targeting bacteria engineered to fight cancer. Nat. Rev. Cancer 2018, 18, 727–743. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.-D.; Shu, L.-Z.; He, R.-S.; Chen, K.-Y.; Deng, Y.-J.; Zhou, Z.-B.; Xiong, Y.; Deng, H. Listeria monocytogenes: A promising vector for tumor immunotherapy. Front. Immunol. 2023, 14, 1278011. [Google Scholar] [CrossRef]
- He, L.; Yang, H.; Tang, J.; Liu, Z.; Chen, Y.; Lu, B.; He, H.; Tang, S.; Sun, Y.; Liu, F. Intestinal probiotics E. coli Nissle 1917 as a targeted vehicle for delivery of p53 and Tum-5 to solid tumors for cancer therapy. J. Biol. Eng. 2019, 13, 58. [Google Scholar] [CrossRef]
- Tan, W.; Duong, M.T.-Q.; Zuo, C.; Qin, Y.; Zhang, Y.; Guo, Y.; Hong, Y.; Zheng, J.H.; Min, J.-J. Targeting of pancreatic cancer cells and stromal cells using engineered oncolytic Salmonella typhimurium. Mol. Ther. 2022, 30, 662–671. [Google Scholar] [CrossRef]
- Bottery, M.J.; Pitchford, J.W.; Friman, V.-P. Ecology and evolution of antimicrobial resistance in bacterial communities. ISME J. 2021, 15, 939–948. [Google Scholar] [CrossRef]
- Singh, R.; Patil, S.; Singh, N.; Gupta, S. Dual functionality nanobioconjugates targeting intracellular bacteria in cancer cells with enhanced antimicrobial activity. Sci. Rep. 2017, 7, 5792. [Google Scholar] [CrossRef]
- Zheng, D.-W.; Dong, X.; Pan, P.; Chen, K.-W.; Fan, J.-X.; Cheng, S.-X.; Zhang, X.-Z. Phage-guided modulation of the gut microbiota of mouse models of colorectal cancer augments their responses to chemotherapy. Nat. Biomed. Eng. 2019, 3, 717–728. [Google Scholar] [CrossRef] [PubMed]
- Gogokhia, L.; Buhrke, K.; Bell, R.; Hoffman, B.; Brown, D.G.; Hanke-Gogokhia, C.; Ajami, N.J.; Wong, M.C.; Ghazaryan, A.; Valentine, J.F. Expansion of bacteriophages is linked to aggravated intestinal inflammation and colitis. Cell Host Microbe 2019, 25, 285–299.e8. [Google Scholar] [CrossRef] [PubMed]
- Feng, F.; Tian, Y.; Xu, G.; Liu, Z.; Liu, S.; Zheng, G.; Guo, M.; Lian, X.; Fan, D.; Zhang, H. Diagnostic and prognostic value of CEA, CA19–9, AFP and CA125 for early gastric cancer. BMC Cancer 2017, 17, 737. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, J.; Zhou, Y.; Sheng, S.; Qian, S.Y.; Huo, X. Evaluation of Serum CEA, CA19-9, CA72-4, CA125 and Ferritin as Diagnostic Markers and Factors of Clinical Parameters for Colorectal Cancer. Sci. Rep. 2018, 8, 2732. [Google Scholar] [CrossRef] [PubMed]
- Derosa, L.; Routy, B.; Thomas, A.M.; Iebba, V.; Zalcman, G.; Friard, S.; Mazieres, J.; Audigier-Valette, C.; Moro-Sibilot, D.; Goldwasser, F.; et al. Intestinal Akkermansia muciniphila predicts clinical response to PD-1 blockade in patients with advanced non-small-cell lung cancer. Nat. Med. 2022, 28, 315–324. [Google Scholar] [CrossRef]
- Guo, S.; Li, L.; Xu, B.; Li, M.; Zeng, Q.; Xiao, H.; Xue, Y.; Wu, Y.; Wang, Y.; Liu, W.; et al. A Simple and Novel Fecal Biomarker for Colorectal Cancer: Ratio of Fusobacterium Nucleatum to Probiotics Populations, Based on Their Antagonistic Effect. Clin. Chem. 2018, 64, 1327–1337. [Google Scholar] [CrossRef]
- Brader, P.; Stritzker, J.; Riedl, C.C.; Zanzonico, P.; Cai, S.; Burnazi, E.M.; Ghani, E.R.; Hricak, H.; Szalay, A.A.; Fong, Y.; et al. Escherichia coli Nissle 1917 Facilitates Tumor Detection by Positron Emission Tomography and Optical Imaging. Clin. Cancer Res. 2008, 14, 2295–2302. [Google Scholar] [CrossRef]
- Dai, J.-H.; Tan, X.-R.; Qiao, H.; Liu, N. Emerging clinical relevance of microbiome in cancer: Promising biomarkers and therapeutic targets. Protein Cell 2024, 15, 239–260. [Google Scholar] [CrossRef]
- Steinberg, G.D.; Shore, N.D.; Redorta, J.P.; Galsky, M.D.; Bedke, J.; Ku, J.H.; Kretkowski, M.; Hu, H.; Penkov, K.; Vermette, J.J.; et al. CREST: Phase III study of sasanlimab and Bacillus Calmette-Guérin for patients with Bacillus Calmette-Guérin-naïve high-risk non-muscle-invasive bladder cancer. Future Oncol. 2024, 20, 891–901. [Google Scholar] [CrossRef] [PubMed]
- Luke, J.J.; Piha-Paul, S.A.; Medina, T.; Verschraegen, C.F.; Varterasian, M.; Brennan, A.M.; Riese, R.J.; Sokolovska, A.; Strauss, J.; Hava, D.L.; et al. Phase I Study of SYNB1891, an Engineered E. coli Nissle Strain Expressing STING Agonist, with and without Atezolizumab in Advanced Malignancies. Clin. Cancer Res. 2023, 29, 2435–2444. [Google Scholar] [CrossRef] [PubMed]
- Janku, F.; Zhang, H.H.; Pezeshki, A.; Goel, S.; Murthy, R.; Wang-Gillam, A.; Shepard, D.R.; Helgason, T.; Masters, T.; Hong, D.S.; et al. Intratumoral Injection of Clostridium novyi-NT Spores in Patients with Treatment-refractory Advanced Solid Tumors. Clin. Cancer Res. 2021, 27, 96–106. [Google Scholar] [CrossRef] [PubMed]
| Tumor Types | Intratumoral Bacteria | Mechanism of Tumor Occurrence | References |
|---|---|---|---|
| Colorectal cancer | Fusobacterium nucleatum | Dysregulation of signaling pathways; Inactivation of tumor suppressor genes; Abnormal epigenetic regulation; Immune escape; Polarization of immune cell phenotypes; Genome damage; Chronic inflammation. | [43,61,62,63,64] |
| Escherichia coli | Dysregulation of signaling pathways; Abnormal epigenetic regulation; Immune escape; Polarization of immune cell phenotypes; Genome damage; Chronic inflammation. | [51,65,66,67] | |
| Phylum Proteobacteria | Dysregulation of signaling pathways; Abnormal epigenetic regulation; Immune escape; Genome damage; Chronic inflammation. | [68,69] | |
| Porphyromonas gingivalis | Dysregulation of signaling pathways; Inactivation of tumor suppressor genes; Abnormal epigenetic regulation; Immune escape; Polarization of immune cell phenotypes; Genome damage. | [70] | |
| Prevotella | Dysregulation of signaling pathways; Abnormal epigenetic regulation; Immune escape; Polarization of immune cell phenotypes; Direct DNA damage. | [68] | |
| Peptostreptococcus | Dysregulation of signaling pathways; Abnormal epigenetic regulation; Immune escape; Genome damage. | [68] | |
| Prevotella intermedia | Dysregulation of signaling pathways; Abnormal epigenetic regulation; Immune escape; Polarization of immune cell phenotypes; Genome damage. | [64,71] | |
| Fusobacterium necrophorum | Dysregulation of signaling pathways; Immune escape; Polarization of immune cell phenotypes; Direct DNA damage. | [64] | |
| Breast cancer | Methylobacterium radiotolerans | Dysregulation of signaling pathways; Abnormal epigenetic regulation; Immune escape; Polarization of immune cell phenotypes; Genome damage. | [72] |
| Fusobacterium nucleatum | Dysregulation of signaling pathways; Abnormal epigenetic regulation; Immune escape; Polarization of immune cell phenotypes; Genome damage. | [73] | |
| Bacteroides fragilis | Dysregulation of signaling pathways; Abnormal epigenetic regulation; Immune escape; Polarization of immune cell phenotypes; Direct DNA damage. | [74] | |
| Escherichia coli | Dysregulation of signaling pathways; Abnormal epigenetic regulation; Immune escape; Genome damage. | [10] | |
| Staphylococcus epidermidis | Dysregulation of signaling pathways; Abnormal epigenetic regulation; Immune escape; Direct DNA damage. | [10] | |
| Pancreatic cancer | Fusobacterium nucleatum | Dysregulation of signaling pathways; Chronic inflammation; Immune escape. | [75] |
| Porphyromonas gingivalis | Dysregulation of signaling pathways; Immune escape; Polarization of immune cell phenotypes. | [76] | |
| Pseudomonadaceae | Direct DNA damage; Chronic inflammation. | [60] | |
| Enterobacteriaceae | Immunosuppressive cell infiltration; Direct DNA damage. | [60] | |
| Proteobacteria | Immunosuppression; Chronic inflammation; Genome damage. | [60] | |
| Helicobacter pylori | Dysregulation of signaling pathways; Genome damage; Chronic inflammation. | [77] | |
| Citrobacter freundii | Dysregulation of signaling pathways; Chronic inflammation. | [78] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, J.; Zhang, A.; Sun, J.; Fu, Y.; Li, W.; Wang, Y. Bacteria-Based Roles in Solid Tumors: Potential for Prevention and Treatment. Pathogens 2025, 14, 874. https://doi.org/10.3390/pathogens14090874
Huang J, Zhang A, Sun J, Fu Y, Li W, Wang Y. Bacteria-Based Roles in Solid Tumors: Potential for Prevention and Treatment. Pathogens. 2025; 14(9):874. https://doi.org/10.3390/pathogens14090874
Chicago/Turabian StyleHuang, Jianchang, Ailin Zhang, Jialin Sun, Yuhan Fu, Weinan Li, and Yanhong Wang. 2025. "Bacteria-Based Roles in Solid Tumors: Potential for Prevention and Treatment" Pathogens 14, no. 9: 874. https://doi.org/10.3390/pathogens14090874
APA StyleHuang, J., Zhang, A., Sun, J., Fu, Y., Li, W., & Wang, Y. (2025). Bacteria-Based Roles in Solid Tumors: Potential for Prevention and Treatment. Pathogens, 14(9), 874. https://doi.org/10.3390/pathogens14090874






