Characterization of the Diversity in Host Range of an Extensively Drug-Resistant (XDR) Type IV Secretion System-Encoding Plasmid in Acinetobacter
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Conjugation Assays
2.3. Characterization of Transconjugants
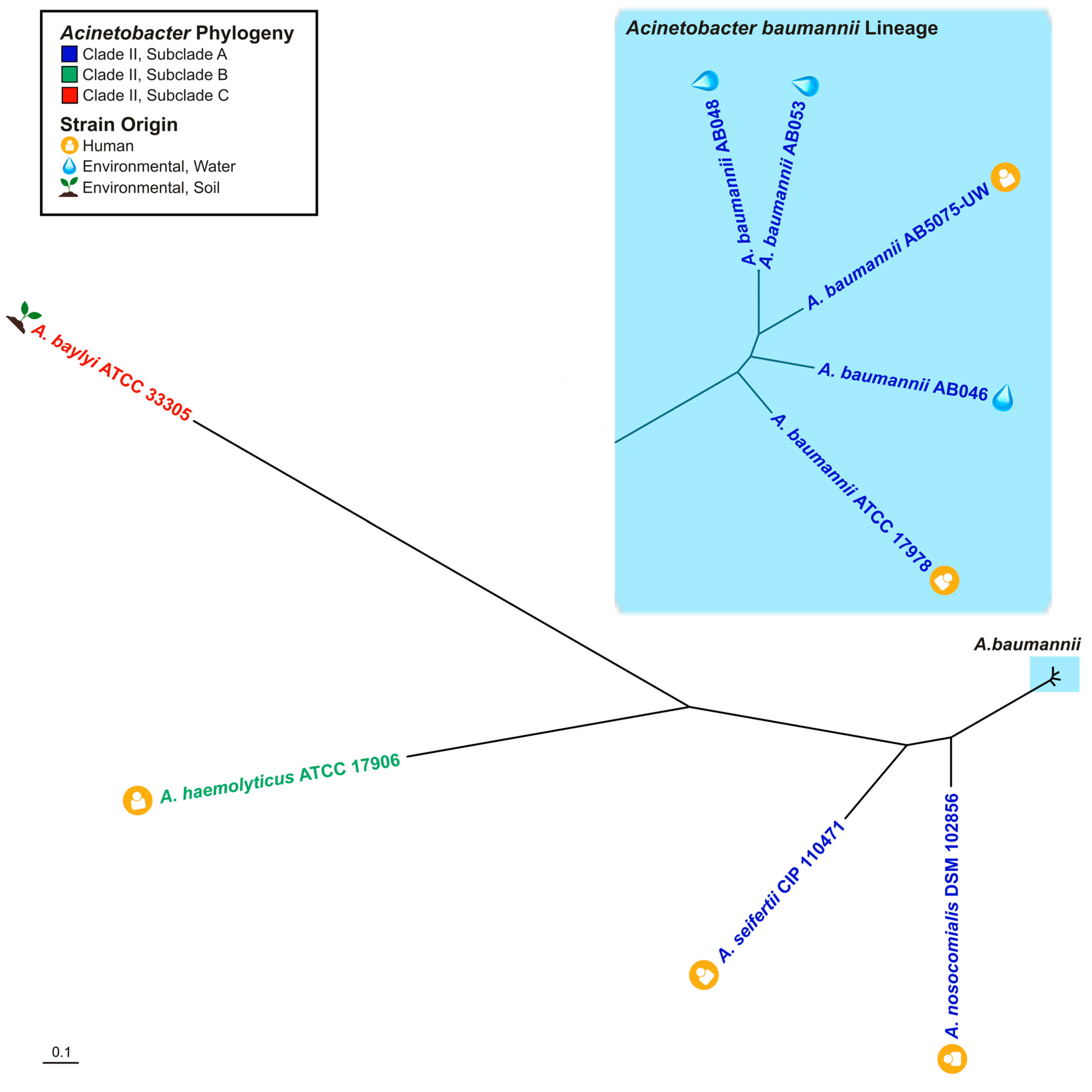
2.4. Phylogenetic Analyses
2.5. Minimum Inhibitory Concentration (MIC) Assays
2.6. Antibiotic Inactivation Assays
3. Results
3.1. Conjugative Transfer of T4SS-Encoding Plasmid to Environmental A. baumannii Strains
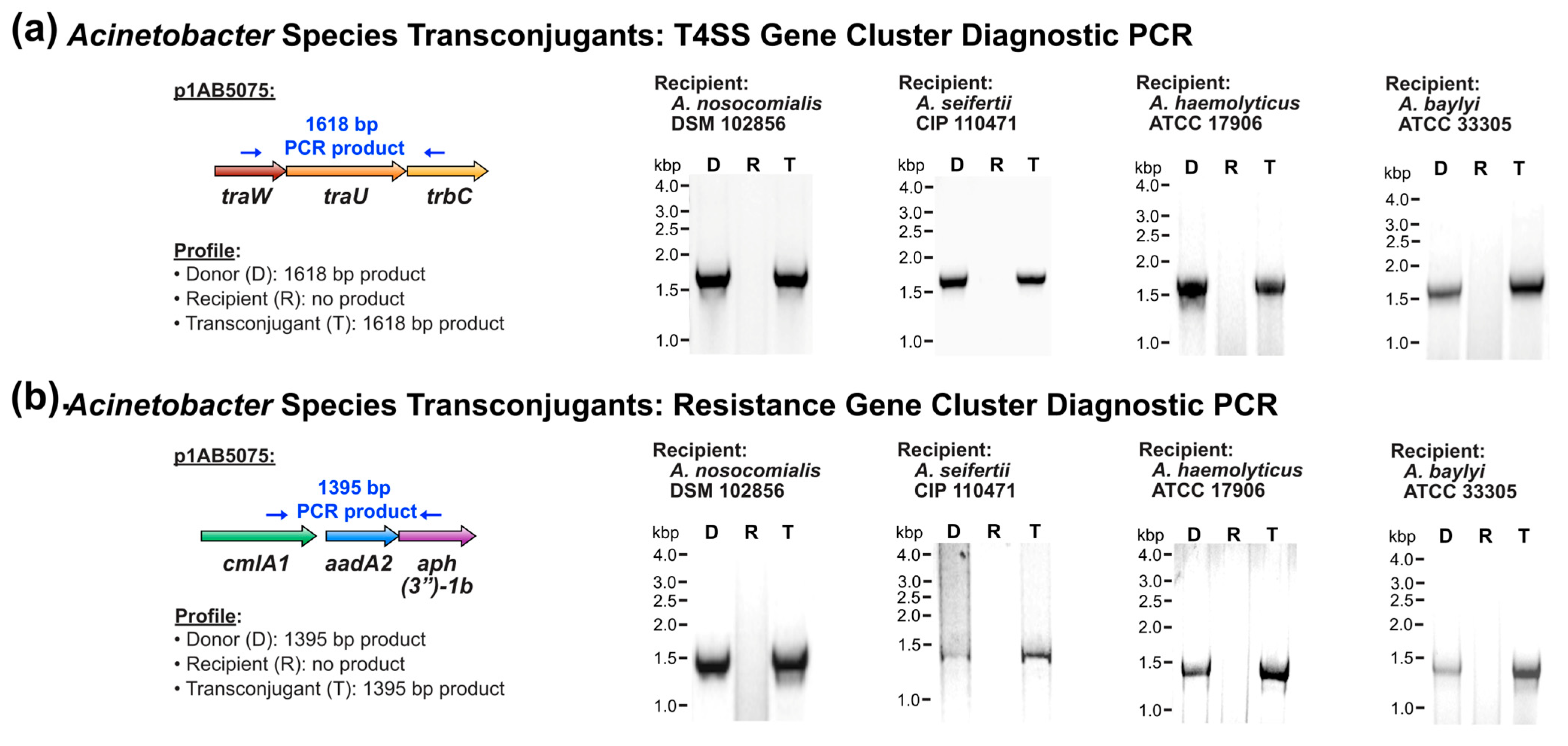
3.2. Conjugative Transfer of T4SS-Encoding Plasmid to More Genetically Divergent Acinetobacter
3.3. Antibiotic Susceptibility of Transconjugant Acinetobacter Strains
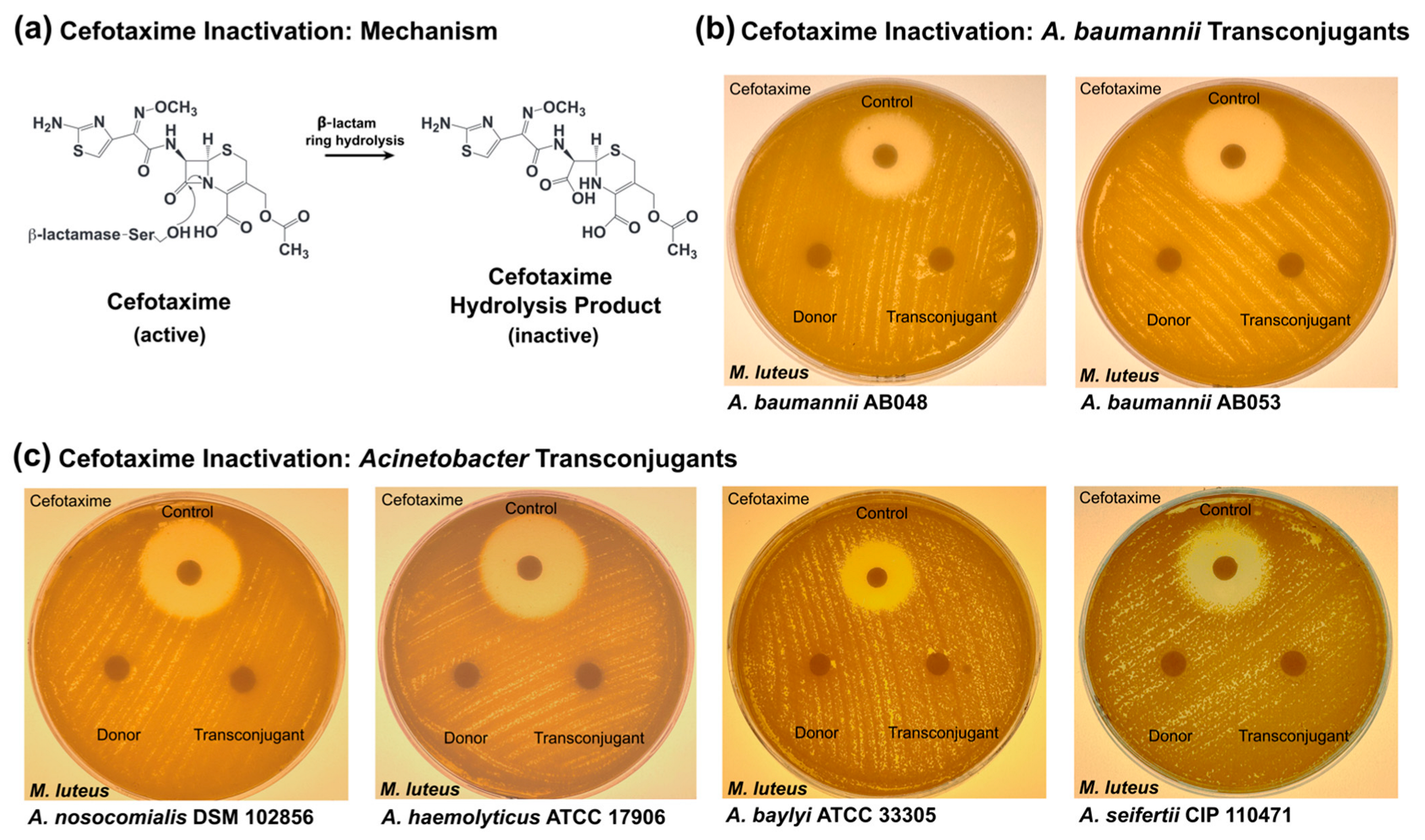
3.4. Cefotaxime Inactivation by Transconjugant Acinetobacter Strains
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics; WHO: Geneva, Switzerland, 2017.
- Dijkshoorn, L.; Nemec, A.; Seifert, H. An increasing threat in hospitals: Multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 2007, 5, 939–951. [Google Scholar] [CrossRef] [PubMed]
- Dexter, C.; Murray, G.L.; Paulsen, I.T.; Peleg, A.Y. Community-acquired Acinetobacter baumannii: Clinical characteristics, epidemiology and pathogenesis. Expert Rev. Anti-Infect. Ther. 2015, 13, 567–573. [Google Scholar] [CrossRef] [PubMed]
- Howard, A.; O’Donoghue, M.; Feeney, A.; Sleator, R.D. Acinetobacter baumannii: An emerging opportunistic pathogen. Virulence 2012, 3, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Ayobami, O.; Willrich, N.; Harder, T.; Okeke, I.N.; Eckmanns, T.; Markwart, R. The incidence and prevalence of hospital-acquired (carbapenem-resistant) Acinetobacter baumannii in Europe, Eastern Mediterranean and Africa: A systematic review and meta-analysis. Emerg. Microbes Infect. 2019, 8, 1747–1759. [Google Scholar] [CrossRef]
- Luyt, C.E.; Hekimian, G.; Koulenti, D.; Chastre, J. Microbial cause of ICU-acquired pneumonia: Hospital-acquired pneumonia versus ventilator-associated pneumonia. Curr. Opin. Crit. Care 2018, 24, 332–338. [Google Scholar] [CrossRef]
- Inchai, J.; Pothirat, C.; Liwsrisakun, C.; Deesomchok, A.; Kositsakulchai, W.; Chalermpanchai, N. Ventilator-associated pneumonia: Epidemiology and prognostic indicators of 30-day mortality. Jpn. J. Infect. Dis. 2015, 68, 181–186. [Google Scholar] [CrossRef]
- Lin, M.F.; Lan, C.Y. Antimicrobial resistance in Acinetobacter baumannii: From bench to bedside. World J. Clin. Cases 2014, 2, 787–814. [Google Scholar] [CrossRef]
- Stietz, M.S.; Ramirez, M.S.; Vilacoba, E.; Merkier, A.K.; Limansky, A.S.; Centron, D.; Catalano, M. Acinetobacter baumannii extensively drug resistant lineages in Buenos Aires hospitals differ from the international clones I-III. Infect. Genet. Evol. 2013, 14, 294–301. [Google Scholar] [CrossRef]
- Valcek, A.; Nesporova, K.; Whiteway, C.; De Pooter, T.; De Coster, W.; Strazisar, M.; Van der Henst, C. Genomic Analysis of a Strain Collection Containing Multidrug-, Extensively Drug-, Pandrug-, and Carbapenem-Resistant Modern Clinical Isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2022, 66, e0089222. [Google Scholar] [CrossRef]
- Kanaan, M.H.G.; Khashan, H.T. Molecular typing, virulence traits and risk factors of pandrug-resistant Acinetobacter baumannii spread in intensive care unit centers of Baghdad city, Iraq. Rev. Res. Med. Microbiol. 2022, 33, 51–55. [Google Scholar] [CrossRef]
- Villalon, P.; Ortega, M.; Saez-Nieto, J.A.; Carrasco, G.; Medina-Pascual, M.J.; Garrido, N.; Valdezate, S. Dynamics of a Sporadic Nosocomial Acinetobacter calcoaceticus—Acinetobacter baumannii Complex Population. Front. Microbiol. 2019, 10, 593. [Google Scholar] [CrossRef]
- Nemec, A.; Krizova, L.; Maixnerova, M.; Sedo, O.; Brisse, S.; Higgins, P.G. Acinetobacter seifertii sp. nov., a member of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex isolated from human clinical specimens. Int. J. Syst. Evol. Microbiol. 2015, 65, 934–942. [Google Scholar] [CrossRef] [PubMed]
- Dahal, U.; Paul, K.; Gupta, S. The multifaceted genus Acinetobacter: From infection to bioremediation. J. Appl. Microbiol. 2023, 134, lxad145. [Google Scholar] [CrossRef] [PubMed]
- Adewoyin, M.A.; Okoh, A.I. The natural environment as a reservoir of pathogenic and non-pathogenic Acinetobacter species. Rev. Environ. Health 2018, 33, 265–272. [Google Scholar] [CrossRef]
- Castillo-Ramirez, S. The importance of Acinetobacter baumannii from non-human sources. Lancet Microbe 2023, 4, e761–e762. [Google Scholar] [CrossRef]
- Rafei, R.; Hamze, M.; Pailhories, H.; Eveillard, M.; Marsollier, L.; Joly-Guillou, M.L.; Dabboussi, F.; Kempf, M. Extrahuman epidemiology of Acinetobacter baumannii in Lebanon. Appl. Environ. Microbiol. 2015, 81, 2359–2367. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, A.; Hanke, D.; Luschow, D.; Schwarz, S.; Higgins, P.G.; Fessler, A.T. Acinetobacter baumannii from Samples of Commercially Reared Turkeys: Genomic Relationships, Antimicrobial and Biocide Susceptibility. Microorganisms 2023, 11, 759. [Google Scholar] [CrossRef]
- Mateo-Estrada, V.; Tyrrell, C.; Evans, B.A.; Aguilar-Vera, A.; Drissner, D.; Castillo-Ramirez, S.; Walsh, F. Acinetobacter baumannii from grass: Novel but non-resistant clones. Microb. Genom. 2023, 9, mgen001054. [Google Scholar] [CrossRef]
- Al Atrouni, A.; Joly-Guillou, M.L.; Hamze, M.; Kempf, M. Reservoirs of Non-baumannii Acinetobacter Species. Front. Microbiol. 2016, 7, 49. [Google Scholar] [CrossRef]
- Castro-Jaimes, S.; Bello-Lopez, E.; Velazquez-Acosta, C.; Volkow-Fernandez, P.; Lozano-Zarain, P.; Castillo-Ramirez, S.; Cevallos, M.A. Chromosome Architecture and Gene Content of the Emergent Pathogen Acinetobacter haemolyticus. Front. Microbiol. 2020, 11, 926. [Google Scholar] [CrossRef]
- Jing, L.; Xu, Z.; Zhang, Y.; Li, D.; Song, Y.; Hu, H.; Fang, Y.; Zhu, W. Metagenomic Insights into Pathogenic Characterization of ST410 Acinetobacter nosocomialis Prevalent in China. Pathogens 2022, 11, 838. [Google Scholar] [CrossRef]
- Mittal, S.; Sharma, M.; Yadav, A.; Bala, K.; Chaudhary, U. Acinetobacter lwoffii an emerging pathogen in neonatal ICU. Infect. Disord. Drug Targets 2015, 15, 184–188. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Gaitan, M.P.; Montoya-Moncada, A.D.; Suescun-Vargas, J.M.; Pinzon-Salamanca, J.Y.; Aguirre-Borrero, B.L. Emerging species in pediatrics: A case of Acinetobacter johnsonii meningitis. Bol. Med. Hosp. Infant. Mex. 2022, 79, 51–55. [Google Scholar] [CrossRef]
- Yaikhan, T.; Chukamnerd, A.; Singkhamanan, K.; Nokchan, N.; Chintakovid, N.; Chusri, S.; Pomwised, R.; Wonglapsuwan, M.; Surachat, K. Genomic Characterization of Mobile Genetic Elements Associated with Multidrug-Resistant Acinetobacter Non-baumannii Species from Southern Thailand. Antibiotics 2024, 13, 149. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Leclercq, S.O.; Tian, J.; Wang, C.; Yahara, K.; Ai, G.; Liu, S.; Feng, J. A new subclass of intrinsic aminoglycoside nucleotidyltransferases, ANT(3″)-II, is horizontally transferred among Acinetobacter spp. by homologous recombination. PLoS Genet. 2017, 13, e1006602. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.Y.; Chen, C.; Liu, B.T.; He, Q.; Wu, X.T.; Sun, R.Y.; Zhang, Y.; Cui, Z.H.; Guo, W.Y.; Jia, Q.L.; et al. Co-occurrence of Plasmid-Mediated Tigecycline and Carbapenem Resistance in Acinetobacter spp. from Waterfowls and Their Neighboring Environment. Antimicrob. Agents Chemother. 2020, 64, e02502-19. [Google Scholar] [CrossRef]
- He, T.; Li, R.; Wei, R.; Liu, D.; Bai, L.; Zhang, L.; Gu, J.; Wang, R.; Wang, Y. Characterization of Acinetobacter indicus co-harbouring tet(X3) and blaNDM-1 of dairy cow origin. J. Antimicrob. Chemother. 2020, 75, 2693–2696. [Google Scholar] [CrossRef]
- Furlan, J.P.R.; de Almeida, O.G.G.; De Martinis, E.C.P.; Stehling, E.G. Characterization of an Environmental Multidrug-Resistant Acinetobacter seifertii and Comparative Genomic Analysis Reveals Co-occurrence of Antimicrobial Resistance and Metal Tolerance Determinants. Front. Microbiol. 2019, 10, 2151. [Google Scholar] [CrossRef]
- Matsui, M.; Suzuki, S.; Yamane, K.; Suzuki, M.; Konda, T.; Arakawa, Y.; Shibayama, K. Distribution of carbapenem resistance determinants among epidemic and non-epidemic types of Acinetobacter species in Japan. J. Med. Microbiol. 2014, 63, 870–877. [Google Scholar] [CrossRef]
- Park, K.H.; Shin, J.H.; Lee, S.Y.; Kim, S.H.; Jang, M.O.; Kang, S.J.; Jung, S.I.; Chung, E.K.; Ko, K.S.; Jang, H.C. The clinical characteristics, carbapenem resistance, and outcome of Acinetobacter bacteremia according to genospecies. PLoS ONE 2014, 8, e65026. [Google Scholar] [CrossRef]
- Sheck, E.; Romanov, A.; Shapovalova, V.; Shaidullina, E.; Martinovich, A.; Ivanchik, N.; Mikotina, A.; Skleenova, E.; Oloviannikov, V.; Azizov, I.; et al. Acinetobacter Non-baumannii Species: Occurrence in Infections in Hospitalized Patients, Identification, and Antibiotic Resistance. Antibiotics 2023, 12, 1301. [Google Scholar] [CrossRef]
- Salimizand, H.; Zomorodi, A.R.; Mansury, D.; Khakshoor, M.; Azizi, O.; Khodaparast, S.; Baseri, Z.; Karami, P.; Zamanlou, S.; Farsiani, H.; et al. Diversity of aminoglycoside modifying enzymes and 16S rRNA methylases in Acinetobacter baumannii and Acinetobacter nosocomialis species in Iran; wide distribution of aadA1 and armA. Infect. Genet. Evol. 2018, 66, 195–199. [Google Scholar] [CrossRef]
- World Health Organization. A One Health Priority Research Agenda for Antimicrobial Resistance; WHO: Geneva, Switzerland, 2023.
- Virolle, C.; Goldlust, K.; Djermoun, S.; Bigot, S.; Lesterlin, C. Plasmid Transfer by Conjugation in Gram-Negative Bacteria: From the Cellular to the Community Level. Genes 2020, 11, 1239. [Google Scholar] [CrossRef]
- Huang, T.W.; Lauderdale, T.L.; Liao, T.L.; Hsu, M.C.; Chang, F.Y.; Chang, S.C.; Khong, W.X.; Ng, O.T.; Chen, Y.T.; Kuo, S.C.; et al. Effective transfer of a 47 kb NDM-1-positive plasmid among Acinetobacter species. J. Antimicrob. Chemother. 2015, 70, 2734–2738. [Google Scholar] [CrossRef]
- Nasser, F.; Gaudreau, A.; Lubega, S.; Zaker, A.; Xia, X.; Mer, A.S.; D’Costa, V.M. Characterization of the diversity of type IV secretion system-encoding plasmids in Acinetobacter. Emerg. Microbes. Infect. 2024, 13, 2320929. [Google Scholar] [CrossRef]
- Tian, C.; Song, J.; Ren, L.; Huang, D.; Wang, S.; Fu, L.; Zhao, Y.; Bai, Y.; Fan, X.; Ma, T.; et al. Complete genetic characterization of carbapenem-resistant Acinetobacter johnsonii, co-producing NDM-1, OXA-58, and PER-1 in a patient source. Front. Cell Infect. Microbiol. 2023, 13, 1227063. [Google Scholar] [CrossRef]
- Espinal, P.; Mosqueda, N.; Telli, M.; van der Reijden, T.; Rolo, D.; Fernandez-Orth, D.; Dijkshoorn, L.; Roca, I.; Vila, J. Identification of NDM-1 in a Putatively Novel Acinetobacter Species (“NB14”) Closely Related to Acinetobacter pittii. Antimicrob. Agents Chemother. 2015, 59, 6657–6660. [Google Scholar] [CrossRef]
- Zou, D.; Huang, Y.; Zhao, X.; Liu, W.; Dong, D.; Li, H.; Wang, X.; Huang, S.; Wei, X.; Yan, X.; et al. A novel New Delhi metallo-beta-lactamase variant, NDM-14, isolated in a Chinese Hospital possesses increased enzymatic activity against carbapenems. Antimicrob. Agents Chemother. 2015, 59, 2450–2453. [Google Scholar] [CrossRef]
- Jones, L.S.; Carvalho, M.J.; Toleman, M.A.; White, P.L.; Connor, T.R.; Mushtaq, A.; Weeks, J.L.; Kumarasamy, K.K.; Raven, K.E.; Torok, M.E.; et al. Characterization of plasmids in extensively drug-resistant Acinetobacter strains isolated in India and Pakistan. Antimicrob. Agents Chemother. 2015, 59, 923–929. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Hu, Y.; Pan, Y.; Liang, H.; Wang, H.; Wang, X.; Hao, Q.; Yang, X.; Yang, X.; Xiao, X.; et al. Novel plasmid and its variant harboring both a bla(NDM-1) gene and type IV secretion system in clinical isolates of Acinetobacter lwoffii. Antimicrob. Agents Chemother. 2012, 56, 1698–1702. [Google Scholar] [CrossRef] [PubMed]
- Oinuma, K.I.; Suzuki, M.; Sakiyama, A.; Tsubouchi, T.; Saeki, K.; Sato, K.; Niki, M.; Yamada, K.; Shibayama, K.; Kakeya, H.; et al. Genomic characterization of triple-carbapenemase-producing Acinetobacter baumannii. JAC Antimicrob. Resist. 2021, 3, dlab191. [Google Scholar] [CrossRef]
- Zhang, W.J.; Lu, Z.; Schwarz, S.; Zhang, R.M.; Wang, X.M.; Si, W.; Yu, S.; Chen, L.; Liu, S. Complete sequence of the bla(NDM-1)-carrying plasmid pNDM-AB from Acinetobacter baumannii of food animal origin. J. Antimicrob. Chemother. 2013, 68, 1681–1682. [Google Scholar] [CrossRef]
- Li, P.; Zhang, S.; Wang, J.; Al-Shamiri, M.M.; Han, B.; Chen, Y.; Han, S.; Han, L. Uncovering the Secretion Systems of Acinetobacter baumannii: Structures and Functions in Pathogenicity and Antibiotic Resistance. Antibiotics 2023, 12, 195. [Google Scholar] [CrossRef]
- Liu, C.C.; Kuo, H.Y.; Tang, C.Y.; Chang, K.C.; Liou, M.L. Prevalence and mapping of a plasmid encoding a type IV secretion system in Acinetobacter baumannii. Genomics 2014, 104, 215–223. [Google Scholar] [CrossRef]
- Iacono, M.; Villa, L.; Fortini, D.; Bordoni, R.; Imperi, F.; Bonnal, R.J.; Sicheritz-Ponten, T.; De Bellis, G.; Visca, P.; Cassone, A.; et al. Whole-genome pyrosequencing of an epidemic multidrug-resistant Acinetobacter baumannii strain belonging to the European clone II group. Antimicrob. Agents Chemother. 2008, 52, 2616–2625. [Google Scholar] [CrossRef]
- Salgado-Camargo, A.D.; Castro-Jaimes, S.; Gutierrez-Rios, R.M.; Lozano, L.F.; Altamirano-Pacheco, L.; Silva-Sanchez, J.; Perez-Oseguera, A.; Volkow, P.; Castillo-Ramirez, S.; Cevallos, M.A. Structure and Evolution of Acinetobacter baumannii Plasmids. Front. Microbiol. 2020, 11, 1283. [Google Scholar] [CrossRef]
- Gallagher, L.A.; Ramage, E.; Weiss, E.J.; Radey, M.; Hayden, H.S.; Held, K.G.; Huse, H.K.; Zurawski, D.V.; Brittnacher, M.J.; Manoil, C. Resources for Genetic and Genomic Analysis of Emerging Pathogen Acinetobacter baumannii. J. Bacteriol. 2015, 197, 2027–2035. [Google Scholar] [CrossRef]
- Jacobs, A.C.; Thompson, M.G.; Black, C.C.; Kessler, J.L.; Clark, L.P.; McQueary, C.N.; Gancz, H.Y.; Corey, B.W.; Moon, J.K.; Si, Y.; et al. AB5075, a Highly Virulent Isolate of Acinetobacter baumannii, as a Model Strain for the Evaluation of Pathogenesis and Antimicrobial Treatments. mBio 2014, 5, e01076-14. [Google Scholar] [CrossRef]
- Kalivoda, E.J.; Horzempa, J.; Stella, N.A.; Sadaf, A.; Kowalski, R.P.; Nau, G.J.; Shanks, R.M. New vector tools with a hygromycin resistance marker for use with opportunistic pathogens. Mol. Biotechnol. 2011, 48, 7–14. [Google Scholar] [CrossRef][Green Version]
- Fernando, D.M.; Khan, I.U.; Patidar, R.; Lapen, D.R.; Talbot, G.; Topp, E.; Kumar, A. Isolation and Characterization of Acinetobacter baumannii Recovered from Campylobacter Selective Medium. Front. Microbiol. 2016, 7, 1871. [Google Scholar] [CrossRef]
- Di Venanzio, G.; Moon, K.H.; Weber, B.S.; Lopez, J.; Ly, P.M.; Potter, R.F.; Dantas, G.; Feldman, M.F. Multidrug-resistant plasmids repress chromosomally encoded T6SS to enable their dissemination. Proc. Natl. Acad. Sci. USA 2019, 116, 1378–1383. [Google Scholar] [CrossRef]
- Weber, B.S.; Ly, P.M.; Irwin, J.N.; Pukatzki, S.; Feldman, M.F. A multidrug resistance plasmid contains the molecular switch for type VI secretion in Acinetobacter baumannii. Proc. Natl. Acad. Sci. USA 2015, 112, 9442–9447. [Google Scholar] [CrossRef]
- Salto, I.P.; Torres Tejerizo, G.; Wibberg, D.; Puhler, A.; Schluter, A.; Pistorio, M. Comparative genomic analysis of Acinetobacter spp. plasmids originating from clinical settings and environmental habitats. Sci. Rep. 2018, 8, 7783. [Google Scholar] [CrossRef]
- Diancourt, L.; Passet, V.; Nemec, A.; Dijkshoorn, L.; Brisse, S. The population structure of Acinetobacter baumannii: Expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS ONE 2010, 5, e10034. [Google Scholar] [CrossRef]
- Heuer, H.; Krsek, M.; Baker, P.; Smalla, K.; Wellington, E.M. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 1997, 63, 3233–3241. [Google Scholar] [CrossRef]
- Hao, W.; Golding, G.B. Uncovering rate variation of lateral gene transfer during bacterial genome evolution. BMC Genom. 2008, 9, 235. [Google Scholar] [CrossRef][Green Version]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- CSLI. M100 Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- D’Costa, V.M.; McGrann, K.M.; Hughes, D.W.; Wright, G.D. Sampling the antibiotic resistome. Science 2006, 311, 374–377. [Google Scholar] [CrossRef]
- Almeida, O.G.G.; Furlan, J.P.R.; Stehling, E.G.; De Martinis, E.C.P. Comparative phylo-pangenomics reveals generalist lifestyles in representative Acinetobacter species and proposes candidate gene markers for species identification. Gene 2021, 791, 145707. [Google Scholar] [CrossRef]
- Alattraqchi, A.G.; Mohd Rani, F.; Ni, A.R.; Ismail, S.; Cleary, D.W.; Clarke, S.C.; Yeo, C.C. Complete Genome Sequencing of Acinetobacter baumannii AC1633 and Acinetobacter nosocomialis AC1530 Unveils a Large Multidrug-Resistant Plasmid Encoding the NDM-1 and OXA-58 Carbapenemases. mSphere 2021, 6, e01076-20. [Google Scholar] [CrossRef]
- Zhou, S.; Chen, X.; Meng, X.; Zhang, G.; Wang, J.; Zhou, D.; Guo, X. “Roar” of blaNDM-1 and “silence” of blaOXA-58 co-exist in Acinetobacter pittii. Sci. Rep. 2015, 5, 8976. [Google Scholar] [CrossRef]
- Yusuf, E.; Tompa, M.; Strepis, N.; Klaassen, C.H.W.; Goessens, W.H.F. High Prevalence of ST502 Carrying an OXA-24 Carbapenemase gene in Carbapenem-Nonsusceptible Acinetobacter baumannii-calcoaceticus Isolates in Romania. Microb. Drug Resist. 2022, 28, 636–644. [Google Scholar] [CrossRef]
- Cox, G.; Stogios, P.J.; Savchenko, A.; Wright, G.D. Structural and molecular basis for resistance to aminoglycoside antibiotics by the adenylyltransferase ANT(2″)-Ia. mBio 2015, 6, e02180-14. [Google Scholar] [CrossRef]
- Page, M.I. The reactivity of beta-lactams, the mechanism of catalysis and the inhibition of beta-lactamases. Curr. Pharm. Des. 1999, 5, 895–913. [Google Scholar] [CrossRef]
- Singh, B.; Agri, H.; Karthikeyan, R.; Jayakumar, V. Common Bacterial Causes of Septicaemia in Ani-mals and Birds Detected in Heart Blood Samples of Refer-red Cases of Mortality in Northern India. J. Clin. Med. Images 2023, 7, 1–14. [Google Scholar]
- Maboni, G.; Seguel, M.; Lorton, A.; Sanchez, S. Antimicrobial resistance patterns of Acinetobacter spp. of animal origin reveal high rate of multidrug resistance. Vet. Microbiol. 2020, 245, 108702. [Google Scholar] [CrossRef]
- Singh, B.R.; Karthikeyan, R.; Sinha, D.K.; Vinodhkumar, O.; Jaykumar, V.; Yadav, A.; Agri, H. Potentially Pathogenic Bacteria in Water Bodies and Drinking Water Supplies in and Around Bareilly, India. Acta Sci. Microbiol. 2022, 5, 113–126. [Google Scholar] [CrossRef]
- Alderliesten, J.B.; Duxbury, S.J.N.; Zwart, M.P.; de Visser, J.; Stegeman, A.; Fischer, E.A.J. Effect of donor-recipient relatedness on the plasmid conjugation frequency: A meta-analysis. BMC Microbiol. 2020, 20, 135. [Google Scholar] [CrossRef]
- Shintani, M.; Matsui, K.; Takemura, T.; Yamane, H.; Nojiri, H. Behavior of the IncP-7 carbazole-degradative plasmid pCAR1 in artificial environmental samples. Appl. Microbiol. Biotechnol. 2008, 80, 485–497. [Google Scholar] [CrossRef]
- Wang, Y.; Kou, S.; Jiang, Q.; Xu, B.; Liu, X.; Xiao, J.; Tian, Y.; Zhou, C.; Zhang, C.; Xiao, M. Factors affecting transfer of degradative plasmids between bacteria in soils. Appl. Soil Ecol. 2014, 84, 254–261. [Google Scholar] [CrossRef]
- Seoane, J.; Yankelevich, T.; Dechesne, A.; Merkey, B.; Sternberg, C.; Smets, B.F. An individual-based approach to explain plasmid invasion in bacterial populations. FEMS Microbiol. Ecol. 2011, 75, 17–27. [Google Scholar] [CrossRef]
- Gao, C.; Jin, X.; Ren, J.; Fang, H.; Yu, Y. Bioaugmentation of DDT-contaminated soil by dissemination of the catabolic plasmid pDOD. J. Environ. Sci. 2015, 27, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wang, B.; Cao, Z.; Yu, Y. Plasmid-mediated bioaugmentation for the degradation of chlorpyrifos in soil. J. Hazard Mater. 2012, 221–222, 178–184. [Google Scholar] [CrossRef] [PubMed]
- DiGiovanni, G.D.; Neilson, J.W.; Pepper, I.L.; Sinclair, N.A. Gene transfer of Alcaligenes eutrophus JMP134 plasmid pJP4 to indigenous soil recipients. Appl. Environ. Microbiol. 1996, 62, 2521–2526. [Google Scholar] [CrossRef]
- Ikuma, K.; Gunsch, C.K. Genetic bioaugmentation as an effective method for in situ bioremediation: Functionality of catabolic plasmids following conjugal transfers. Bioengineered 2012, 3, 236–241. [Google Scholar] [CrossRef]
- Krol, J.E.; Nguyen, H.D.; Rogers, L.M.; Beyenal, H.; Krone, S.M.; Top, E.M. Increased transfer of a multidrug resistance plasmid in Escherichia coli biofilms at the air-liquid interface. Appl. Environ. Microbiol. 2011, 77, 5079–5088. [Google Scholar] [CrossRef]
- Pinilla-Redondo, R.; Riber, L.; Sorensen, S.J. Fluorescence Recovery Allows the Implementation of a Fluorescence Reporter Gene Platform Applicable for the Detection and Quantification of Horizontal Gene Transfer in Anoxic Environments. Appl. Environ. Microbiol. 2018, 84, e02507-17. [Google Scholar] [CrossRef]
- Skolimowski, M.; Nielsen, M.W.; Emneus, J.; Molin, S.; Taboryski, R.; Sternberg, C.; Dufva, M.; Geschke, O. Microfluidic dissolved oxygen gradient generator biochip as a useful tool in bacterial biofilm studies. Lab Chip 2010, 10, 2162–2169. [Google Scholar] [CrossRef]
- Guiney, D.G.; Hasegawa, P.; Davis, C.E. Plasmid transfer from Escherichia coli to Bacteroides fragilis: Differential expression of antibiotic resistance phenotypes. Proc. Natl. Acad. Sci. USA 1984, 81, 7203–7206. [Google Scholar] [CrossRef]
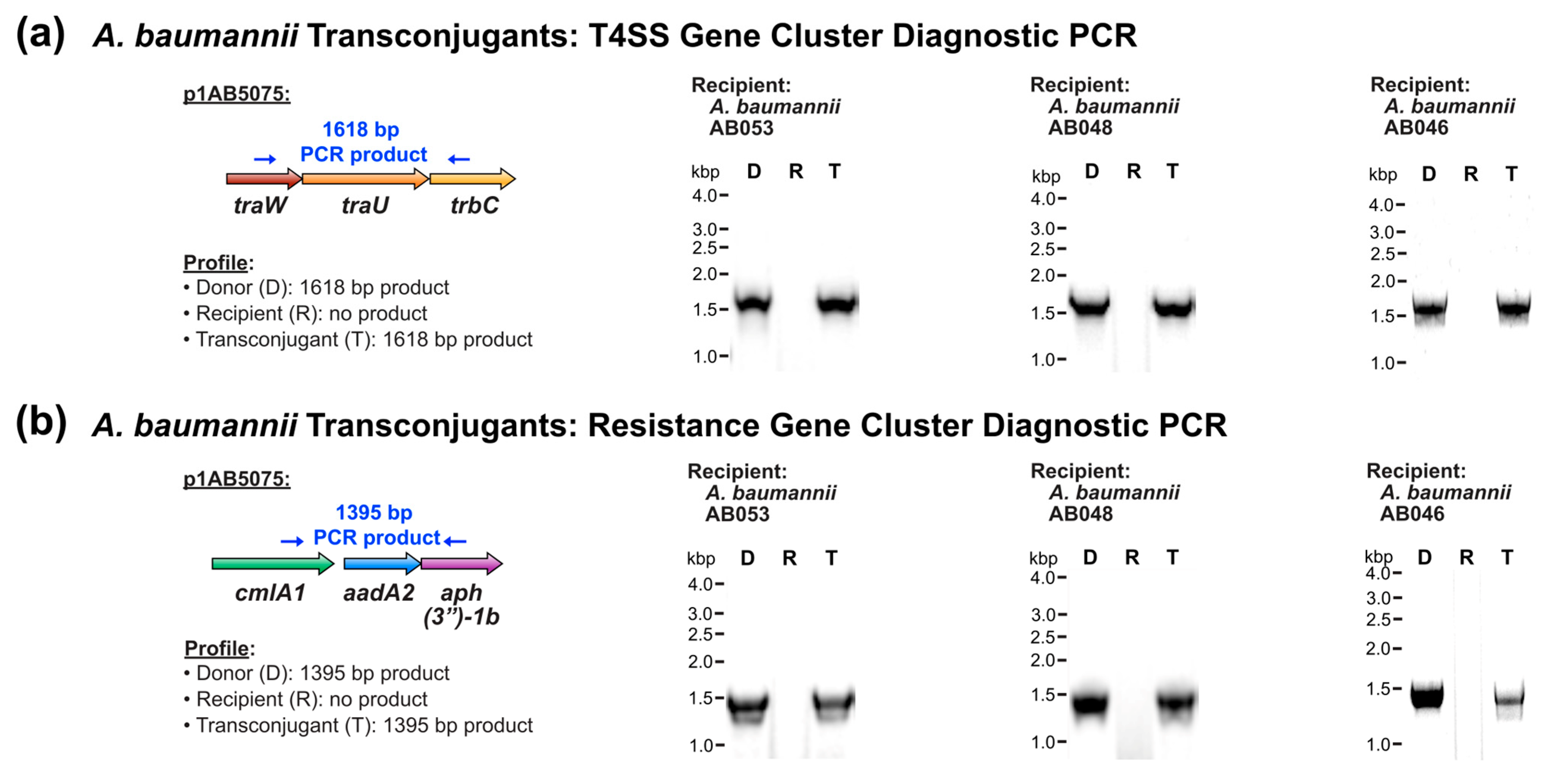

| Recipient Strain | MLST Sequence Type 1 | Strain Origin 2 | Plasmid Transfer | Conjugation Frequency 3 |
|---|---|---|---|---|
| A. baumannii AB053 | 1039 | E, stream | Positive | L 5 |
| A. baumannii AB052 | unknown 4 | E, stream | Negative | N/A 6 |
| A. baumannii AB048 | 1039 | E, river | Positive | L 5 |
| A. baumannii AB047 | 2251 | E, river | Negative | N/A 6 |
| A. baumannii AB046 | 2250 | E, river | Positive | 5.8 × 10−5 |
| Recipient Strain | Phylogenetic Designation (Clade, Subclade (If Applicable)) 1 | Strain Origin 2 | Plasmid Transfer | Conjugation Frequency 3 |
|---|---|---|---|---|
| A. nosocomialis DSM 102856 | II, A | C, human | Positive | 5.6 × 10−2 |
| A. seifertii CIP 110471 | II, A | C, human | Positive | L 4 |
| A. haemolyticus ATCC 17906 | II, B | C, human | Positive | 4.7 × 10−3 |
| A. junii ATCC 17908 | II, B | C, human | Negative | N/A 5 |
| A. baylyi ATCC 33305 | II, C | E, soil | Positive | 3.6 × 10−9 |
| Strain 1 | MIC (μg/mL) | |||
|---|---|---|---|---|
| Kanamycin | Tobramycin | Cefotaxime | Ciprofloxacin | |
| A. baumannii AB5075-UW | >256 | 64 | >256 | 64 |
| A. baumannii AB053-R | 2 | ≤1 | 16 | ≤1 |
| A. baumannii AB053-T | >256 | 16 | >256 | ≤1 |
| A. baumannii AB048-R | 2 | ≤1 | 16 | ≤1 |
| A. baumannii AB048-T | >256 | 8 | >256 | ≤1 |
| A. baumannii AB046-R | 2 | ≤1 | 8 | ≤1 |
| A. baumannii AB046-T | >256 | ≤1 | 8 | ≤1 |
| Strain | MIC (μg/mL) | |||
|---|---|---|---|---|
| Kanamycin | Tobramycin | Cefotaxime | Ciprofloxacin | |
| A. baumannii AB5075-UW | >256 | 64 | >256 | 64 |
| A. nosocomialis DSM 102856-R | 8 | 2 | 16 | ≤1 |
| A. nosocomialis DSM 102856-T | >256 | 32 | >256 | ≤1 |
| A. seifertii CIP 110471-R | 2 | ≤1 | 8 | ≤1 |
| A. seifertii CIP 110471-T | >256 | 16 | >256 | ≤1 |
| A. haemolyticus ATCC 17906-R | 16 | 16 | 8 | ≤1 |
| A. haemolyticus ATCC 17906-T | >256 | 64 | >256 | ≤1 |
| A. baylyi ATCC 33305-R | ≤1 | ≤1 | 4 | ≤1 |
| A. baylyi ATCC 33305-T | >256 | 16 | >256 | ≤1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martz, K.; Alomar, D.; Karim, M.; Knezevic, S.; D’Costa, V.M. Characterization of the Diversity in Host Range of an Extensively Drug-Resistant (XDR) Type IV Secretion System-Encoding Plasmid in Acinetobacter. Pathogens 2025, 14, 606. https://doi.org/10.3390/pathogens14060606
Martz K, Alomar D, Karim M, Knezevic S, D’Costa VM. Characterization of the Diversity in Host Range of an Extensively Drug-Resistant (XDR) Type IV Secretion System-Encoding Plasmid in Acinetobacter. Pathogens. 2025; 14(6):606. https://doi.org/10.3390/pathogens14060606
Chicago/Turabian StyleMartz, Kailey, Dalya Alomar, Marisha Karim, Sara Knezevic, and Vanessa M. D’Costa. 2025. "Characterization of the Diversity in Host Range of an Extensively Drug-Resistant (XDR) Type IV Secretion System-Encoding Plasmid in Acinetobacter" Pathogens 14, no. 6: 606. https://doi.org/10.3390/pathogens14060606
APA StyleMartz, K., Alomar, D., Karim, M., Knezevic, S., & D’Costa, V. M. (2025). Characterization of the Diversity in Host Range of an Extensively Drug-Resistant (XDR) Type IV Secretion System-Encoding Plasmid in Acinetobacter. Pathogens, 14(6), 606. https://doi.org/10.3390/pathogens14060606





