High Prevalence of Virulence-Associated Genes and Length Polymorphism in actA and inlB Genes Identified in Listeria monocytogenes Isolates from Meat Products and Meat-Processing Environments in Poland
Abstract
1. Introduction
2. Aim of This Study
3. Materials and Methods
3.1. Bacterial Isolates and Genetic Material
3.2. Detection of Virulence-Associated Genes
3.3. Hemolysis Assay
4. Results
4.1. Presence of Virulence-Associated Genes
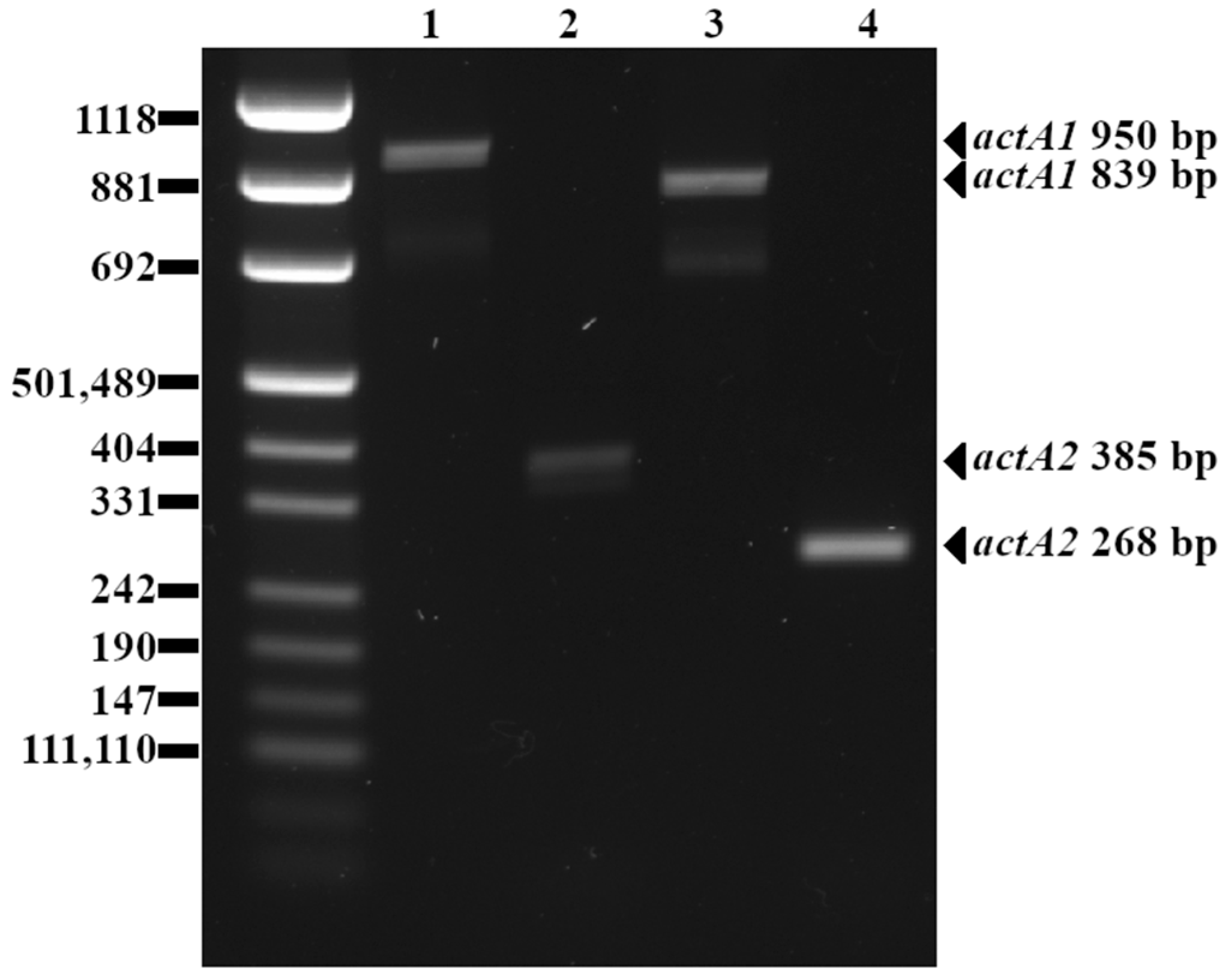
4.2. actA Polymorphism
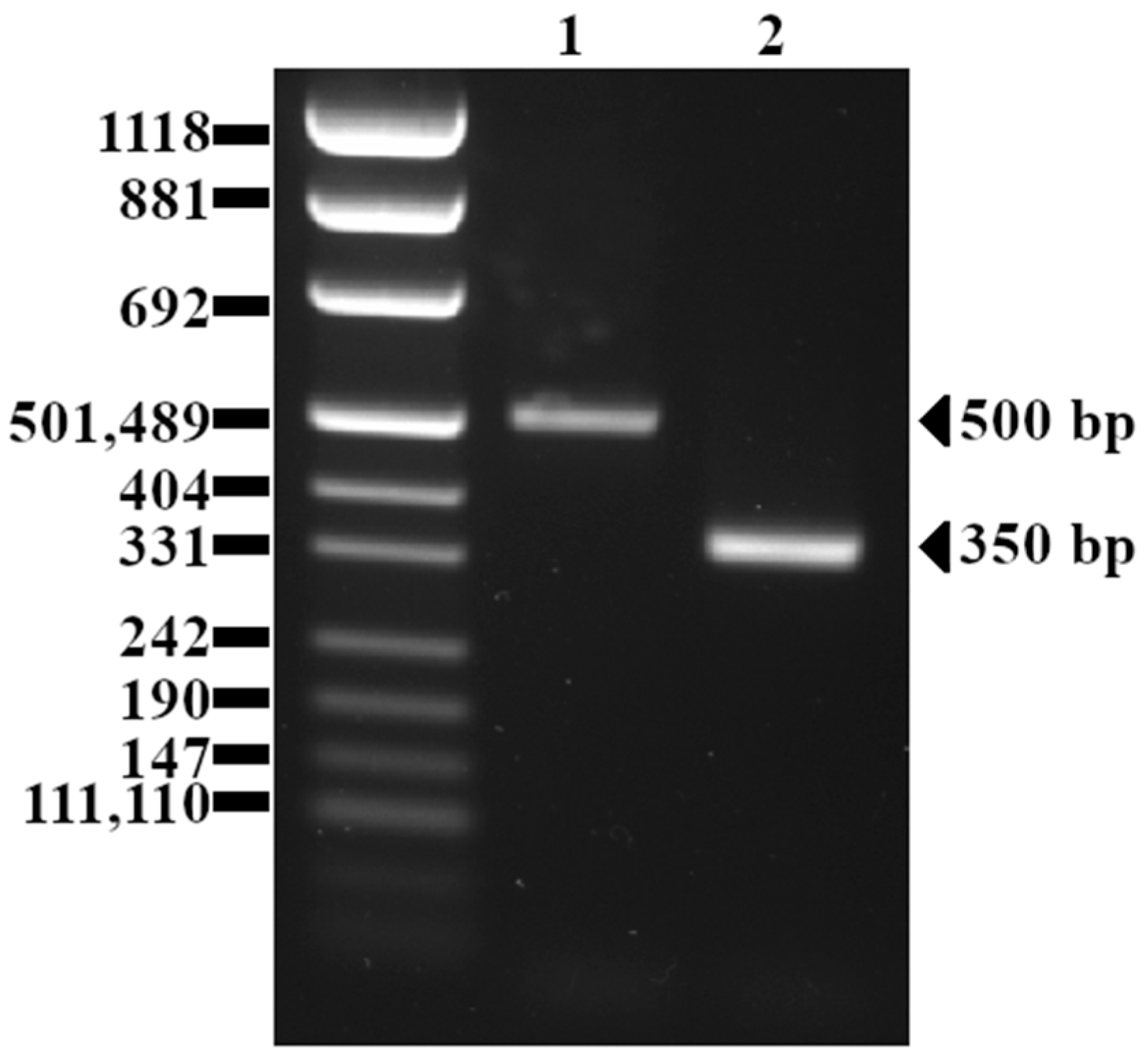
4.3. inlB Polymorphism
4.4. Hemolysis
5. Discussion
5.1. General Prevalence of Virulence-Associated Genes
5.2. actA Polymorphism
5.3. inlB Polymorphism
5.4. Hemolysis
6. Summary and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Murray, E.G.D. A Characterization of Listeriosis in Man and Other Animals. Can. Med. Assoc. J. 1955, 72, 99–103. [Google Scholar] [PubMed]
- Osek, J.; Lachtara, B.; Wieczorek, K. Listeria monocytogenes in Foods-From Culture Identification to Whole-Genome Characteristics. Food Sci. Nutr. 2022, 10, 2825–2854. [Google Scholar] [CrossRef] [PubMed]
- Reed, R.W.; Gavin, W.F.; Crosby, J.; Dobson, P. Listeriosis in Man. Can. Med. Assoc. J. 1955, 73, 400–402. [Google Scholar] [PubMed]
- Rogalla, D.; Bomar, P.A. Listeria monocytogenes. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2020. [Google Scholar]
- Wiktorczyk-Kapischke, N.; Skowron, K.; Wałecka-Zacharska, E. Genomic and Pathogenicity Islands of Listeria monocytogenes-Overview of Selected Aspects. Front. Mol. Biosci. 2023, 10, 1161486. [Google Scholar] [CrossRef] [PubMed]
- Lungu, B.; Ricke, S.C.; Johnson, M.G. Growth, Survival, Proliferation and Pathogenesis of Listeria monocytogenes under Low Oxygen or Anaerobic Conditions: A Review. Anaerobe 2009, 15, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Sibanda, T.; Buys, E.M. Listeria monocytogenes Pathogenesis: The Role of Stress Adaptation. Microorganisms 2022, 10, 1522. [Google Scholar] [CrossRef] [PubMed]
- Osek, J.; Wieczorek, K. Listeria monocytogenes-How This Pathogen Uses Its Virulence Mechanisms to Infect the Hosts. Pathogens 2022, 11, 1491. [Google Scholar] [CrossRef] [PubMed]
- Quereda, J.J.; Morón-García, A.; Palacios-Gorba, C.; Dessaux, C.; García-del Portillo, F.; Pucciarelli, M.G.; Ortega, A.D. Pathogenicity and Virulence of Listeria monocytogenes: A Trip from Environmental to Medical Microbiology. Virulence 2021, 12, 2509–2545. [Google Scholar] [CrossRef]
- Guariglia-Oropeza, V.; Orsi, R.; Wiedmann, M.; Yu, H.; Boor, K.; Guldimann, C. Regulatory Network Features in Listeria monocytogenes—Changing the Way We Talk. Front. Cell. Infect. Microbiol. 2014, 4, 14. [Google Scholar] [CrossRef]
- Ollinger, J.; Bowen, B.; Wiedmann, M.; Boor, K.J.; Bergholz, T.M. Listeria monocytogenes sigmaB Modulates PrfA-Mediated Virulence Factor Expression. Infect. Immun. 2009, 77, 2113–2124. [Google Scholar] [CrossRef]
- Lopes-Luz, L.; Mendonça, M.; Bernardes Fogaça, M.; Kipnis, A.; Bhunia, A.K.; Bührer-Sékula, S. Listeria monocytogenes: Review of Pathogenesis and Virulence Determinants-Targeted Immunological Assays. Crit. Rev. Microbiol. 2021, 47, 647–666. [Google Scholar] [CrossRef] [PubMed]
- Mir, S.A. Structure and Function of the Important Internalins of Listeria monocytogenes. Curr. Protein Pept. Sci. 2021, 22, 620–628. [Google Scholar] [CrossRef] [PubMed]
- Wiśniewski, P.; Zakrzewski, A.J.; Zadernowska, A.; Chajęcka-Wierzchowska, W. Antimicrobial Resistance and Virulence Characterization of Listeria monocytogenes Strains Isolated from Food and Food Processing Environments. Pathogens 2022, 11, 1099. [Google Scholar] [CrossRef] [PubMed]
- Zakrzewski, A.J.; Chajęcka-Wierzchowska, W.; Zadernowska, A.; Podlasz, P. Virulence Characterization of Listeria monocytogenes, Listeria Innocua, and Listeria Welshimeri Isolated from Fish and Shrimp Using In Vivo Early Zebrafish Larvae Models and Molecular Study. Pathogens 2020, 9, E1028. [Google Scholar] [CrossRef]
- Swetha, C.S.; Porteen, K.; Elango, A.; Ronald, B.S.M.; Senthil Kumar, T.M.A.; Milton, A.P.; Sureshkannan, S. Genetic Diversity, Virulence and Distribution of Antimicrobial Resistance among Listeria monocytogenes Isolated from Milk, Beef, and Bovine Farm Environment. Iran. J. Vet. Res. 2021, 22, 1–8. [Google Scholar] [CrossRef]
- Fernandez-Garayzabal, J.F.; Delgado, C.; Blanco, M.; Vazquez-Boland, J.A.; Briones, V.; Suarez, G.; Dominguez, L. Role of Potassium Tellurite and Brain Heart Infusion in Expression of the Hemolytic Phenotype of Listeria Spp. on Agar Plates. Appl. Environ. Microbiol. 1992, 58, 434–438. [Google Scholar] [CrossRef] [PubMed]
- Fox, E.M.; Bierne, H.; Stessl, B. (Eds.) Listeria monocytogenes: Methods and Protocols, 2nd ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2021; ISBN 978-1-07-160981-1. [Google Scholar]
- Hitchins, A.D.; Jinneman, K.; Chen, Y. Chapter 10: Detection of Listeria monocytogenes in Foods and Environmental Samples, and Enumeration of Listeria monocytogenes in Foods. In FDA Bacteriological Analytical Manual; US Food Drug Administration: Washington, DC, USA, 2017. [Google Scholar]
- Maury, M.M.; Chenal-Francisque, V.; Bracq-Dieye, H.; Han, L.; Leclercq, A.; Vales, G.; Moura, A.; Gouin, E.; Scortti, M.; Disson, O.; et al. Spontaneous Loss of Virulence in Natural Populations of Listeria monocytogenes. Infect. Immun. 2017, 85, e00541-17. [Google Scholar] [CrossRef]
- Paillard, D.; Dubois, V.; Duran, R.; Nathier, F.; Guittet, C.; Caumette, P.; Quentin, C. Rapid Identification of Listeria Species by Using Restriction Fragment Length Polymorphism of PCR-Amplified 23S rRNA Gene Fragments. AEM 2003, 69, 6386–6392. [Google Scholar] [CrossRef]
- Li, F.; Ye, Q.; Chen, M.; Zhou, B.; Xiang, X.; Wang, C.; Shang, Y.; Zhang, J.; Pang, R.; Wang, J.; et al. Mining of Novel Target Genes through Pan-Genome Analysis for Multiplex PCR Differentiation of the Major Listeria monocytogenes Serotypes. Int. J. Food Microbiol. 2021, 339, 109026. [Google Scholar] [CrossRef]
- Kawacka, I.; Olejnik-Schmidt, A. Genoserotyping of Listeria monocytogenes Strains Originating from Meat Products and Meat Processing Environments. ŻNTJ 2022, 2, 34–44. [Google Scholar] [CrossRef]
- D’Agostino, M.; Wagner, M.; Vazquez-Boland, J.A.; Kuchta, T.; Karpiskova, R.; Hoorfar, J.; Novella, S.; Scortti, M.; Ellison, J.; Murray, A.; et al. A Validated PCR-Based Method to Detect Listeria monocytogenes Using Raw Milk as a Food Model--towards an International Standard. J. Food Prot. 2004, 67, 1646–1655. [Google Scholar] [CrossRef]
- Bae, D.; Liu, C.; Zhang, T.; Jones, M.; Peterson, S.N.; Wang, C. Global Gene Expression of Listeria monocytogenes to Salt Stress. J. Food Prot. 2012, 75, 906–912. [Google Scholar] [CrossRef]
- Notermans, S.H.; Dufrenne, J.; Leimeister-Wächter, M.; Domann, E.; Chakraborty, T. Phosphatidylinositol-Specific Phospholipase C Activity as a Marker to Distinguish between Pathogenic and Nonpathogenic Listeria Species. Appl. Environ. Microbiol. 1991, 57, 2666–2670. [Google Scholar] [CrossRef]
- Nishibori, T.; Cooray, K.; Xiong, H.; Kawamura, I.; Fujita, M.; Mitsuyama, M. Correlation between the Presence of Virulence-Associated Genes as Determined by PCR and Actual Virulence to Mice in Various Strains of Listeria spp. Microbiol. Immunol. 1995, 39, 343–349. [Google Scholar] [CrossRef]
- Osman, K.M.; Zolnikov, T.R.; Samir, A.; Orabi, A. Prevalence, Pathogenic Capability, Virulence Genes, Biofilm Formation, and Antibiotic Resistance of Listeria in Goat and Sheep Milk Confirms Need of Hygienic Milking Conditions. Pathog. Glob. Health 2014, 108, 21–29. [Google Scholar] [CrossRef]
- Jallewar, P.K.; Kalorey, D.R.; Kurkure, N.V.; Pande, V.V.; Barbuddhe, S.B. Genotypic Characterization of Listeria spp. Isolated from Fresh Water Fish. Int. J. Food Microbiol. 2007, 114, 120–123. [Google Scholar] [CrossRef]
- Jaradat, Z.W.; Schutze, G.E.; Bhunia, A.K. Genetic Homogeneity among Listeria monocytogenes Strains from Infected Patients and Meat Products from Two Geographic Locations Determined by Phenotyping, Ribotyping and PCR Analysis of Virulence Genes. Int. J. Food Microbiol. 2002, 76, 1–10. [Google Scholar] [CrossRef]
- Liu, D.; Lawrence, M.L.; Austin, F.W.; Ainsworth, A.J. A Multiplex PCR for Species- and Virulence-Specific Determination of Listeria monocytogenes. J. Microbiol. Methods 2007, 71, 133–140. [Google Scholar] [CrossRef]
- Zhang, W.; Knabel, S.J. Multiplex PCR Assay Simplifies Serotyping and Sequence Typing of Listeria monocytogenes Associated with Human Outbreaks. J. Food Prot. 2005, 68, 1907–1910. [Google Scholar] [CrossRef]
- Furrer, B.; Candrian, U.; Hoefelein, C.; Luethy, J. Detection and Identification of Listeria monocytogenes in Cooked Sausage Products and in Milk by in Vitro Amplification of Haemolysin Gene Fragments. J. Appl. Bacteriol. 1991, 70, 372–379. [Google Scholar] [CrossRef]
- Borucki, M.K.; Call, D.R. Listeria monocytogenes Serotype Identification by PCR. J. Clin. Microbiol. 2003, 41, 5537–5540. [Google Scholar] [CrossRef]
- Morgulis, A.; Coulouris, G.; Raytselis, Y.; Madden, T.L.; Agarwala, R.; Schäffer, A.A. Database Indexing for Production MegaBLAST Searches. Bioinformatics 2008, 24, 1757–1764. [Google Scholar] [CrossRef]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A Greedy Algorithm for Aligning DNA Sequences. J. Comput. Biol. 2000, 7, 203–214. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Doumith, M.; Buchrieser, C.; Glaser, P.; Jacquet, C.; Martin, P. Differentiation of the Major Listeria monocytogenes Serovars by Multiplex PCR. J. Clin. Microbiol. 2004, 42, 3819–3822. [Google Scholar] [CrossRef]
- Kawacka, I.; Pietrzak, B.; Schmidt, M.; Olejnik-Schmidt, A. Listeria monocytogenes Isolates from Meat Products and Processing Environment in Poland Are Sensitive to Commonly Used Antibiotics, with Rare Cases of Reduced Sensitivity to Ciprofloxacin. Life 2023, 13, 821. [Google Scholar] [CrossRef]
- Kurpas, M.; Osek, J.; Moura, A.; Leclercq, A.; Lecuit, M.; Wieczorek, K. Genomic Characterization of Listeria monocytogenes Isolated from Ready-to-Eat Meat and Meat Processing Environments in Poland. Front. Microbiol. 2020, 11, 1412. [Google Scholar] [CrossRef]
- Parra-Flores, J.; Holý, O.; Bustamante, F.; Lepuschitz, S.; Pietzka, A.; Contreras-Fernández, A.; Castillo, C.; Ovalle, C.; Alarcón-Lavín, M.P.; Cruz-Córdova, A.; et al. Virulence and Antibiotic Resistance Genes in Listeria monocytogenes Strains Isolated From Ready-to-Eat Foods in Chile. Front. Microbiol. 2022, 12, 796040. [Google Scholar] [CrossRef]
- Bania, J.; Żarczyńska, A.; Molenda, J.; Dąbrowska, A.; Kosek-Paszkowska, K.; Więckowska-Szakiel, M.; Różalska, B. Subtyping of Listeria monocytogenes Isolates by actA Gene Sequencing, PCR-Fingerprinting, and Cell-Invasion Assay. Folia Microbiol. 2009, 54, 17–24. [Google Scholar] [CrossRef]
- Zhou, X.; Jiao, X.; Wiedmann, M. Listeria monocytogenes in the Chinese Food System: Strain Characterization through Partial actA Sequencing and Tissue-Culture Pathogenicity Assays. J. Med. Microbiol. 2005, 54, 217–224. [Google Scholar] [CrossRef]
- Portnoy, D.A.; Chakraborty, T.; Goebel, W.; Cossart, P. Molecular Determinants of Listeria monocytogenes Pathogenesis. Infect. Immun. 1992, 60, 1263–1267. [Google Scholar] [CrossRef]
- Moriishi, K.; Terao, M.; Koura, M.; Inoue, S. Sequence Analysis of the actA Gene of Listeria monocytogenes Isolated from Human. Microbiol. Immunol. 1998, 42, 129–132. [Google Scholar] [CrossRef]
- Conter, M.; Vergara, A.; Di Ciccio, P.; Zanardi, E.; Ghidini, S.; Ianieri, A. Polymorphism of actA Gene Is Not Related to in Vitro Virulence of Listeria monocytogenes. Int. J. Food Microbiol. 2010, 137, 100–105. [Google Scholar] [CrossRef]
- Nightingale, K.K.; Milillo, S.R.; Ivy, R.A.; Ho, A.J.; Oliver, H.F.; Wiedmann, M. Listeria monocytogenes F2365 Carries Several Authentic Mutations Potentially Leading to Truncated Gene Products, Including inlB, and Demonstrates Atypical Phenotypic Characteristics. J. Food Prot. 2007, 70, 482–488. [Google Scholar] [CrossRef]
- Quereda, J.J.; Rodríguez-Gómez, I.M.; Meza-Torres, J.; Gómez-Laguna, J.; Nahori, M.A.; Dussurget, O.; Carrasco, L.; Cossart, P.; Pizarro-Cerdá, J. Reassessing the Role of Internalin B in Listeria monocytogenes Virulence Using the Epidemic Strain F2365. Clin. Microbiol. Infect. 2019, 25, 252.e1–252.e4. [Google Scholar] [CrossRef]
- Chiba, S.; Nagai, T.; Hayashi, T.; Baba, Y.; Nagai, S.; Koyasu, S. Listerial Invasion Protein Internalin B Promotes Entry into Ileal Peyer’s Patches in Vivo. Microbiol. Immunol. 2011, 55, 123–129. [Google Scholar] [CrossRef]
- Roche, S.M.; Grépinet, O.; Corde, Y.; Teixeira, A.P.; Kerouanton, A.; Témoin, S.; Mereghetti, L.; Brisabois, A.; Velge, P. A Listeria monocytogenes Strain Is Still Virulent despite Nonfunctional Major Virulence Genes. J. Infect. Dis. 2009, 200, 1944–1948. [Google Scholar] [CrossRef]
- Sobyanin, K.; Sysolyatina, E.; Krivozubov, M.; Chalenko, Y.; Karyagina, A.; Ermolaeva, S. Naturally Occurring InlB Variants That Support Intragastric Listeria monocytogenes Infection in Mice. FEMS Microbiol. Lett. 2017, 364, fnx011. [Google Scholar] [CrossRef] [PubMed]
- Kaur, S.; Malik, S.V.S.; Vaidya, V.M.; Barbuddhe, S.B. Listeria monocytogenes in Spontaneous Abortions in Humans and Its Detection by Multiplex PCR. J. Appl. Microbiol. 2007, 103, 1889–1896. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.S.; Duarte, E.A.; Oliveira, T.A.; Evangelista-Barreto, N.S. Identification of Listeria monocytogenes in Cattle Meat Using Biochemical Methods and Amplification of the Hemolysin Gene. An. Acad. Bras. Ciênc. 2020, 92, e20180557. [Google Scholar] [CrossRef]
- Wieczorek, K.; Osek, J. Prevalence, Genetic Diversity and Antimicrobial Resistance of Listeria monocytogenes Isolated from Fresh and Smoked Fish in Poland. Food Microbiol. 2017, 64, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Kawacka, I.; Olejnik-Schmidt, A.; Schmidt, M. Nonhemolytic Listeria monocytogenes-Prevalence Rate, Reasons Underlying Atypical Phenotype, and Methods for Accurate Hemolysis Assessment. Microorganisms 2022, 10, 483. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.S. The Investigation of Molecular Characterization of Presumptive Listeria monocytogenes Isolates from a Food-Processing Environment. Iran. J. Vet. Res. 2019, 20, 46–50. [Google Scholar] [PubMed]
- Bou-m’handi, N.; Jacquet, C.; El Marrakchi, A.; Martin, P. Phenotypic and Molecular Characterization of Listeria monocytogenes Strains Isolated from a Marine Environment in Morocco. Foodborne Pathog. Dis. 2007, 4, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Lindbäck, T.; Secic, I.; Rørvik, L.M. A Contingency Locus in prfA in a Listeria monocytogenes Subgroup Allows Reactivation of the PrfA Virulence Regulator during Infection in Mice. Appl. Environ. Microbiol. 2011, 77, 3478–3483. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Milillo, S.R.; Stout, J.C.; Hanning, I.B.; Clement, A.; Fortes, E.D.; den Bakker, H.C.; Wiedmann, M.; Ricke, S.C. Listeria monocytogenes and Hemolytic Listeria innocua in Poultry. Poult. Sci. 2012, 91, 2158–2163. [Google Scholar] [CrossRef] [PubMed]
- Moreno, L.Z.; Paixão, R.; de Gobbi, D.D.S.; Raimundo, D.C.; Porfida Ferreira, T.S.; Micke Moreno, A.; Hofer, E.; dos Reis, C.M.F.; Matté, G.R.; Matté, M.H. Phenotypic and Genotypic Characterization of Atypical Listeria monocytogenes and Listeria innocua Isolated from Swine Slaughterhouses and Meat Markets. Biomed. Res. Int. 2014, 2014, 742032. [Google Scholar] [CrossRef] [PubMed]
- Moura, A.; Disson, O.; Lavina, M.; Thouvenot, P.; Huang, L.; Leclercq, A.; Fredriksson-Ahomaa, M.; Eshwar, A.K.; Stephan, R.; Lecuit, M. Atypical Hemolytic Listeria innocua Isolates Are Virulent, Albeit Less than Listeria monocytogenes. Infect. Immun. 2019, 87, e00758-18. [Google Scholar] [CrossRef]
- Palerme, J.-S.; Pan, P.C.; Parsons, C.T.; Kathariou, S.; Ward, T.J.; Jacob, M.E. Isolation and Characterization of Atypical Listeria monocytogenes Associated with a Canine Urinary Tract Infection. J. Vet. Diagn. Invest. 2016, 28, 604–607. [Google Scholar] [CrossRef]
- Schärer, K.; Stephan, R.; Tasara, T. Cold Shock Proteins Contribute to the Regulation of Listeriolysin O Production in Listeria monocytogenes. Foodborne Pathog. Dis. 2013, 10, 1023–1029. [Google Scholar] [CrossRef]
| Primers Name | Target | Primers’ Sequence | Primer Concentration [µM] | Annealing Temperature [°C] | Amplicon Length (bp) | Reference |
|---|---|---|---|---|---|---|
| prfA | Listeriolysin positive regulatory protein | F: 5′-GATACAGAAACATCGGTTGGC-3′ R: 5′-GTGTAATCTTGATGCCATCAGG-3′ | 0.3 | 49 | 274 | [24] |
| sigB | Sigma factor | F: 5′-TCATCGGTGTCACGGAAGAA-3′ R: 5′-TGACGTTGGATTCTAGACAC-3′ | 0.35 | 51 | 310 | [25] |
| plcA | Phosphatidylinositol-specific phospholipase C | F: 5′-CTGCTTGAGCGTTCATGTCTCATCCCCC-3′ R: 5′-CATGGGTTTCACTCTCCTTCTAC-3′ | 0.5 | 60 | 1484 | [26] |
| plcB | Phosphatidylicholin-specific phospholipase C | F: 5′-GCAAGTGTTCTAGTCTTTCCGG-3′ R: 5′- ACCTGCCAAAGTTTGCTGTGA-3′ | 0.5 | 55 | 795 | [27] |
| hly | Listeriolysin O | F: 5′-GCAGTTGCAAGCGCTTGGAGTGAA-3′ R: 5′-GCAACGTATCCTCCAGAGTGATCG-3′ | 0.3 | 62 | 456 | [28] |
| actA1 | Actin polymerization protein | F: 5′- CGCCGCGGAAATTAAAAAAAGA-3′ R: 5′- ACGAAGGAACCGGGCTGCTAG-3′ | 0.4 | 62 | 839 (or 950) | [29] |
| actA2 | Actin polymerization protein | F: 5′-GACGAAAATCCCGAAGTGAA-3′ R: 5′-CTAGCGAAGGTGCTGTTTCC-3′ | 1.0 | 63 | 268 (or 385) | [30] |
| mpl | Metalloprotease | F: 5′-GGCTCATTTCACTATGACGG-3′ R: 5′- GCTTCCCAAGCTTCAGCAACT-3′ | 0.5 | 60 | 143 | [27] |
| inlA | Internalin A | F: 5′-ACGAGTAACGGGACAAATGC-3′ R: 5′-CCCGACAGTGGTGCTAGATT-3′ | 0.5 | 55 | 800 | [31] |
| inlB | Internalin B | F: 5′- CATGGGAGAGTAACCCAACC-3′ R: 5′- GCGGTAACCCCTTTGTCATA-3′ | 0.75 | 57 | 500 | [32] |
| inlC | Internalin C | F: 5′- CCCACAATCAAATAAGTGACCTT-3′ R: 5′- CTGGGTCTTTGACAGTATTTGTT-3′ | 1.25 | 57 | 400 | [32] |
| inlJ | Internalin J | F: 5′-TGTAACCCCGCTTACACAGTT-3′ R: 5′-AGCGGCTTGGCAGTCTAATA-3′ | 0.5 | 55 | 238 | [31] |
| iap | Invasion associated protein | F: 5′-ACAAGCTGCACCTGTTGCAG-3′ R: 5′-TGACAGCGTGTGTAGTAGCA-3′ | 0.3 | 56 | 131 | [33] * |
| flaA | Flagellin | F: 5′-TTACTAGATCAAACTGCTCC-3′ R: 5′-AAGAAAAGCCCCTCGTCC-3′ | 1.0 | 54 | 538 | [34] |
| Primer Name | Approx. Amplicon Size (bp) | Number of Isolates (%) |
|---|---|---|
| prfA | 274 | 153 (100%) |
| sigB | 310 | 153 (100%) |
| plcA | 1484 | 153 (100%) |
| plcB | 795 | 153 (100%) |
| hlyA | 456 | 153 (100%) |
| actA1 | 839 | 15 (10%) |
| 950 | 120 (78%) | |
| no amplicon | 18 (12%) | |
| actA2 | 268 | 28 (18%) |
| 385 | 125 (82%) | |
| mpl | 143 | 153 (100%) |
| inlA | 800 | 153 (100%) |
| inlB | 500 | 151 (99%) |
| 360 | 2 (1%) | |
| inlC | 400 | 153 (100%) |
| inlJ | 238 | 153 (100%) |
| iap | 131 | 153 (100%) |
| flaA | 538 | 113 (74%) |
| no amplicon | 40 (26%) |
| actA1 Shorter Amplicon (839 bp) | actA1 Longer Amplicon (950 bp) | actA1 No Amplicon | |
|---|---|---|---|
| actA2 shorter amplicon (268 bp) | 15 | 0 | 13 |
| actA2 longer amplicon (385 bp) | 0 | 120 | 5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kawacka, I.; Olejnik-Schmidt, A. High Prevalence of Virulence-Associated Genes and Length Polymorphism in actA and inlB Genes Identified in Listeria monocytogenes Isolates from Meat Products and Meat-Processing Environments in Poland. Pathogens 2024, 13, 444. https://doi.org/10.3390/pathogens13060444
Kawacka I, Olejnik-Schmidt A. High Prevalence of Virulence-Associated Genes and Length Polymorphism in actA and inlB Genes Identified in Listeria monocytogenes Isolates from Meat Products and Meat-Processing Environments in Poland. Pathogens. 2024; 13(6):444. https://doi.org/10.3390/pathogens13060444
Chicago/Turabian StyleKawacka, Iwona, and Agnieszka Olejnik-Schmidt. 2024. "High Prevalence of Virulence-Associated Genes and Length Polymorphism in actA and inlB Genes Identified in Listeria monocytogenes Isolates from Meat Products and Meat-Processing Environments in Poland" Pathogens 13, no. 6: 444. https://doi.org/10.3390/pathogens13060444
APA StyleKawacka, I., & Olejnik-Schmidt, A. (2024). High Prevalence of Virulence-Associated Genes and Length Polymorphism in actA and inlB Genes Identified in Listeria monocytogenes Isolates from Meat Products and Meat-Processing Environments in Poland. Pathogens, 13(6), 444. https://doi.org/10.3390/pathogens13060444






