Fluorescently Tagged Verticillium dahliae to Understand the Infection Process on Cotton (Gossypium hirsutum) and Weed Plant Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Isolates
2.2. Plant Materials Used in this Study
2.3. Vector Construction and Transformation
2.4. Assessment of Fungal Transformants
2.5. Plant Growth and Pathogenicity Assay
2.6. Scoring
2.7. Re-Isolation
2.8. Confocal Microscopy
2.9. Statistical Analysis
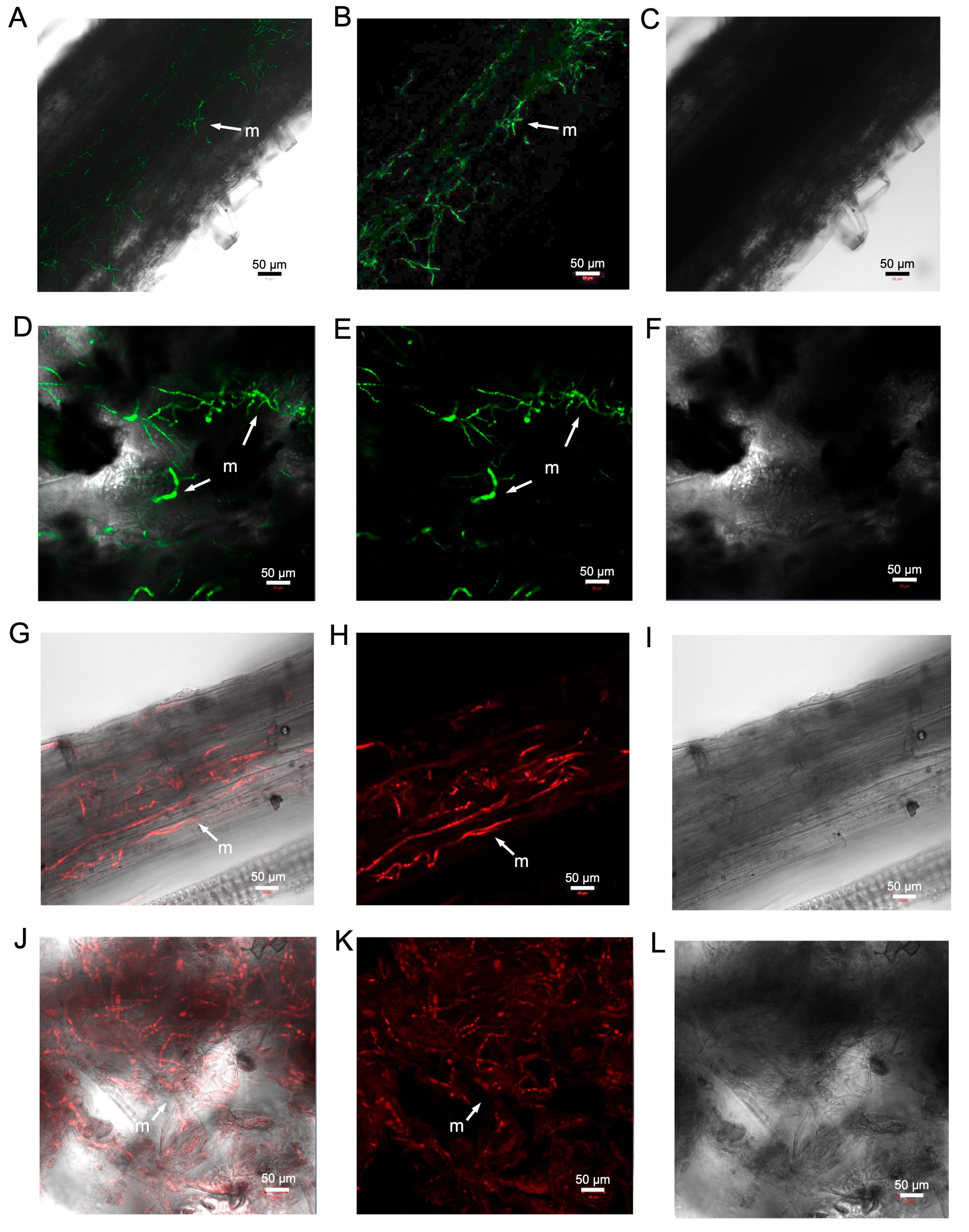
3. Results
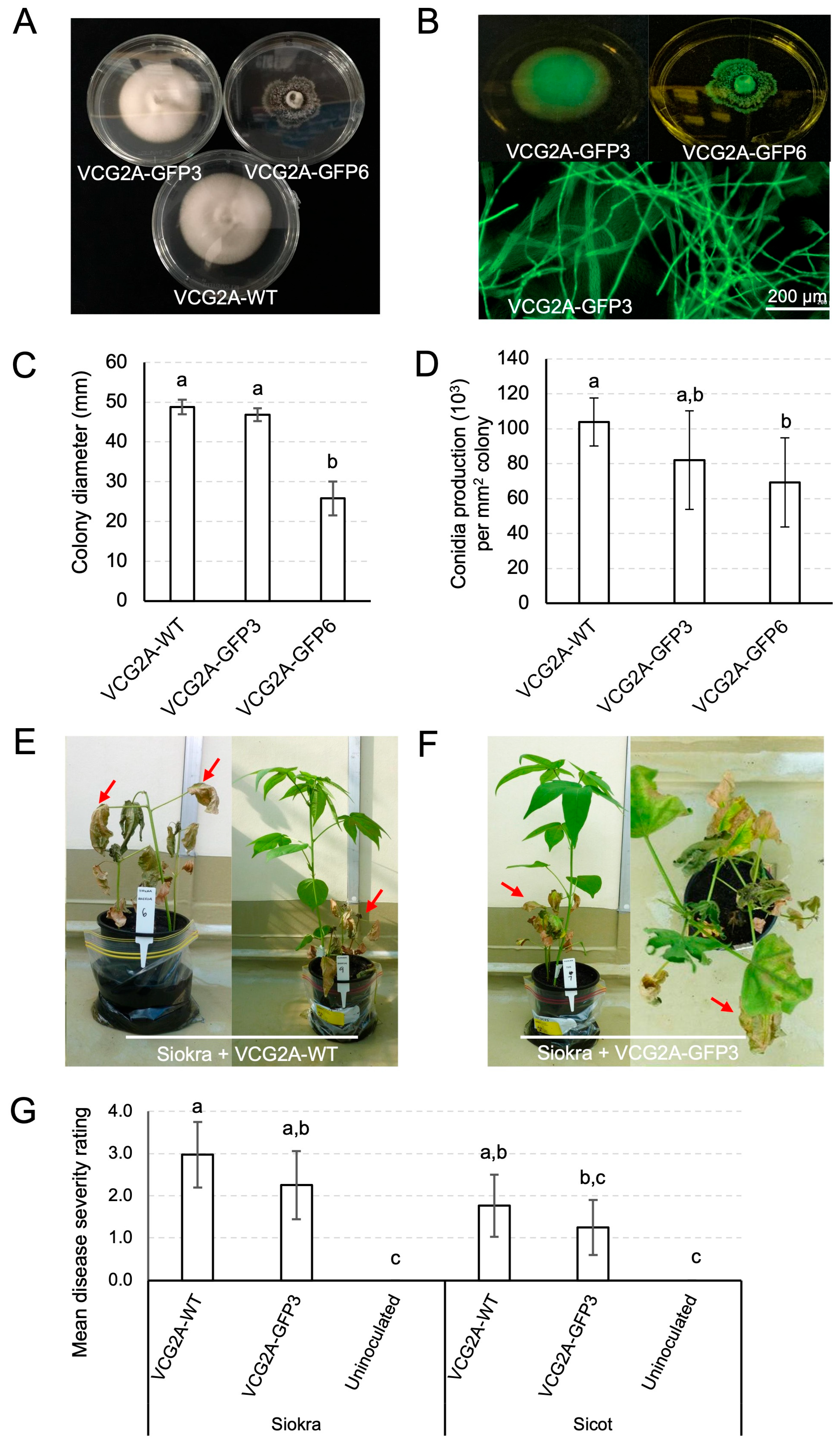
3.1. In Vitro and in Plantae Assessment of VCG 2A Transformants of VCG2A-WT
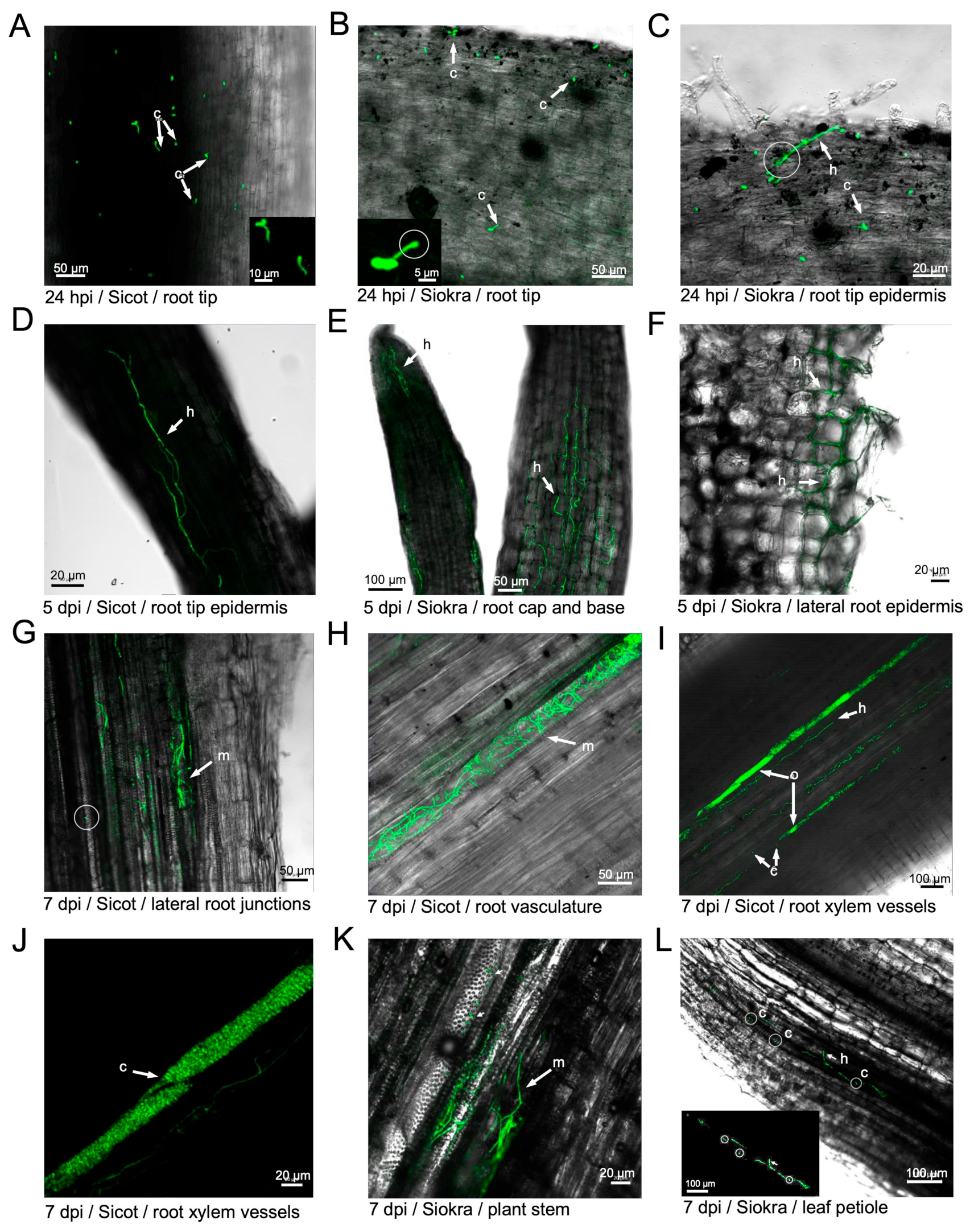
3.2. Infection of Cotton Plants by V. dahliae
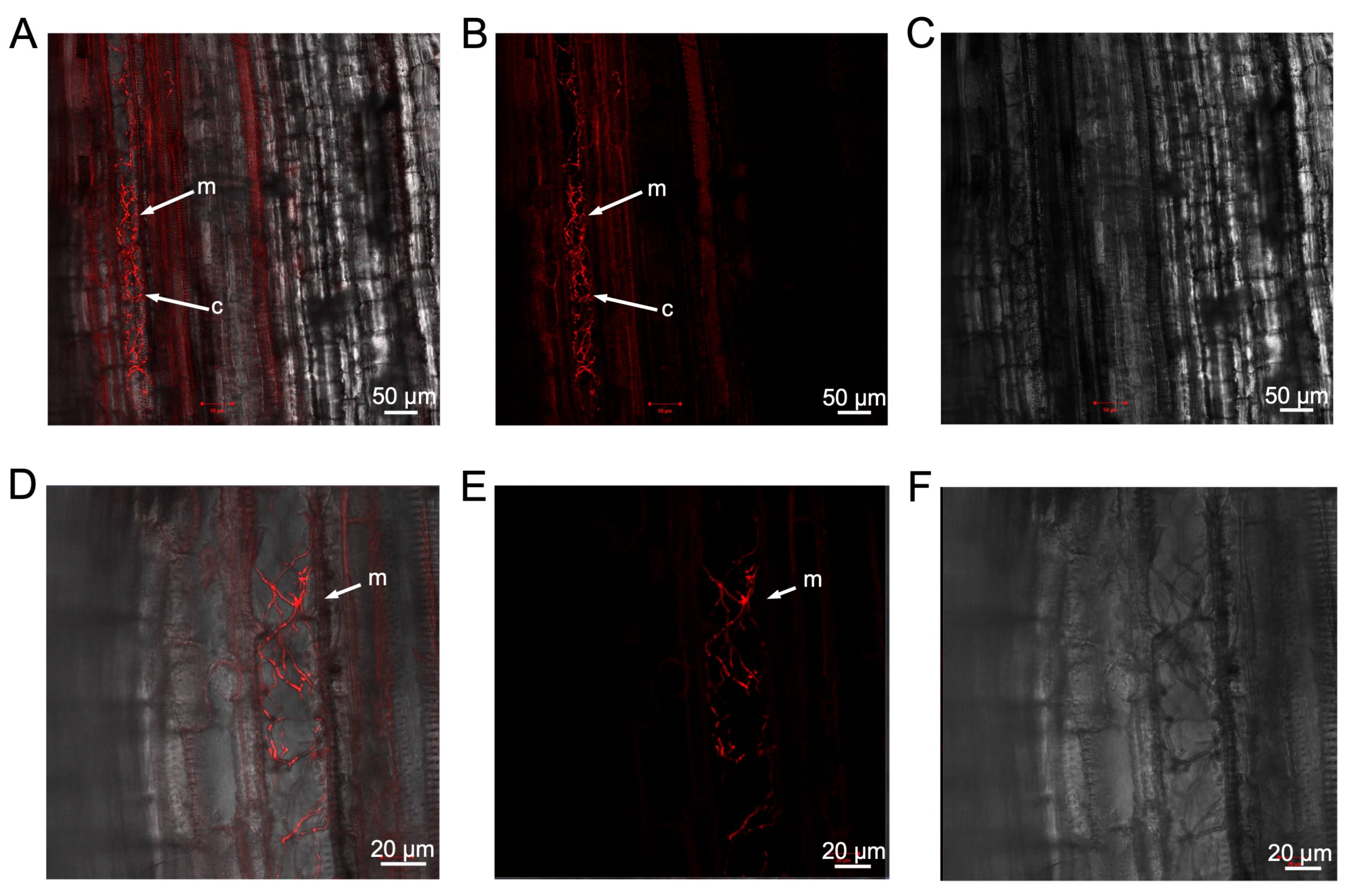
3.3. Development of a Defoliating V. dahliae Strain Expressing mCherry
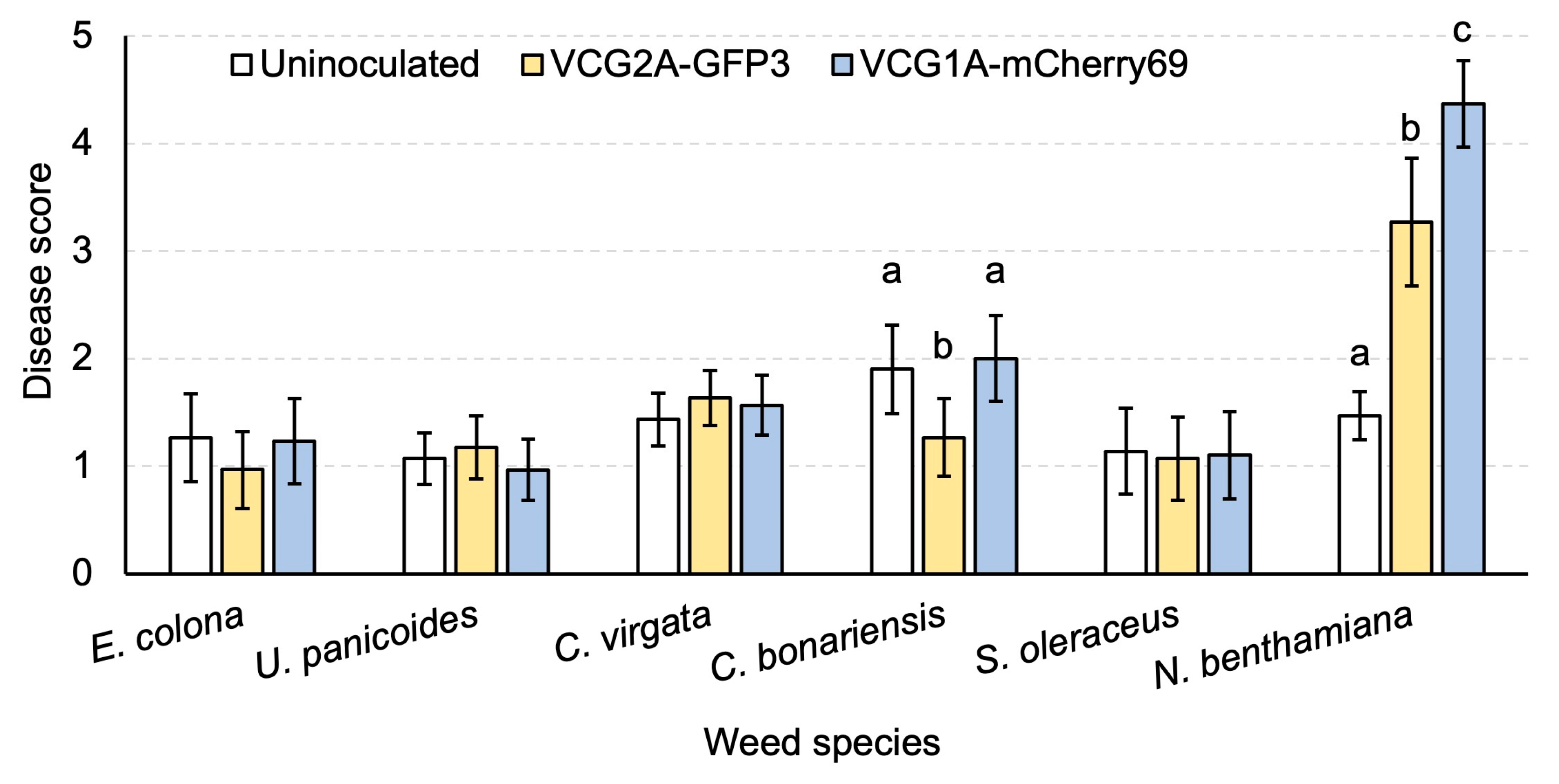
3.4. Both Defoliating and Non-Defoliating V. dahliae Can Colonise Weed Species and Cause Disease in Nicotiana benthamiana
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Oosterhuis, D.M. Growth and Development of a Cotton Plant. In Nitrogen Nutrition of Cotton: Practical Issues; Wiley: Hoboken, NJ, USA, 1990; pp. 1–24. [Google Scholar]
- Hu, Y.; Chen, J.; Fang, L.; Zhang, Z.; Ma, W.; Niu, Y.; Ju, L.; Deng, J.; Zhao, T.; Lian, J.; et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 2019, 51, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Cotton Industry Australia Overview. Available online: https://cottonaustralia.com.au/industry-overview (accessed on 15 February 2024).
- Dadd-Daigle, P.; Kirkby, K.; Chowdhury, P.R.; Labbate, M.; Chapman, T.A. The Verticillium wilt problem in Australian cotton. Australas. Plant Pathol. 2021, 50, 129–135. [Google Scholar] [CrossRef]
- Martín-Sanz, A.; Rueda, S.; García-Carneros, A.B.; González-Fernández, S.; Miranda-Fuentes, P.; Castuera-Santacruz, S.; Molinero-Ruiz, L. Genetics, host range, and molecular and pathogenic characterization of Verticillium dahliae from sunflower reveal two differentiated groups in Europe. Front. Plant Sci. 2018, 9, 288. [Google Scholar] [CrossRef] [PubMed]
- Berlander, I.; Powelson, M.L. Verticillium wilt. Plant Health Instructor. 2000. [Google Scholar] [CrossRef]
- Klimes, A.; Dobinson, K.F.; Thomma, B.P.H.J.; Klosterman, S.J. Genomics spurs rapid advances in our understanding of the biology of vascular wilt pathogens in the genus Verticillium. Annu. Rev. Phytopathol. 2015, 53, 181–198. [Google Scholar] [CrossRef] [PubMed]
- Holman, S.; Kirkby, K.; Smith, L.; Kartnett, H. Vert Update: The Latest in Vert Research. CottonInfo. Available online: https://www.cottoninfo.com.au/publications/disease-vert-update-latest-verticillium-research (accessed on 15 February 2024).
- Klosterman, S.J.; Atallah, Z.K.; Vallad, G.E.; Subbarao, K.V. Diversity, pathogenicity, and management of Verticillium species. Annu. Rev. Phytopathol. 2009, 47, 39–62. [Google Scholar] [CrossRef] [PubMed]
- Bellahcene, M.; Assigbetsé, K.; Fortas, Z.; Geiger, J.-P.; Nicole, M.; Fernandez, D. Genetic diversity of Verticillium dahliae isolates from olive trees in Algeria. Phytopathol. Mediterr. 2005, 44, 266–274. [Google Scholar]
- Baroudy, F.; Putman, A.I.; Habib, W.; Puri, K.D.; Subbarao, K.V.; Nigro, F. Genetic diversity of Verticillium dahliae populations from olive and potato in Lebanon. Plant Dis. 2019, 103, 656–667. [Google Scholar] [CrossRef]
- Pérez-Artés, E.; García-Pedrajas, M.D.; Bejarano-Alcázar, J.; Jiménez-Díaz, R.M. Differentiation of cotton-defoliating and nondefoliating pathotypes of Verticillium dahliae by RAPD and specific PCR analyses. Eur. J. Plant Pathol. 2000, 106, 507–517. [Google Scholar] [CrossRef]
- Collado-Romero, M.; Mercado-Blanco, J.; Olivares-García, C.; Jiménez-Díaz, R.M. Phylogenetic analysis of Verticillium dahliae vegetative compatibility groups. Phytopathology 2008, 98, 1019–1028. [Google Scholar] [CrossRef]
- Chapman, T.A.; Chambers, G.A.; Kirkby, K.; Jiménez-Díaz, R.M. First report of the presence of Verticillium dahliae VCG1A in Australia. Australas. Plant Dis. Notes 2016, 11, 13. [Google Scholar] [CrossRef]
- Keykhasaber, M.; Thomma, B.P.H.J.; Hiemstra, J.A. Verticillium wilt caused by Verticillium dahliae in woody plants with emphasis on olive and shade trees. Eur. J. Plant Pathol. 2018, 150, 21–37. [Google Scholar] [CrossRef]
- Yildiz, A.; Doğan, M.N.; Boz, Ö.; Benlioğlu, S. Weed hosts of Verticillium dahliae in cotton fields in Turkey and characterization of V. dahliae isolates from weeds. Phytoparasitica 2009, 37, 171–178. [Google Scholar] [CrossRef]
- Evans, G. Influence of weed hosts on the ecology of Verticillium dahliae in newly cultivated areas of the Namoi Valley, New South Wales. Ann. Appl. Biol. 1971, 67, 169–175. [Google Scholar] [CrossRef]
- Li, F.; Bibi, N.; Fan, K.; Ni, M.I.; Yuan, S.; Wang, M. Agrobacterium-mediated transformation of Verticillium dahliae with GFP gene to study cotton-pathogen interaction using a novel inoculation method. Pak. J. Bot. 2016, 48, 1219–1227. [Google Scholar]
- Su, X.; Lu, G.; Rehman, L.; Li, X.; Sun, L.; Guo, H.; Cheng, H. mCherry-labeled Verticillium dahliae could be utilized to investigate its pathogenicity process in Nicotiana benthamiana. Genes 2018, 9, 508. [Google Scholar] [CrossRef]
- Sabburg, R.; Gregson, A.; Urquhart, A.S.; Aitken, E.A.B.; Smith, L.; Thatcher, L.F.; Gardiner, D.M. A method for high throughput image based antifungal screening. J. Microbiol. Methods 2021, 190, 106342. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, J.; Gao, J.; Zhang, G.; Yu, Y.; Zhou, H.; Chen, W.; Zhao, J. The Colonization process of sunflower by a green fluorescent protein-tagged isolate of Verticillium dahliae and its seed transmission. Plant Dis. 2018, 102, 1772–1778. [Google Scholar] [CrossRef]
- Vallad, G.E.; Subbarao, K.V. Colonization of resistant and susceptible lettuce cultivars by a green fluorescent protein-tagged isolate of Verticillium dahliae. Phytopathology 2008, 98, 871–885. [Google Scholar] [CrossRef]
- Jiménez-Díaz, R.M.; Olivares-García, C.; Landa, B.B.; del Mar Jiménez-Gasco, M.; Navas-Cortés, J.A. Region-wide analysis of genetic diversity in Verticillium dahliae populations infecting olive in southern Spain and agricultural factors influencing the distribution and prevalence of vegetative compatibility groups and pathotypes. Phytopathology 2011, 101, 304–315. [Google Scholar] [CrossRef]
- Mercado-Blanco, J.; Rodríguez-Jurado, D.; Parrilla-Araujo, S.; Jiménez-Díaz, R. Simultaneous detection of the defoliating and nondefoliating Verticillium dahliae pathotypes in infected olive plants by duplex, nested polymerase chain reaction. Plant Dis. 2003, 87, 1487–1494. [Google Scholar] [CrossRef] [PubMed]
- Gardiner, D.M.; Smith, L.J.; Rusu, A.; Aitken, E.B.A. The genomes of two Australian isolates of Verticillium dahliae reccovered from cotton fields. Australas. Plant Pathol. 2024; submitted. [Google Scholar]
- Crespo-Sempere, A.; Marín, S.; Sanchis, V.; Ramos, A.J. VeA and LaeA transcriptional factors regulate ochratoxin A biosynthesis in Aspergillus carbonarius. Int. J. Food Microbiol. 2013, 166, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Fang, H.; Zhou, H.; Sanogo, S.; Ma, Z. Genetics, breeding, and marker-assisted selection for Verticillium wilt resistance in cotton. Crop Sci. 2014, 54, 1289–1303. [Google Scholar] [CrossRef]
- Cirulli, M.; Ciccarese, F.; Amenduni, M. Progress in the search for Verticillium wilt-resistant eggplant. Phytopathol. Mediterr. 1990, 29, 184–190. [Google Scholar]
- Le, D.P.; Gregson, A.; Tran, T.T.; Jackson, R. Co-occurrence of defoliating and non-defoliating pathotypes of Verticillium dahliae in field-grown cotton plants in New South Wales, Australia. Plants 2020, 9, 750. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.-H.; Yeh, Y.-L.; Shen, W.-C. Fast preparation of fungal DNA for PCR screening. J. Microbiol. Methods 2011, 85, 170–172. [Google Scholar] [CrossRef] [PubMed]
- Lievens, B.; Brouwer, M.; Vanachter, A.C.R.C.; Cammue, B.P.A.; Thomma, B.P.H.J. Real-time PCR for detection and quantification of fungal and oomycete tomato pathogens in plant and soil samples. Plant Sci. 2006, 171, 155–165. [Google Scholar] [CrossRef]
- Zhang, W.-W.; Jiang, T.-F.; Cui, X.; Qi, F.-J.; Jian, G.-L. Colonization in cotton plants by a green fluorescent protein labelled strain of Verticillium dahliae. Eur. J. Plant Pathol. 2013, 135, 867–876. [Google Scholar] [CrossRef]
- Blum, A.; Bressan, M.; Zahid, A.; Trinsoutrot-Gattin, I.; Driouich, A.; Laval, K. Verticillium wilt on fiber flax: Symptoms and pathogen development in planta. Plant Dis. 2018, 102, 2421–2429. [Google Scholar] [CrossRef]
- Eynck, C.; Koopmann, B.; Grunewaldt-Stoecker, G.; Karlovsky, P.; von Tiedemann, A. Differential interactions of Verticillium longisporum and V. dahliae with Brassica napus detected with molecular and histological techniques. Eur. J. Plant Pathol. 2007, 118, 259–274. [Google Scholar] [CrossRef]
- Zhao, P.; Zhao, Y.L.; Jin, Y.; Zhang, T.; Guo, H.S. Colonization process of Arabidopsis thaliana roots by a green fluorescent protein-tagged isolate of Verticillium dahliae. Protein Cell 2014, 5, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.L.; Zhou, T.T.; Guo, H.S. Hyphopodium-specific VdNoxB/VdPls1-dependent ROS-Ca2+ signaling is required for plant infection by Verticillium dahliae. PLoS Pathog. 2016, 12, e1005793. [Google Scholar] [CrossRef] [PubMed]
- Bishop, C.D.; Cooper, R.M. An ultrastructural study of root invasion in three vascular wilt diseases. Physiol. Plant Pathol. 1983, 22, 15-IN13. [Google Scholar] [CrossRef]
- Pegg, G.F.; Brady, B.L. Verticillium Wilts; CABI Publishing: Wallingford, UK, 2002. [Google Scholar]
- Garber, R.; Houston, B. Penetration and development of Verticillium alboatrum in the cotton plant. Phytopathology 1967, 56, 1121–1126. [Google Scholar]
- Frederick, Z.A.; Cummings, T.F.; Johnson, D.A. Susceptibility of weedy hosts from Pacific northwest potato production systems to crop-aggressive isolates of Verticillium dahliae. Plant Dis. 2017, 101, 1500–1506. [Google Scholar] [CrossRef] [PubMed]
- Ligoxigakis, E.K.; Vakalounakis, D.J.; Thanassoulopoulos, C.C. Weed hosts of Verticillium dahliae in crete: Susceptibility, symptomatology and significance. Phytoparasitica 2002, 30, 511–518. [Google Scholar] [CrossRef]
- Thanassoulopoulos, C.C.; Biris, D.A.; Tjamos, E.C. Weed hosts as inoculum source of Verticillium in olive orchards. Phytopathol. Mediterr. 1981, 20, 164–168. [Google Scholar]
- Vallad, G.E.; Bhat, R.G.; Koike, S.T.; Ryder, E.J.; Subbarao, K.V. Weedborne reservoirs and seed transmission of Verticillium dahliae in lettuce. Plant Dis. 2005, 89, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Woolliams, G.E. Host range and symptomatology of Verticillium dahliae in economic, weed, and native plants in interrior British Columbia. Can. J. Plant Sci. 1966, 46, 661–669. [Google Scholar] [CrossRef]
- Busch, L.; Smith, E.A.; Njoh-Elango, F. The effect of weeds on the value of rotation as a practical control for Verticillium wilt of potato. Can. Plant Dis. Surv. 1978, 58, 61. [Google Scholar]

| Isolate | Accession | VCG | Pathotype | Locality |
|---|---|---|---|---|
| V. dahliae “Vd71171” | BRIP71171 | 2A 1 | Non-defoliating | Namoi Valley, NSW, Australia |
| V. dahliae “Vd71181” | BRIP71181 | 1A 2 | Defoliating | Gwydir Valley, NSW, Australia |
| Score | Description of Symptoms |
|---|---|
| 0 | Healthy, no symptoms |
| 1 | 1–20% total leaf area affected |
| 2 | 21–40% total leaf area affected |
| 3 | 41–60% total leaf area affected |
| 4 | 61–80% total leaf area affected |
| 5 | 81–100% total leaf area affected, and or plant death |
| Symptoms | Affected Leaves (%) | Degree of Stunting Compared to Control 1 | ||
|---|---|---|---|---|
| None or Very Slight (<10%) | Moderate (11–50%) | Severe (>50%) | ||
| None | 0 | 0 | - | - |
| Slight leaf chlorosis, flaccidity, necrosis | 1–10 | 1 | 2 | 3 |
| Moderate leaf chlorosis, flaccidity, necrosis, slight defoliation | 2 | 3 | 4 | |
| 11–25 | ||||
| Severe leaf chlorosis, flaccidity, necrosis, moderate defoliation | 26–50 | 3 | 4 | 4 |
| Plants with severe or complete defoliation | >50 | 4 | 4 | 4 |
| Dead plants | – | 5 | 5 | 5 |
| Siokra 1–4 | Siokra 1–4 | Sicot 714B3F | Sicot 714B3F | |
|---|---|---|---|---|
| Treatment | Recovery/Total Diseased 1 | Recovery/Total Symptomless 2 | Recovery/Total Diseased 1 | Recovery/Total Symptomless 2 |
| Uninoculated | 0/0 | 0/48 (0%) | 0/0 | 0/48 (0%) |
| VCG2A-WT 3 | 31/33 (94%) | 1/15 (7%) | 20/24 (83%) | 0/24 (0%) |
| VCG2A-GFP3 3 | 24/24 (100%) | 1/24 (4%) | 7/18 (39%) | 1/30 (3%) |
| Family | Weed Plant Species | Isolate | Frequency 1 |
|---|---|---|---|
| Asteraceae | Conyza bonariensis L. | VCG2A-GFP3 | 4/26 (15.4%) |
| VCG1A-mCherry69 | 1/26 (3.8%) | ||
| Sonchus oleraceus L. | VCG2A-GFP3 | 2/25 (8%) | |
| VCG1A-mCherry69 | 0/25 (0%) | ||
| Poaceae | Avena fatua L. | VCG2A-GFP3 | 0/10 (0%) |
| VCG1A-mCherry69 | 0/10 (0%) | ||
| Chloris truncata R.Br. | VCG2A-GFP3 | 0/26 (0%) | |
| VCG1A-mCherry69 | 0/26 (0%) | ||
| Chloris virgata Sw. | VCG2A-GFP3 | 3/26 (11.5%) | |
| VCG1A-mCherry69 | 0/26 (0%) | ||
| Echinochloa colona L. | VCG2A-GFP3 | 2/26 (7.7%) | |
| VCG1A-mCherry69 | 2/26 (7.7%) | ||
| Urochloa panicoides P.Beauv. | VCG2A-GFP3 VCG1A-mCherry69 | 6/25 (24%) 1/25 (4%) | |
| Solanaceae | Nicotiana benthamiana Domin | VCG2A-GFP3 VCG1A-mCherry69 | 16/26 (61.5%) 13/26 (50%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, A.; Morrison, S.; Gregson, A.; Le, D.P.; Urquhart, A.S.; Smith, L.J.; Aitken, E.A.B.; Gardiner, D.M. Fluorescently Tagged Verticillium dahliae to Understand the Infection Process on Cotton (Gossypium hirsutum) and Weed Plant Species. Pathogens 2024, 13, 442. https://doi.org/10.3390/pathogens13060442
Chen A, Morrison S, Gregson A, Le DP, Urquhart AS, Smith LJ, Aitken EAB, Gardiner DM. Fluorescently Tagged Verticillium dahliae to Understand the Infection Process on Cotton (Gossypium hirsutum) and Weed Plant Species. Pathogens. 2024; 13(6):442. https://doi.org/10.3390/pathogens13060442
Chicago/Turabian StyleChen, Andrew, Sabrina Morrison, Aphrika Gregson, Duy P. Le, Andrew S. Urquhart, Linda J. Smith, Elizabeth A. B. Aitken, and Donald M. Gardiner. 2024. "Fluorescently Tagged Verticillium dahliae to Understand the Infection Process on Cotton (Gossypium hirsutum) and Weed Plant Species" Pathogens 13, no. 6: 442. https://doi.org/10.3390/pathogens13060442
APA StyleChen, A., Morrison, S., Gregson, A., Le, D. P., Urquhart, A. S., Smith, L. J., Aitken, E. A. B., & Gardiner, D. M. (2024). Fluorescently Tagged Verticillium dahliae to Understand the Infection Process on Cotton (Gossypium hirsutum) and Weed Plant Species. Pathogens, 13(6), 442. https://doi.org/10.3390/pathogens13060442






