The Trimeric Autotransporter Adhesin EmaA and Infective Endocarditis †
Abstract
1. Infectious Endocarditis
2. A. actinomycetemcomitans Physiology
3. A. actinomycetemcomitans Interactions with Collagen
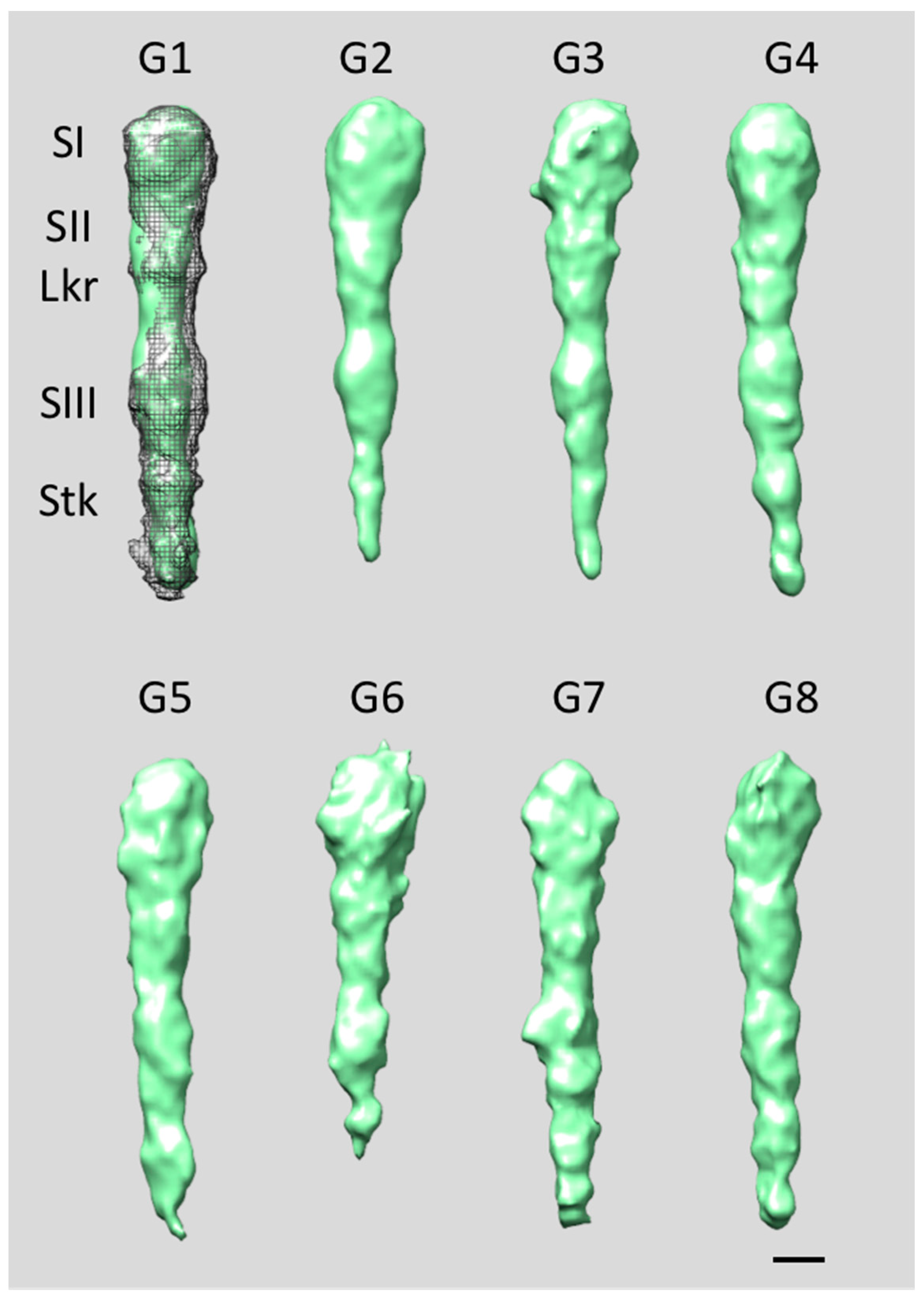
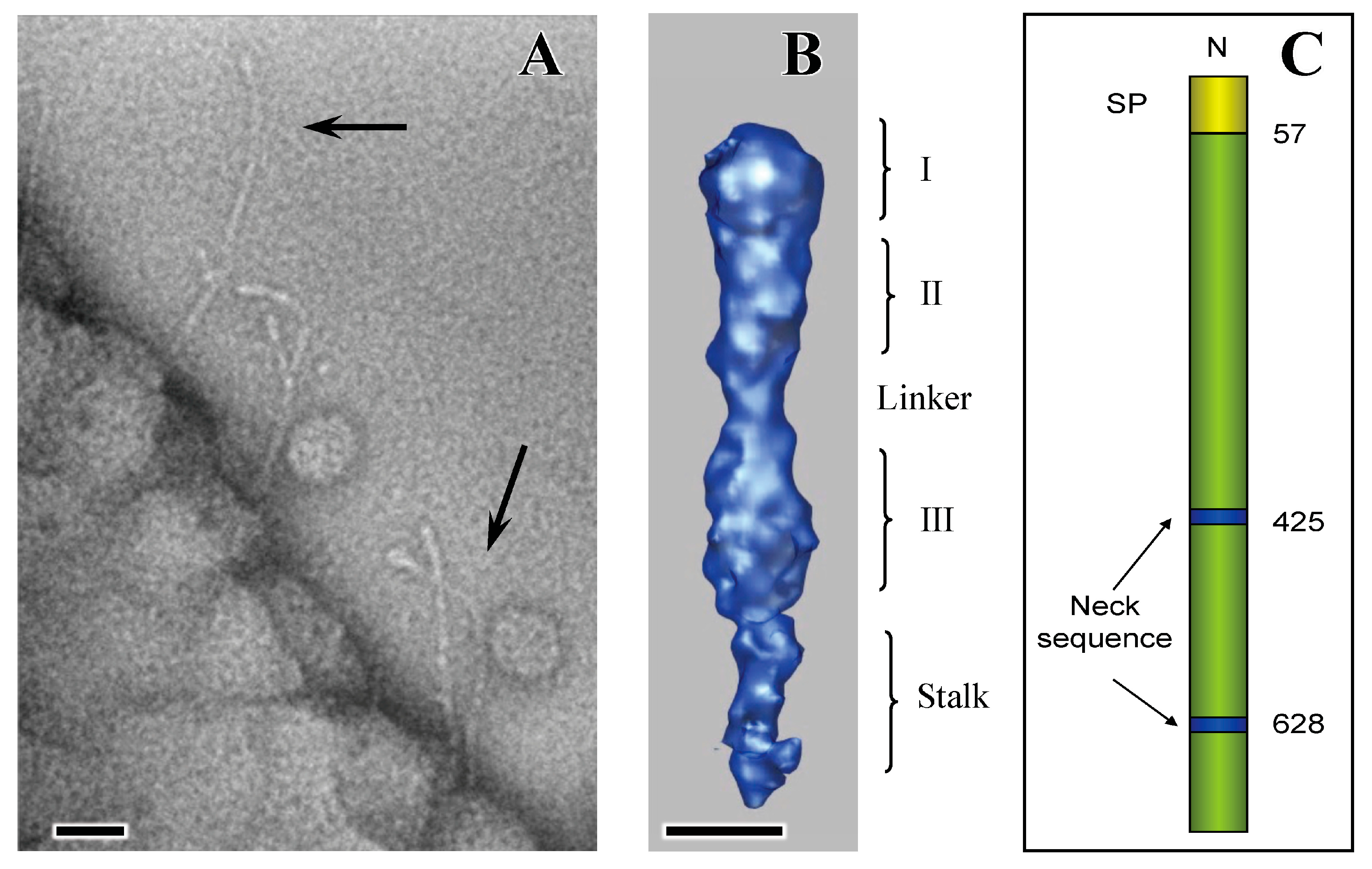
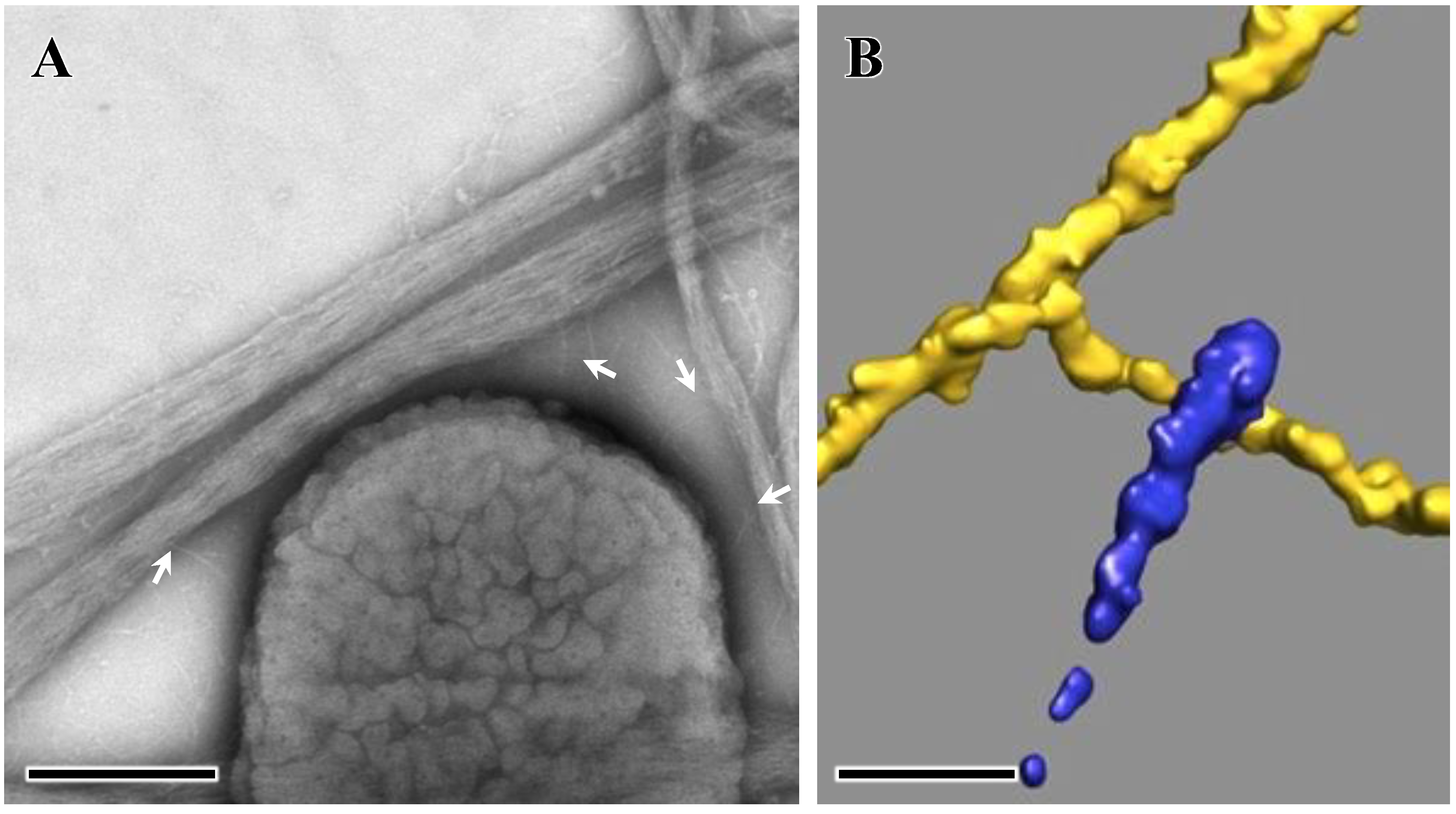
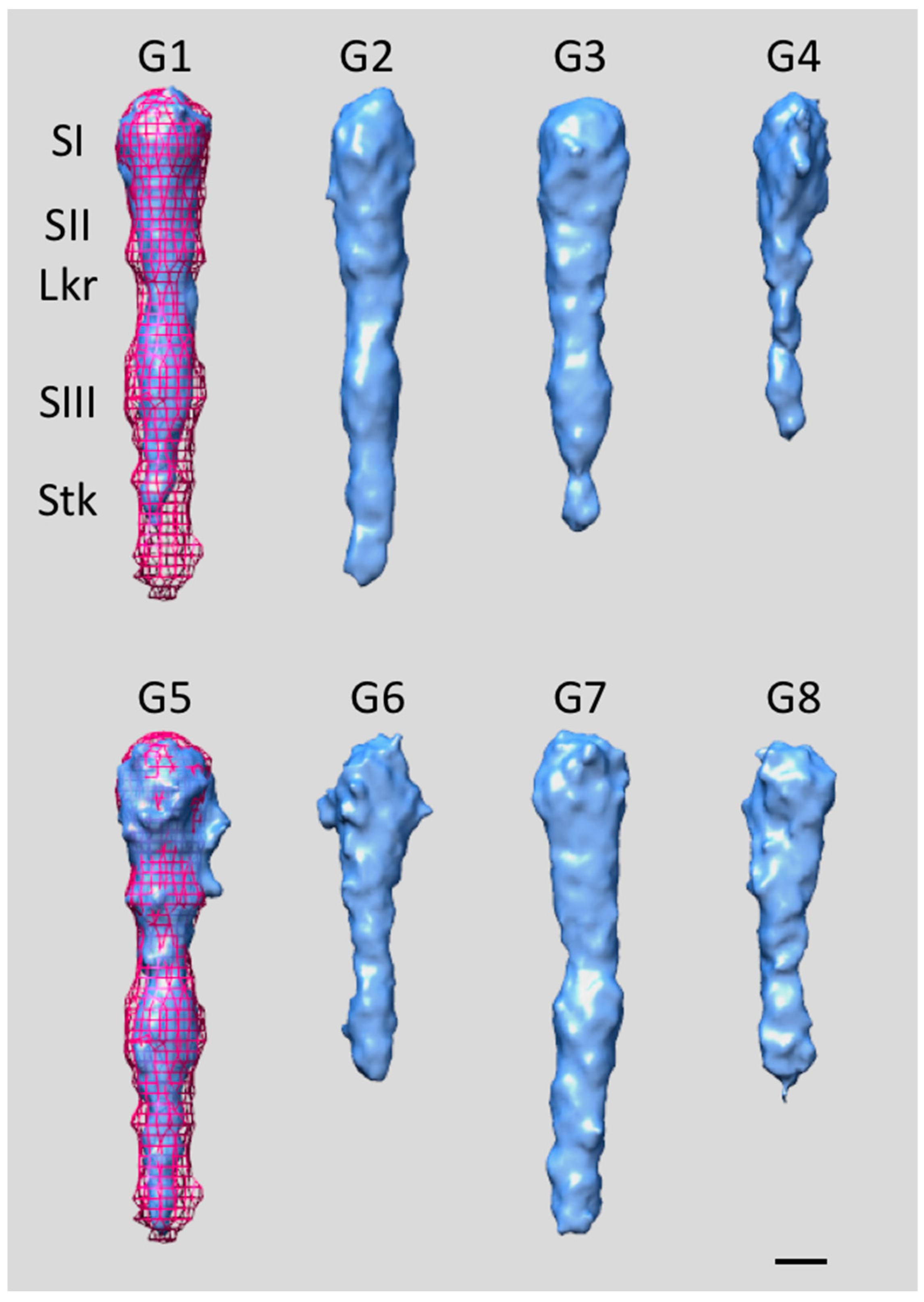
4. Extracellular Matrix Protein Adhesin A (EmaA)
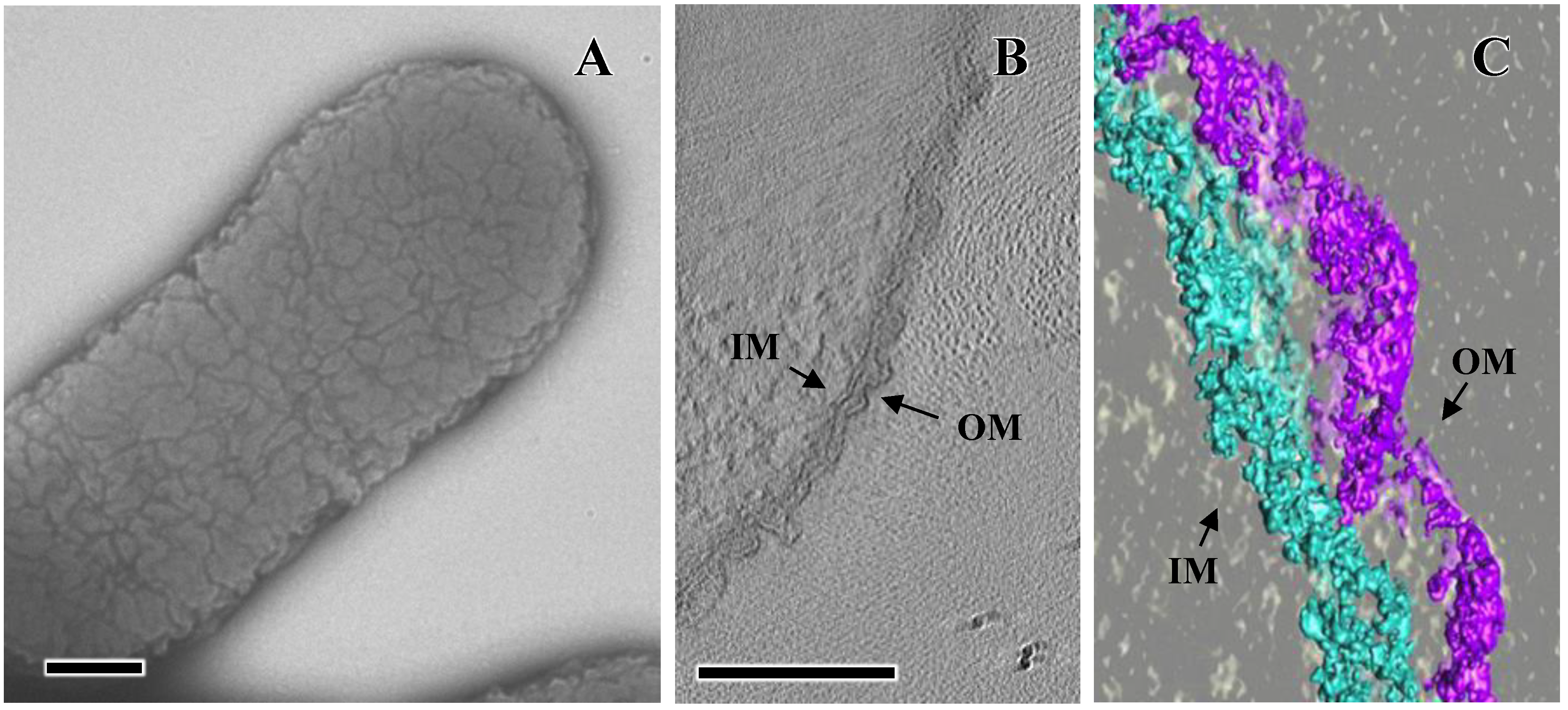
5. EmaA Interactions with Collagen
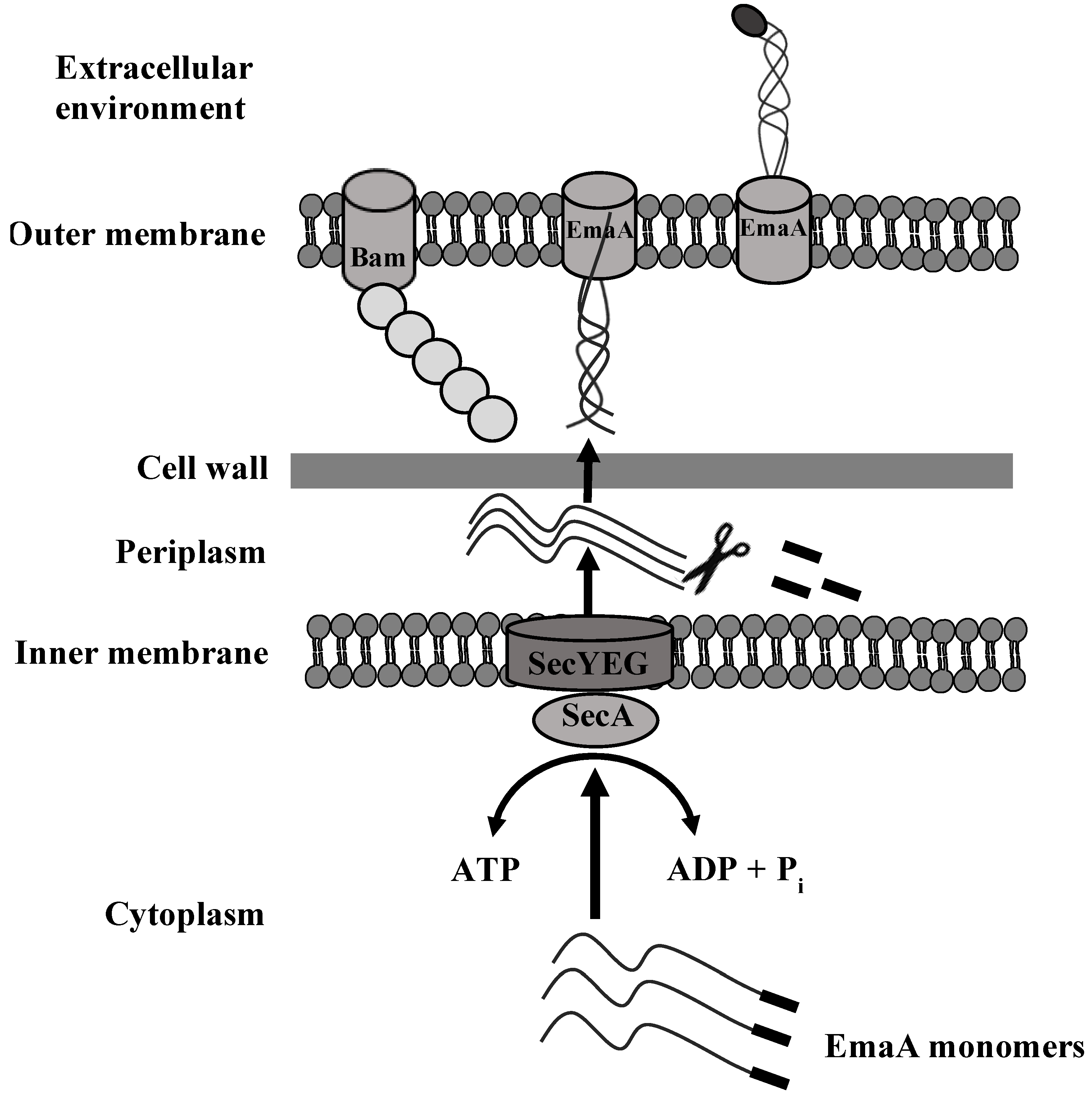
6. Secretion of EmaA and Cell Surface Expression
7. A. actinomycetemcomitans Serotypes and the Molecular Heterogeneity of EmaA
8. EmaA and Biofilm Formation
9. Transcriptional Control of emaA Expression
10. Summary and Conclusions
11. Future Directions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Holland, T.L.; Baddour, L.M.; Bayer, A.S.; Hoen, B.; Miro, J.M.; Fowler, V.G., Jr. Infective endocarditis. Nat. Rev. Dis. Primers 2016, 2, 16059. [Google Scholar] [CrossRef] [PubMed]
- Chambers, S.T.; Murdoch, D.; Morris, A.; Holland, D.; Pappas, P.; Almela, M.; Fernandez-Hidalgo, N.; Almirante, B.; Bouza, E.; Forno, D.; et al. HACEK infective endocarditis: Characteristics and outcomes from a large, multi-national cohort. PLoS ONE 2013, 8, e63181. [Google Scholar] [CrossRef]
- Cahill, T.J.; Prendergast, B.D. Infective endocarditis. Lancet 2016, 387, 882–893. [Google Scholar] [CrossRef] [PubMed]
- Akifusa, S.; Poole, S.; Lewthwaite, J.; Henderson, B.; Nair, S.P. Recombinant Actinobacillus actinomycetemcomitans cytolethal distending toxin proteins are required to interact to inhibit human cell cycle progression and to stimulate human leukocyte cytokine synthesis. Infect. Immun. 2001, 69, 5925–5930. [Google Scholar] [CrossRef] [PubMed]
- Paturel, L.; Casalta, J.P.; Habib, G.; Nezri, M.; Raoult, D. Actinobacillus actinomycetemcomitans endocarditis. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2004, 10, 98–118. [Google Scholar] [CrossRef] [PubMed]
- Petti, C.A.; Bhally, H.S.; Weinstein, M.P.; Joho, K.; Wakefield, T.; Reller, L.B.; Carroll, K.C. Utility of extended blood culture incubation for isolation of Haemophilus, Actinobacillus, Cardiobacterium, Eikenella, and Kingella organisms: A retrospective multicenter evaluation. J. Clin. Microbiol. 2006, 44, 257–259. [Google Scholar] [CrossRef] [PubMed]
- Blackberg, A.; Morenius, C.; Olaison, L.; Berge, A.; Rasmussen, M. Infective endocarditis caused by HACEK group bacteria—A registry-based comparative study. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 1919–1924. [Google Scholar] [CrossRef]
- Yew, H.S.; Chambers, S.T.; Roberts, S.A.; Holland, D.J.; Julian, K.A.; Raymond, N.J.; Beardsley, J.; Read, K.M.; Murdoch, D.R. Association between HACEK bacteraemia and endocarditis. J. Med. Microbiol. 2014, 63, 892–895. [Google Scholar] [CrossRef]
- Azari, F.; Nyland, L.; Yu, C.; Radermacher, M.; Mintz, K.P.; Ruiz, T. Ultrastructural analysis of the rugose cell envelope of a member of the Pasteurellaceae family. J. Bacteriol. 2013, 195, 1680–1688. [Google Scholar] [CrossRef]
- Gallant, C.V.; Sedic, M.; Chicoine, E.A.; Ruiz, T.; Mintz, K.P. Membrane morphology and leukotoxin secretion are associated with a novel membrane protein of Aggregatibacter actinomycetemcomitans. J. Bacteriol. 2008, 190, 5972–5980. [Google Scholar] [CrossRef]
- Smith, K.P.; Voogt, R.D.; Ruiz, T.; Mintz, K.P. The conserved carboxyl domain of MorC, an inner membrane protein of Aggregatibacter actinomycetemcomitans, is essential for membrane function. Mol. Oral. Microbiol. 2016, 31, 43–58. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.P.; Fields, J.G.; Voogt, R.D.; Deng, B.; Lam, Y.W.; Mintz, K.P. Alteration in abundance of specific membrane proteins of Aggregatibacter actinomycetemcomitans is attributed to deletion of the inner membrane protein MorC. Proteomics 2015, 15, 1859–1867. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.P.; Ruiz, T.; Mintz, K.P. Inner-membrane protein MorC is involved in fimbriae production and biofilm formation in Aggregatibacter actinomycetemcomitans. Microbiology 2016, 162, 513–525. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.P.; Fields, J.G.; Voogt, R.D.; Deng, B.; Lam, Y.W.; Mintz, K.P. The cell envelope proteome of Aggregatibacter actinomycetemcomitans. Mol. Oral. Microbiol. 2015, 30, 97–110. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, P.B.; Brennan, M.T.; Thornhill, M.; Michalowicz, B.S.; Noll, J.; Bahrani-Mougeot, F.K.; Sasser, H.C. Poor oral hygiene as a risk factor for infective endocarditis-related bacteremia. J. Am. Dent. Assoc. 2009, 140, 1238–1244. [Google Scholar] [CrossRef]
- Shoulders, M.D.; Raines, R.T. Collagen structure and stability. Annu. Rev. Biochem. 2009, 78, 929–958. [Google Scholar] [CrossRef]
- Halper, J. Basic components of connective tissues and extracellular matrix: Fibronectin, fibrinogen, laminin, elastin, fibrillins, fibulins, matrilins, tenascins and thrombospondins. Adv. Exp. Med. Biol. 2021, 1348, 105–126. [Google Scholar] [CrossRef]
- Westerlund, B.; Korhonen, T.K. Bacterial proteins binding to the mammalian extracellular matrix. Mol. Microbiol. 1993, 9, 687–694. [Google Scholar] [CrossRef]
- Carranza, F.A., Jr.; Saglie, R.; Newman, M.G.; Valentin, P.L. Scanning and transmission electron microscopic study of tissue-invading microorganisms in localized juvenile periodontitis. J. Periodontol. 1983, 54, 598–617. [Google Scholar] [CrossRef]
- Gillett, R.; Johnson, N.W. Bacterial invasion of the periodontium in a case of juvenile periodontitis. J. Clin. Periodontol. 1982, 9, 93–100. [Google Scholar] [CrossRef]
- Mintz, K.P.; Fives-Taylor, P.M. Binding of the periodontal pathogen Actinobacillus actinomycetemcomitans to extracellular matrix proteins. Oral. Microbiol. Immunol. 1999, 14, 109–116. [Google Scholar] [CrossRef]
- Alugupalli, K.R.; Kalfas, S.; Forsgren, A. Laminin binding to a heat-modifiable outer membrane protein of Actinobacillus actinomycetemcomitans. Oral. Microbiol. Immunol. 1996, 11, 326–331. [Google Scholar] [CrossRef] [PubMed]
- Cole, W.G.; Chan, D.; Hickey, A.J.; Wilcken, D.E. Collagen composition of normal and myxomatous human mitral heart valves. Biochem. J. 1984, 219, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Mintz, K.P. Identification of an extracellular matrix protein adhesin, EmaA, which mediates the adhesion of Actinobacillus actinomycetemcomitans to collagen. Microbiology 2004, 150, 2677–2688. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, N.; Tiller, M.; Anding, G.; Roggenkamp, A.; Heesemann, J.; Akifusa, S.; Poole, S.; Lewthwaite, J.; Henderson, B.; Nair, S.P. Contribution of trimeric autotransporter C-terminal domains of oligomeric coiled-coil adhesin (Oca) family members YadA, UspA1, EibA, and Hia to translocation of the YadA passenger domain and virulence of Yersinia enterocolitica. J. Bacteriol. 2008, 190, 5031–5043. [Google Scholar] [CrossRef]
- Chauhan, N.; Hatlem, D.; Orwick-Rydmark, M.; Schneider, K.; Floetenmeyer, M.; van Rossum, B.; Leo, J.C.; Linke, D. Insights into the autotransport process of a trimeric autotransporter, Yersinia Adhesin A (YadA). Mol. Microbiol. 2019, 111, 844–862. [Google Scholar] [CrossRef]
- Leo, J.C.; Grin, I.; Linke, D. Type V secretion: Mechanism(s) of autotransport through the bacterial outer membrane. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2012, 367, 1088–1101. [Google Scholar] [CrossRef]
- Hoiczyk, E.; Roggenkamp, A.; Reichenbecher, M.; Lupas, A.; Heesemann, J. Structure and sequence analysis of Yersinia YadA and Moraxella UspAs reveal a novel class of adhesins. EMBO J. 2000, 19, 5989–5999. [Google Scholar] [CrossRef]
- Laarmann, S.; Cutter, D.; Juehne, T.; Barenkamp, S.J.; St Geme, J.W. The Haemophilus influenzae Hia autotransporter harbours two adhesive pockets that reside in the passenger domain and recognize the same host cell receptor. Mol. Microbiol. 2002, 46, 731–743. [Google Scholar] [CrossRef]
- Lafontaine, E.R.; Cope, L.D.; Aebi, C.; Latimer, J.L.; McCracken, G.H., Jr.; Hansen, E.J. The UspA1 protein and a second type of UspA2 protein mediate adherence of Moraxella catarrhalis to human epithelial cells in vitro. J. Bacteriol. 2000, 182, 1364–1373. [Google Scholar] [CrossRef]
- Riess, T.; Andersson, S.G.; Lupas, A.; Schaller, M.; Schafer, A.; Kyme, P.; Martin, J.; Walzlein, J.H.; Ehehalt, U.; Lindroos, H.; et al. Bartonella adhesin A mediates a proangiogenic host cell response. J. Exp. Med. 2004, 200, 1267–1278. [Google Scholar] [CrossRef] [PubMed]
- Tahir, Y.E.; Kuusela, P.; Skurnik, M. Functional mapping of the Yersinia enterocolitica adhesin YadA. Identification Of eight NSVAIG—S motifs in the amino-terminal half of the protein involved in collagen binding. Mol. Microbiol. 2000, 37, 192–206. [Google Scholar] [CrossRef] [PubMed]
- Azari, F.; Radermacher, M.; Mintz, K.P.; Ruiz, T. Correlation of the amino-acid sequence and the 3D structure of the functional domain of EmaA from Aggregatibacter actinomycetemcomitans. J. Struct. Biol. 2012, 177, 439–446. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ruiz, T.; Lenox, C.; Radermacher, M.; Mintz, K.P. Novel surface structures are associated with the adhesion of Actinobacillus actinomycetemcomitans to collagen. Infect. Immun. 2006, 74, 6163–6170. [Google Scholar] [CrossRef]
- Yu, C.; Ruiz, T.; Lenox, C.; Mintz, K.P. Functional mapping of an oligomeric autotransporter adhesin of Aggregatibacter actinomycetemcomitans. J. Bacteriol. 2008, 190, 3098–3109. [Google Scholar] [CrossRef]
- Yu, C.; Mintz, K.P.; Ruiz, T. Investigation of the three-dimensional architecture of the collagen adhesin EmaA of Aggregatibacter actinomycetemcomitans by electron tomography. J. Bacteriol. 2009, 191, 6253–6261. [Google Scholar] [CrossRef]
- Tang, G.; Ruiz, T.; Barrantes-Reynolds, R.; Mintz, K.P. Molecular heterogeneity of EmaA, an oligomeric autotransporter adhesin of Aggregatibacter (Actinobacillus) actinomycetemcomitans. Microbiology 2007, 153, 2447–2457. [Google Scholar] [CrossRef][Green Version]
- Tang, G.; Kitten, T.; Munro, C.L.; Wellman, G.C.; Mintz, K.P. EmaA, a potential virulence determinant of Aggregatibacter actinomycetemcomitans in infective endocarditis. Infect. Immun. 2008, 76, 2316–2324. [Google Scholar] [CrossRef]
- Paik, S.; Senty, L.; Das, S.; Noe, J.C.; Munro, C.L.; Kitten, T. Identification of virulence determinants for endocarditis in Streptococcus sanguinis by signature-tagged mutagenesis. Infect. Immun. 2005, 73, 6064–6074. [Google Scholar] [CrossRef]
- Hook, E.W., 3rd; Sande, M.A. Role of the vegetation in experimental Streptococcus viridans endocarditis. Infect. Immun. 1974, 10, 1433–1438. [Google Scholar] [CrossRef]
- Bayer, A.S.; Coulter, S.N.; Stover, C.K.; Schwan, W.R. Impact of the high-affinity proline permease gene (putP) on the virulence of Staphylococcus aureus in experimental endocarditis. Infect. Immun. 1999, 67, 740–744. [Google Scholar] [CrossRef] [PubMed]
- Veltrop, M.H.; Bancsi, M.J.; Bertina, R.M.; Thompson, J. Role of monocytes in experimental Staphylococcus aureus endocarditis. Infect. Immun. 2000, 68, 4818–4821. [Google Scholar] [CrossRef]
- Azari, F.; Radermacher, M.; Mintz, K.P.; Ruiz, T. Interactions between the trimeric autotransporter adhesin EmaA and collagen revealed by three-dimensional electron tomography. J. Bacteriol. 2019, 201, e00297-19. [Google Scholar] [CrossRef] [PubMed]
- Ponnuraj, K.; Bowden, M.G.; Davis, S.; Gurusiddappa, S.; Moore, D.; Choe, D.; Xu, Y.; Hook, M.; Narayana, S.V. A "dock, lock, and latch" structural model for a staphylococcal adhesin binding to fibrinogen. Cell 2003, 115, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Zong, Y.; Xu, Y.; Liang, X.; Keene, D.R.; Hook, A.; Gurusiddappa, S.; Hook, M.; Narayana, S.V. A ‘Collagen Hug’ model for Staphylococcus aureus CNA binding to collagen. EMBO J. 2005, 24, 4224–4236. [Google Scholar] [CrossRef]
- Nummelin, H.; Merckel, M.C.; Leo, J.C.; Lankinen, H.; Skurnik, M.; Goldman, A. The Yersinia adhesin YadA collagen-binding domain structure is a novel left-handed parallel beta-roll. EMBO J. 2004, 23, 701–711. [Google Scholar] [CrossRef] [PubMed]
- Delepelaire, P. Type I secretion in gram-negative bacteria. Biochim. Biophys. Acta 2004, 1694, 149–161. [Google Scholar] [CrossRef]
- Pugsley, A.P. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 1993, 57, 50–108. [Google Scholar] [CrossRef]
- Zwizinski, C.; Wickner, W. Purification and characterization of leader (signal) peptidase from Escherichia coli. J. Biol. Chem. 1980, 255, 7973–7977. [Google Scholar] [CrossRef]
- Silhavy, T.J.; Kahne, D.; Walker, S. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000414. [Google Scholar] [CrossRef]
- Leyton, D.L.; Rossiter, A.E.; Henderson, I.R. From self sufficiency to dependence: Mechanisms and factors important for autotransporter biogenesis. Nat. Rev. Microbiol. 2012, 10, 213–225. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, H.D. Looks can be deceiving: Recent insights into the mechanism of protein secretion by the autotransporter pathway. Mol. Microbiol. 2015, 97, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Rossiter, A.E.; Leyton, D.L.; Tveen-Jensen, K.; Browning, D.F.; Sevastsyanovich, Y.; Knowles, T.J.; Nichols, K.B.; Cunningham, A.F.; Overduin, M.; Schembri, M.A.; et al. The essential beta-barrel assembly machinery complex components BamD and BamA are required for autotransporter biogenesis. J. Bacteriol. 2011, 193, 4250–4253. [Google Scholar] [CrossRef] [PubMed]
- Owji, H.; Nezafat, N.; Negahdaripour, M.; Hajiebrahimi, A.; Ghasemi, Y. A comprehensive review of signal peptides: Structure, roles, and applications. Eur. J. Cell Biol. 2018, 97, 422–441. [Google Scholar] [CrossRef] [PubMed]
- von Heijne, G. The signal peptide. J. Membr. Biol. 1990, 115, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Ruiz, T.; Mintz, K.P. The extended signal peptide of the trimeric autotransporter EmaA of Aggregatibacter actinomycetemcomitans modulates secretion. J. Bacteriol. 2011, 193, 6983–6994. [Google Scholar] [CrossRef]
- Hiss, J.A.; Schneider, G. Domain organization of long autotransporter signal sequences. Bioinform. Biol. Insights 2009, 3, 189–204. [Google Scholar] [CrossRef]
- Szabady, R.L.; Peterson, J.H.; Skillman, K.M.; Bernstein, H.D. An unusual signal peptide facilitates late steps in the biogenesis of a bacterial autotransporter. Proc. Natl. Acad. Sci. USA 2005, 102, 221–226. [Google Scholar] [CrossRef]
- Jiang, X.; Ruiz, T.; Mintz, K.P. Characterization of the secretion pathway of the collagen adhesin EmaA of Aggregatibacter actinomycetemcomitans. Mol. Oral. Microbiol. 2012, 27, 382–396. [Google Scholar] [CrossRef]
- Takada, K.; Saito, M.; Tsuzukibashi, O.; Kawashima, Y.; Ishida, S.; Hirasawa, M. Characterization of a new serotype g isolate of Aggregatibacter actinomycetemcomitans. Mol. Oral. Microbiol. 2010, 25, 200–206. [Google Scholar] [CrossRef]
- Kittichotirat, W.; Bumgarner, R.E.; Chen, C. Evolutionary divergence of Aggregatibacter actinomycetemcomitans. J. Dent. Res. 2016, 95, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Tang-Siegel, G.G.; Danforth, D.R.; Tristano, J.; Ruiz, T.; Mintz, K.P. The serotype a-EmaA adhesin of Aggregatibacter actinomycetemcomitans does not require O-PS synthesis for collagen binding activity. Microbiology 2022, 168, 001191. [Google Scholar] [CrossRef] [PubMed]
- Bertani, B.; Ruiz, N. Function and biogenesis of lipopolysaccharides. EcoSal Plus 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Tang, G.; Mintz, K.P. Glycosylation of the collagen adhesin EmaA of Aggregatibacter actinomycetemcomitans is dependent upon the lipopolysaccharide biosynthetic pathway. J. Bacteriol. 2010, 192, 1395–1404. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, Y.; Nakano, Y.; Nezu, T.; Yamashita, Y.; Koga, T. A novel NDP-6-deoxyhexosyl-4-ulose reductase in the pathway for the synthesis of thymidine diphosphate-D-fucose. J. Biol. Chem. 1999, 274, 16933–16939. [Google Scholar] [CrossRef]
- Yoshida, Y.; Nakano, Y.; Yamashita, Y.; Koga, T. Identification of a genetic locus essential for serotype b-specific antigen synthesis in Actinobacillus actinomycetemcomitans. Infect. Immun. 1998, 66, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Danforth, D.R.; Melloni, M.; Thorpe, R.; Cohen, A.; Voogt, R.; Tristano, J.; Mintz, K.P. Dual function of the O-antigen WaaL ligase of Aggregatibacter actinomycetemcomitans. Mol. Oral. Microbiol. 2023, 38, 471–488. [Google Scholar] [CrossRef]
- Tang, G.; Ruiz, T.; Mintz, K.P. O-polysaccharide glycosylation is required for stability and function of the collagen adhesin EmaA of Aggregatibacter actinomycetemcomitans. Infect. Immun. 2012, 80, 2868–2877. [Google Scholar] [CrossRef]
- Brooks, C.J.; Mintz, K.P.; Radermacher, M.; Ruiz, T. 3D structural analysis and classification of EmaA, a collagen binding adhesin. Microsc. Microanal. 2017, 23, 1260–1261. [Google Scholar] [CrossRef][Green Version]
- Brooks, C.J.; Ruiz, T.; Radermacher, M. The alignment and classification of 3D reconstructions of rod-like molecules obtained by electron tomography. Microsc. Microanal. 2017, 23, 1118–1119. [Google Scholar] [CrossRef][Green Version]
- Radermacher, M. A new environment for modular image reconstruction and data analysis. Microsc. Microanal. 2013, 19, 762–763. [Google Scholar] [CrossRef]
- Yu, L.; Snapp, R.R.; Ruiz, T.; Radermacher, M. Probabilistic principal component analysis with expectation maximization (PPCA-EM) facilitates volume classification and estimates the missing data. J. Struct. Biol. 2010, 171, 18–30. [Google Scholar] [CrossRef]
- Watson, A.; Naughton, H.; Radermacher, M.; Mintz, K.P.; Ruiz, T. Tomographic analysis of EmaA adhesin glycosylation in Aggregatibacter actinomycetemcomitans. Microsc. Microanal. 2015, 21, 899–900. [Google Scholar] [CrossRef]
- Watson, A.; Tang-Siegel, G.; Brooks, C.J.; Radermacher, M.; Mintz, K.P.; Ruiz, T. Structural significance of EmaA glycosylation in A. actinomycetemcomitans. Microsc. Microanal. 2016, 22, 1132–1133. [Google Scholar] [CrossRef]
- Tang, G.; Kawai, T.; Komatsuzawa, H.; Mintz, K.P. Lipopolysaccharides mediate leukotoxin secretion in Aggregatibacter actinomycetemcomitans. Mol. Oral. Microbiol. 2012, 27, 70–82. [Google Scholar] [CrossRef]
- Danforth, D.R.; Tang-Siegel, G.; Ruiz, T.; Mintz, K.P. A non-fimbrial adhesin of Aggregatibacter actinomycetemcomitans mediates biofilm biogenesis. Infect. Immun. 2018, 87, 70–82. [Google Scholar] [CrossRef]
- Fine, D.H.; Velliyagounder, K.; Furgang, D.; Kaplan, J.B. The Actinobacillus actinomycetemcomitans autotransporter adhesin Aae exhibits specificity for buccal epithelial cells from humans and old world primates. Infect. Immun. 2005, 73, 1947–1953. [Google Scholar] [CrossRef]
- Rose, J.E.; Meyer, D.H.; Fives-Taylor, P.M. Aae, an autotransporter involved in adhesion of Actinobacillus actinomycetemcomitans to epithelial cells. Infect. Immun. 2003, 71, 2384–2393. [Google Scholar] [CrossRef]
- Asakawa, R.; Komatsuzawa, H.; Kawai, T.; Yamada, S.; Goncalves, R.B.; Izumi, S.; Fujiwara, T.; Nakano, Y.; Suzuki, N.; Uchida, Y.; et al. Outer membrane protein 100, a versatile virulence factor of Actinobacillus actinomycetemcomitans. Mol. Microbiol. 2003, 50, 1125–1139. [Google Scholar] [CrossRef]
- Yue, G.; Kaplan, J.B.; Furgang, D.; Mansfield, K.G.; Fine, D.H. A second Aggregatibacter actinomycetemcomitans autotransporter adhesin exhibits specificity for buccal epithelial cells in humans and Old World primates. Infect. Immun. 2007, 75, 4440–4448. [Google Scholar] [CrossRef]
- Danforth, D.R.; Melloni, M.; Tristano, J.; Mintz, K.P. Contribution of adhesion proteins to Aggregatibacter actinomycetemcomitans biofilm formation. Mol. Oral. Microbiol. 2021, 36, 243–253. [Google Scholar] [CrossRef]
- Tristano, J.; Danforth, D.R.; Wargo, M.J.; Mintz, K.P. Regulation of adhesin synthesis in Aggregatibacter actinomycetemcomitans. Mol. Oral. Microbiol. 2023, 38, 237–250. [Google Scholar] [CrossRef]
- Price, N.L.; Raivio, T.L. Characterization of the Cpx regulon in Escherichia coli strain MC4100. J. Bacteriol. 2009, 191, 1798–1815. [Google Scholar] [CrossRef]
- Raivio, T.L.; Silhavy, T.J. Periplasmic stress and ECF sigma factors. Annu. Rev. Microbiol. 2001, 55, 591–624. [Google Scholar] [CrossRef]
- Danese, P.N.; Silhavy, T.J. The sigma(E) and the Cpx signal transduction systems control the synthesis of periplasmic protein-folding enzymes in Escherichia coli. Genes. Dev. 1997, 11, 1183–1193. [Google Scholar] [CrossRef]
- Danese, P.N.; Snyder, W.B.; Cosma, C.L.; Davis, L.J.; Silhavy, T.J. The Cpx two-component signal transduction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic protease, DegP. Genes. Dev. 1995, 9, 387–398. [Google Scholar] [CrossRef]
- Dorel, C.; Lejeune, P.; Rodrigue, A. The Cpx system of Escherichia coli, a strategic signaling pathway for confronting adverse conditions and for settling biofilm communities? Res. Microbiol. 2006, 157, 306–314. [Google Scholar] [CrossRef]
- Dorel, C.; Vidal, O.; Prigent-Combaret, C.; Vallet, I.; Lejeune, P. Involvement of the Cpx signal transduction pathway of E. coli in biofilm formation. FEMS Microbiol. Lett. 1999, 178, 169–175. [Google Scholar] [CrossRef]
- Jubelin, G.; Vianney, A.; Beloin, C.; Ghigo, J.M.; Lazzaroni, J.C.; Lejeune, P.; Dorel, C. CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J. Bacteriol. 2005, 187, 2038–2049. [Google Scholar] [CrossRef]
- Kershaw, C.J.; Brown, N.L.; Constantinidou, C.; Patel, M.D.; Hobman, J.L. The expression profile of Escherichia coli K-12 in response to minimal, optimal and excess copper concentrations. Microbiology 2005, 151, 1187–1198. [Google Scholar] [CrossRef] [PubMed]
- Nevesinjac, A.Z.; Raivio, T.L. The Cpx envelope stress response affects expression of the type IV bundle-forming pili of enteropathogenic Escherichia coli. J. Bacteriol. 2005, 187, 672–686. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, A.F.; Georgellis, D.; Brown, A.N.; Anderson, M.T.; Bachman, M.A.; Mobley, H.L.T. In vitro and in vivo analysis of the ArcB/A redox signaling pathway. Methods Enzymol. 2010, 471, 205–228. [Google Scholar] [CrossRef]
- Brown, A.N.; Anderson, M.T.; Bachman, M.A.; Mobley, H.L.T. The ArcAB two-component system: Function in metabolism, redox control, and infection. Microbiol. Mol. Biol. Rev. 2022, 86, e0011021. [Google Scholar] [CrossRef]
- Longo, P.; Ota-Tsuzuki, C.; Nunes, A.; Fernandes, B.; Mintz, K.; Fives-Taylor, P.; Mayer, M. Aggregatibacter actinomycetemcomitans arcB influences hydrophobic properties, biofilm formation and adhesion to hydroxyapatite. Braz. J. Microbiol. 2009, 40, 550–562. [Google Scholar] [CrossRef]
- Shalel-Levanon, S.; San, K.Y.; Bennett, G.N. Effect of ArcA and FNR on the expression of genes related to the oxygen regulation and the glycolysis pathway in Escherichia coli under microaerobic growth conditions. Biotechnol. Bioeng. 2005, 92, 147–159. [Google Scholar] [CrossRef]
- Pena-Sandoval, G.R.; Georgellis, D. The ArcB sensor kinase of Escherichia coli autophosphorylates by an intramolecular reaction. J. Bacteriol. 2010, 192, 1735–1739. [Google Scholar] [CrossRef]
- Sun, H.; Song, Y.; Chen, F.; Zhou, C.; Liu, P.; Fan, Y.; Zheng, Y.; Wan, X.; Feng, L. An ArcA-modulateds RNA in pathogenic Escherichia coli K1. Front. Microbiol. 2020, 11, 574833. [Google Scholar] [CrossRef]
- Xi, D.; Yang, S.; Liu, Q.; Li, Y.; Li, Y.; Yan, J.; Wang, X.; Ning, K.; Cao, B. The response regulator ArcA enhances biofilm formation in the vpsT manner under the anaerobic condition in Vibrio cholerae. Microb. Pathog. 2020, 144, 104197. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Li, Y.; Guo, X.; Wang, X.; Liu, F.; Li, A.; Cao, B. The effect of ArcA on the growth, motility, biofilm formation, and virulence of Plesiomonas shigelloides. BMC Microbiol. 2021, 21, 266. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Storz, G. Redox sensing by prokaryotic transcription factors. Biochem. Pharmacol. 2000, 59, 1–6. [Google Scholar] [CrossRef]
- Zheng, M.; Aslund, F.; Storz, G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 1998, 279, 1718–1721. [Google Scholar] [CrossRef]
- Figurski, D.H.; Fine, D.H.; Perez-Cheeks, B.A.; Grosso, V.W.; Kram, K.E.; Hua, J.; Xu, K.; Hedhli, J. Targeted mutagenesis in the study of the tightadherence (tad) locus of Aggregatibacter actinomycetemcomitans. In Genetic Manipulation of DNA and Protein; David, F., Ed.; IntechOpen: Rijeka, Croatia, 2013; p. Ch. 3. [Google Scholar]
- Ramsey, M.M.; Whiteley, M. Polymicrobial interactions stimulate resistance to host innate immunity through metabolite perception. Proc. Natl. Acad. Sci. USA 2009, 106, 1578–1583. [Google Scholar] [CrossRef]
- Valentin-Hansen, P.; Hojrup, P.; Short, S. The primary structure of the DeoR repressor from Escherichia coli K-12. Nucleic Acids Res. 1985, 13, 5927–5936. [Google Scholar] [CrossRef]
- Ishihama, A. Prokaryotic genome regulation: Multifactor promoters, multitarget regulators and hierarchic networks. FEMS Microbiol. Rev. 2010, 34, 628–645. [Google Scholar] [CrossRef]
- Saglie, F.R.; Carranza, F.A., Jr.; Newman, M.G.; Cheng, L.; Lewin, K.J. Identification of tissue-invading bacteria in human periodontal disease. J. Periodontal Res. 1982, 17, 452–455. [Google Scholar] [CrossRef]
- Chen, Y.C.; Chang, S.C.; Luh, K.T.; Hsieh, W.C. Actinobacillus actinomycetemcomitans endocarditis: A report of four cases and review of the literature. QJM An. Int. J. Med. 1991, 81, 871–878. [Google Scholar] [CrossRef]
- Mintz, K.P.; Fives-Taylor, P.M. Adhesion of Actinobacillus actinomycetemcomitans to a human oral cell line. Infect. Immun. 1994, 62, 3672–3678. [Google Scholar] [CrossRef]
- Schreiner, H.C.; Sinatra, K.; Kaplan, J.B.; Furgang, D.; Kachlany, S.C.; Planet, P.J.; Perez, B.A.; Figurski, D.H.; Fine, D.H. Tight-adherence genes of Actinobacillus actinomycetemcomitans are required for virulence in a rat model. Proc. Natl. Acad. Sci. USA 2003, 100, 7295–7300. [Google Scholar] [CrossRef]
- Fine, D.H.; Goncharoff, P.; Schreiner, H.; Chang, K.M.; Furgang, D.; Figurski, D. Colonization and persistence of rough and smooth colony variants of Actinobacillus actinomycetemcomitans in the mouths of rats. Arch. Oral. Biol. 2001, 46, 1065–1078. [Google Scholar] [CrossRef] [PubMed]
- Kram, K.E.; Hovel-Miner, G.A.; Tomich, M.; Figurski, D.H. Transcriptional regulation of the tad locus in Aggregatibacter actinomycetemcomitans: A termination cascade. J. Bacteriol. 2008, 190, 3859–3868. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mintz, K.P.; Danforth, D.R.; Ruiz, T. The Trimeric Autotransporter Adhesin EmaA and Infective Endocarditis. Pathogens 2024, 13, 99. https://doi.org/10.3390/pathogens13020099
Mintz KP, Danforth DR, Ruiz T. The Trimeric Autotransporter Adhesin EmaA and Infective Endocarditis. Pathogens. 2024; 13(2):99. https://doi.org/10.3390/pathogens13020099
Chicago/Turabian StyleMintz, Keith P., David R. Danforth, and Teresa Ruiz. 2024. "The Trimeric Autotransporter Adhesin EmaA and Infective Endocarditis" Pathogens 13, no. 2: 99. https://doi.org/10.3390/pathogens13020099
APA StyleMintz, K. P., Danforth, D. R., & Ruiz, T. (2024). The Trimeric Autotransporter Adhesin EmaA and Infective Endocarditis. Pathogens, 13(2), 99. https://doi.org/10.3390/pathogens13020099