Novel Antigenic Variant Infectious Bursal Disease Virus Outbreaks in Japan from 2014 to 2023 and Characterization of an Isolate from Chicken
Abstract
1. Introduction
2. Materials and Methods
2.1. Detection of IBDV Strains in Japan
2.2. Chickens and Eggs
2.3. Viral Nucleic Acid Extraction
2.4. IBDV Detection Using PCR and Segment A Sequencing of IBDV
2.5. Calculation of EID50 in Embryonating Hen Eggs
2.6. Isolation and Propagation of IBDV B2977 Isolate from a Field Sample
2.7. Sequence Alignment and Phylogenetic Tree Analysis of IBDV
2.8. Pathogenesis of Novel Variant IBDV B2977CE2C3 Strain in SPF Chickens
2.9. Serological Test for Anti-IBDV Antibodies
3. Results
3.1. Detection of Novel Antigenic Variant IBDV Strains in Japan
3.2. Isolation of Novel Variant IBDV
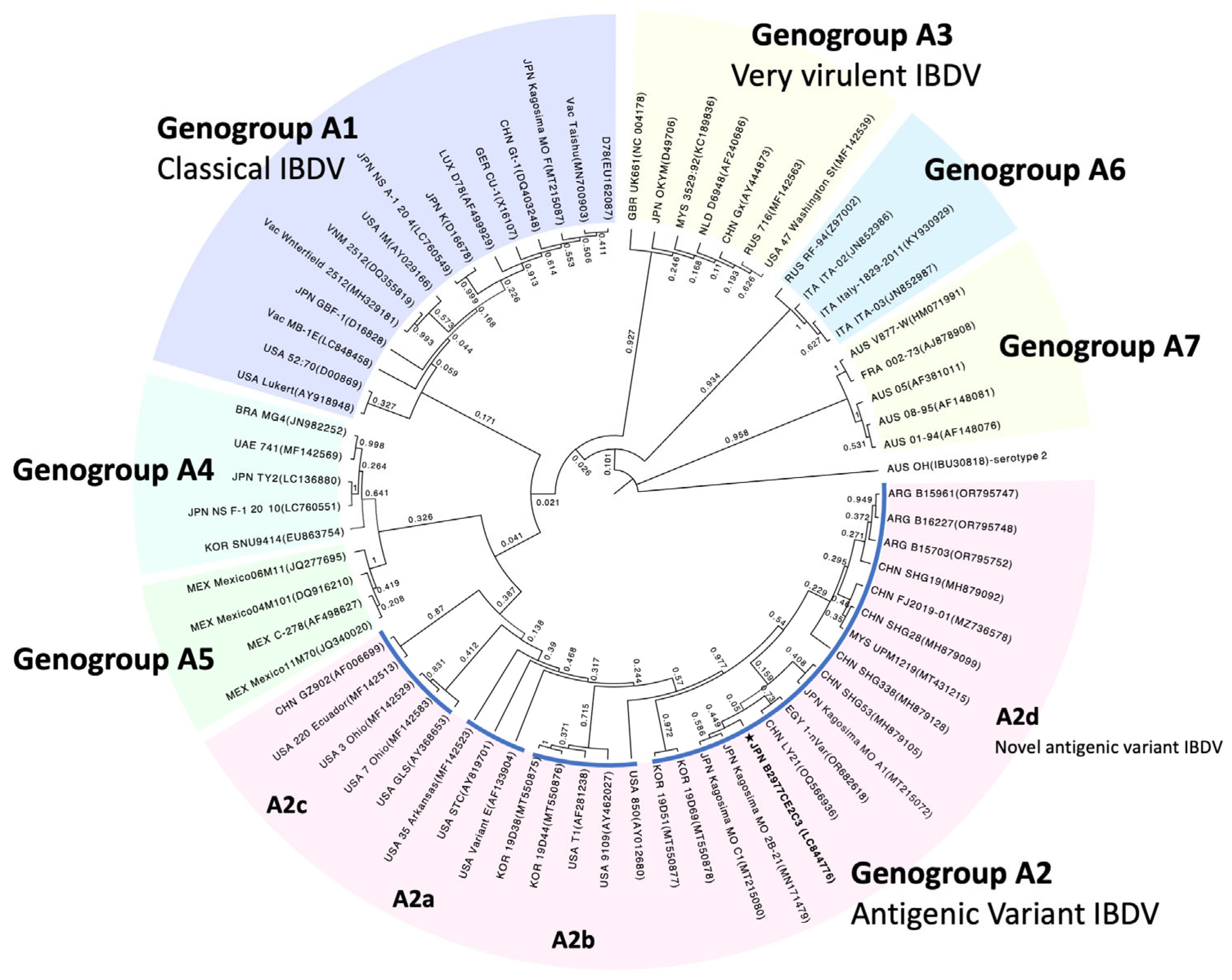
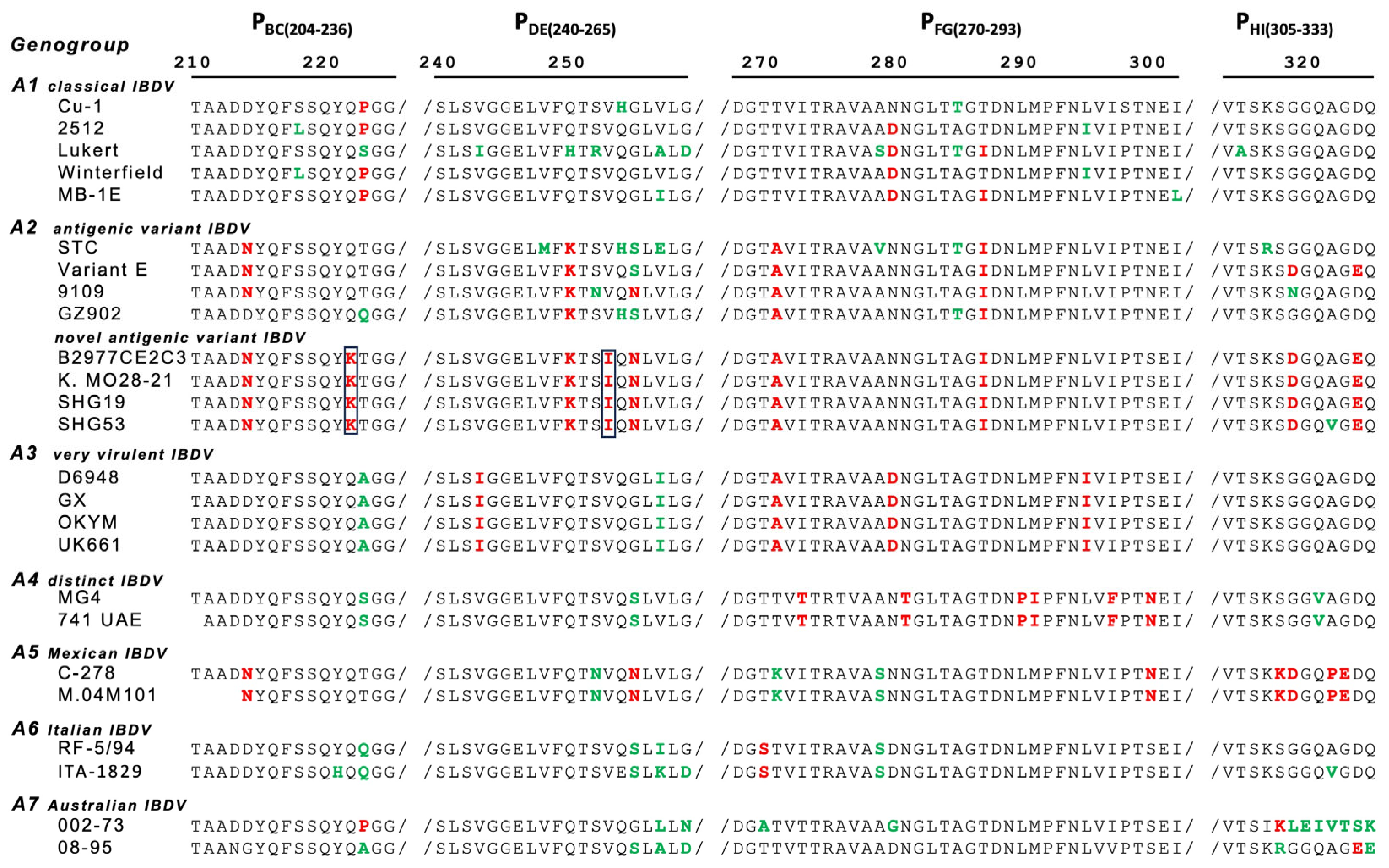
3.3. Segment A Polyprotein ORF Sequence and Phylogenic Tree Analysis
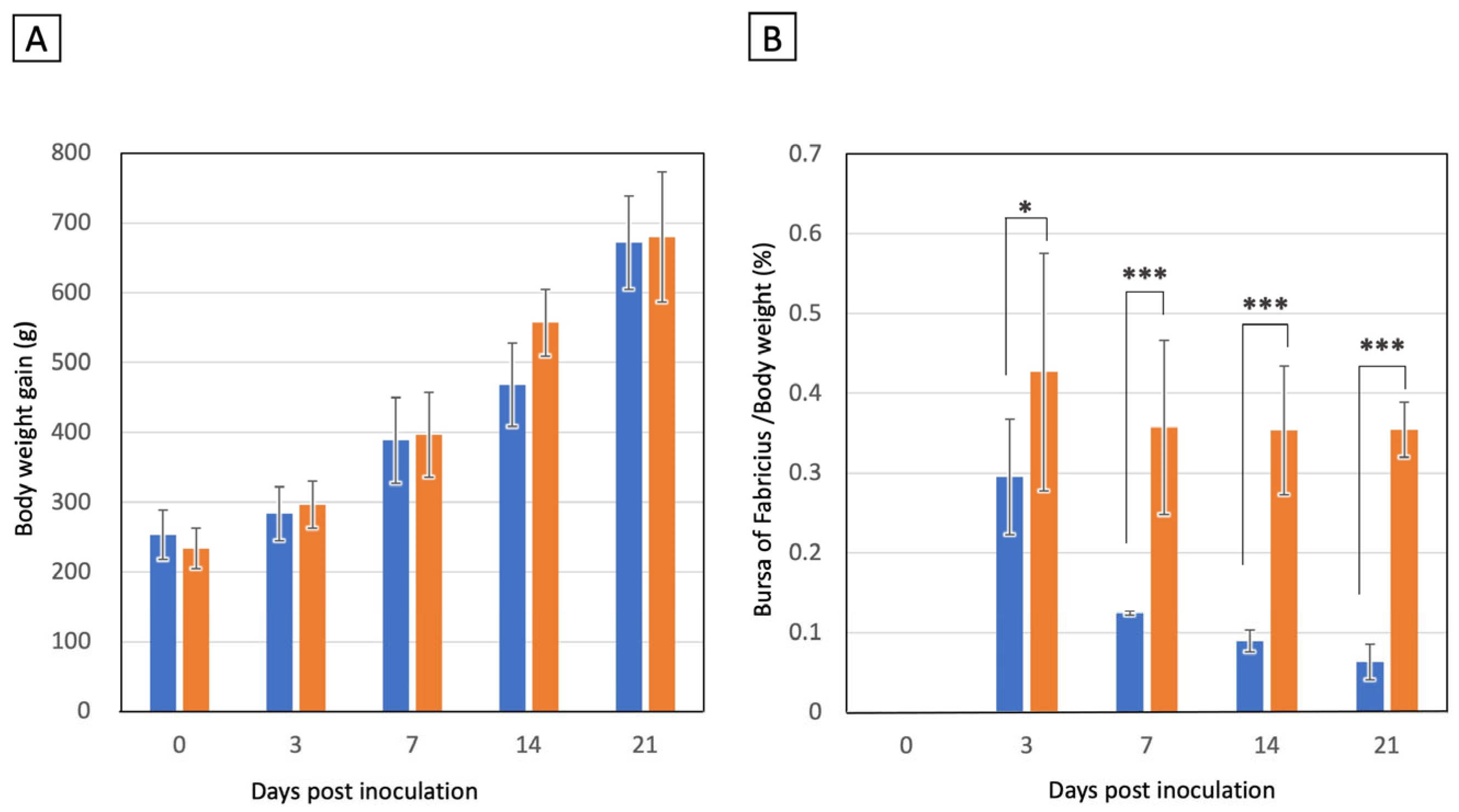
3.4. Experimental Chicken Infection of IBDV B2977CE2C3 Strain
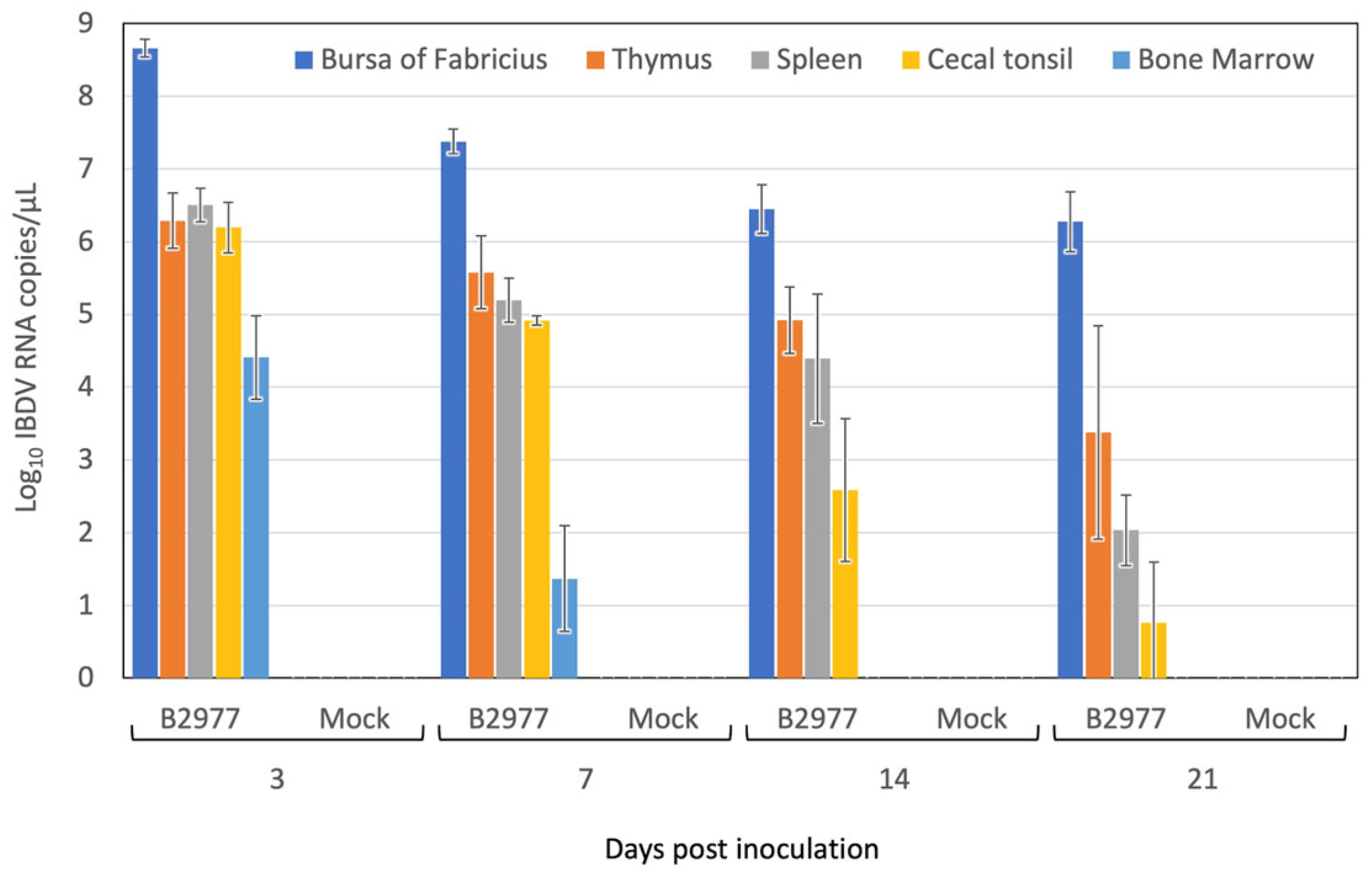
3.5. IBDV Replication in the Chickens
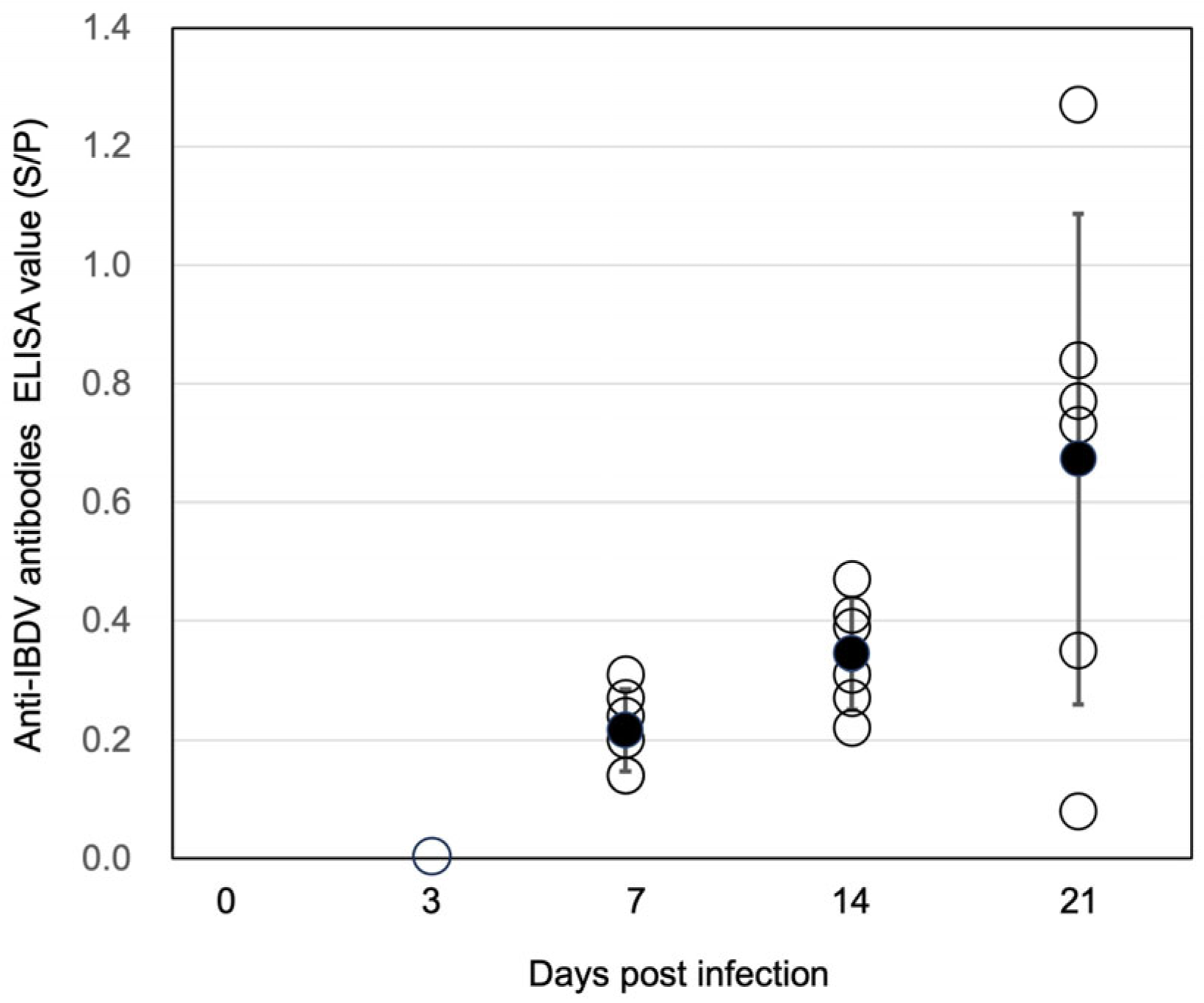
3.6. Anti-IBDV Antibodies in the Chickens
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lasher, H.N.; Davis, V.S. History of Infectious Bursal Disease in the U.S.A.: The First Two Decades. Avian Dis. 1997, 41, 11. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.U.; Li, Y.; Ouyang, W.; Wang, Z.; Zuo, J.; Shi, S.; Yu, Q.; Lin, J.; Yang, Q. From Nasal to Basal: Single-Cell Sequencing of the Bursa of Fabricius Highlights the IBDV Infection Mechanism in Chickens. Cell Biosci. 2021, 11, 212. [Google Scholar] [CrossRef]
- Dobos, P.; Hill, B.J.; Hallett, R.; Kells, D.T.; Becht, H.; Teninges, D. Biophysical and Biochemical Characterization of Five Animal Viruses with Bisegmented Double-Stranded RNA Genomes. J. Virol. 1979, 32, 593–605. [Google Scholar] [CrossRef] [PubMed]
- Fahey, K.J.; Erny, K.; Crooks, J. A Conformational Immunogen on VP-2 of Infectious Bursal Disease Virus That Induces Virus-Neutralizing Antibodies That Passively Protect Chickens. J. Gen. Virol. 1989, 70, 1473–1481. [Google Scholar] [CrossRef]
- Bao, K.; Qi, X.; Li, Y.; Gong, M.; Wang, X.; Zhu, P. Cryo-EM Structures of Infectious Bursal Disease Viruses with Different Virulences Provide Insights into Their Assembly and Invasion. Sci. Bull. 2022, 67, 646–654. [Google Scholar] [CrossRef] [PubMed]
- Coulibaly, F.; Chevalier, C.; Gutsche, I.; Pous, J.; Navaza, J.; Bressanelli, S.; Delmas, B.; Rey, F.A. The Birnavirus Crystal Structure Reveals Structural Relationships among Icosahedral Viruses. Cell 2005, 120, 761–772. [Google Scholar] [CrossRef]
- Huang, Y.; Shu, G.; Huang, C.; Han, J.; Li, J.; Chen, H.; Chen, Z. Characterization and Pathogenicity of a Novel Variant Infectious Bursal Disease Virus in China. Front. Microbiol. 2022, 13, 1039259. [Google Scholar] [CrossRef]
- Islam, M.R.; Nooruzzaman, M.; Rahman, T.; Mumu, T.T.; Rahman, M.M.; Chowdhury, E.H.; Eterradossi, N.; Müller, H. A Unified Genotypic Classification of Infectious Bursal Disease Virus Based on Both Genome Segments. Avian Pathol. 2021, 50, 190–206. [Google Scholar] [CrossRef] [PubMed]
- Michel, L.O.; Jackwood, D.J. Classification of Infectious Bursal Disease Virus into Genogroups. Arch. Virol. 2017, 162, 3661–3670. [Google Scholar] [CrossRef]
- Mata, C.P.; Mertens, J.; Fontana, J.; Luque, D.; Allende-Ballestero, C.; Reguera, D.; Trus, B.L.; Steven, A.C.; Carrascosa, J.L.; Castón, J.R. The RNA-Binding Protein of a Double-Stranded RNA Virus Acts like a Scaffold Protein. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Ganguly, B.; Rastogi, S.K. Structural and Functional Modeling of Viral Protein 5 of Infectious Bursal Disease Virus. Virus Res. 2018, 247, 55–60. [Google Scholar] [CrossRef]
- Hirai, K.; Calnek, B.W. In Vitro Replication of Infectious Bursal Disease Virus in Established Lymphoid Cell Lines and Chicken B Lymphocytes. Infect. Immun. 1979, 25, 964–970. [Google Scholar] [CrossRef]
- Armstrong, L.D.; Tabel, H.; Riddell, C. Subclinical Infectious Bursal Disease in Commercial Broiler Flocks in Saskatchewan. Can. J. Comp. Med. 1981, 45, 26–33. [Google Scholar] [PubMed]
- Nunoya, T.; Otaki, Y.; Tajima, M.; Hiraga, M.; Saito, T. Occurrence of Acute Infectious Bursal Disease with High Mortality in Japan and Pathogenicity of Field Isolates in Specific-Pathogen-Free Chickens. Avian Dis. 1992, 36, 597. [Google Scholar] [CrossRef] [PubMed]
- Van Den Berg, T.P.D. Acute Infectious Bursal Disease in Poultry: A Review. Avian Pathol. 2000, 29, 175–194. [Google Scholar] [CrossRef] [PubMed]
- Khatri, M.; Palmquist, J.M.; Cha, R.M.; Sharma, J.M. Infection and Activation of Bursal Macrophages by Virulent Infectious Bursal Disease Virus. Virus Res. 2005, 113, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Inoue, M.; Yamamoto, H.; Matuo, K.; Hihara, H. Susceptibility of Chicken Monocytic Cell Lines to Infectious Bursal Disease Virus. J. Vet. Med. Sci. 1992, 54, 575–577. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.-W.; Lo, C.-W.; Lai, S.-Y.; Fan, R.-J.; Lo, C.-J.; Chou, Y.; Thiruvengadam, R.; Wang, A.H.-J.; Wang, M.-Y. Chicken Heat Shock Protein 90 Is a Component of the Putative Cellular Receptor Complex of Infectious Bursal Disease Virus. J. Virol. 2007, 81, 8730–8741. [Google Scholar] [CrossRef] [PubMed]
- Delgui, L.; Oña, A.; Gutiérrez, S.; Luque, D.; Navarro, A.; Castón, J.R.; Rodríguez, J.F. The Capsid Protein of Infectious Bursal Disease Virus Contains a Functional A4β1 Integrin Ligand Motif. Virology 2009, 386, 360–372. [Google Scholar] [CrossRef] [PubMed]
- Chi, J.; You, L.; Li, P.; Teng, M.; Zhang, G.; Luo, J.; Wang, A. Surface IgM λ Light Chain Is Involved in the Binding and Infection of Infectious Bursal Disease Virus (IBDV) to DT40 Cells. Virus Genes 2018, 54, 236–245. [Google Scholar] [CrossRef]
- Luo, J.; Zhang, H.; Teng, M.; Fan, J.M.; You, L.M.; Xiao, Z.J.; Yi, M.L.; Zhi, Y.B.; Li, X.W.; Zhang, G.P. Surface IgM on DT40 Cells May Be a Component of the Putative Receptor Complex Responsible for the Binding of Infectious Bursal Disease Virus. Avian Pathol. 2010, 39, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Pan, Q.; Li, Y.; Yan, N.; Wang, J.; Yang, B.; Chen, Z.; Qi, X.; Gao, Y.; Gao, L.; et al. Identification of Chicken CD74 as a Novel Cellular Attachment Receptor for Infectious Bursal Disease Virus in Bursa B Lymphocytes. J. Virol. 2020, 94. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Pan, Q.; Wang, S.; Zhang, Y.; Li, Y.; Wang, Y.; Qi, X.; Gao, L.; Liu, C.; Zhang, Y.; et al. Identification of Chicken CD44 as a Novel B Lymphocyte Receptor for Infectious Bursal Disease Virus. J. Virol. 2022, 96, e0011322. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wang, X.; Gao, Y.; Qi, X. The Over-40-Years-Epidemic of Infectious Bursal Disease Virus in China. Viruses 2022, 14, 2253. [Google Scholar] [CrossRef]
- Fan, L.; Wu, T.; Hussain, A.; Gao, Y.; Zeng, X.; Wang, Y.; Gao, L.; Li, K.; Wang, Y.; Liu, C.; et al. Novel Variant Strains of Infectious Bursal Disease Virus Isolated in China. Vet. Microbiol. 2019, 230, 212–220. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; He, X.; Zhang, Y.; Qiao, Y.; Shi, J.; Chen, R.; Chen, J.; Xiang, Y.; Wang, Z.; Chen, G.; et al. Analysis of the Global Origin, Evolution and Transmission Dynamics of the Emerging Novel Variant IBDV (A2dB1b): The Accumulation of Critical Aa-Residue Mutations and Commercial Trade Contributes to the Emergence and Transmission of Novel Variants. Transbound. Emerg. Dis. 2022, 69, e2832–e2851. [Google Scholar] [CrossRef]
- Thai, T.N.; Jang, I.; Kim, H.A.; Kim, H.S.; Kwon, Y.K.; Kim, H.R. Characterization of Antigenic Variant Infectious Bursal Disease Virus Strains Identified in South Korea. Avian Pathol. 2021, 50, 174–181. [Google Scholar] [CrossRef]
- Myint, O.; Suwanruengsri, M.; Araki, K.; Izzati, U.Z.; Pornthummawat, A.; Nueangphuet, P.; Fuke, N.; Hirai, T.; Jackwood, D.J.; Yamaguchi, R. Bursa Atrophy at 28 Days Old Caused by Variant Infectious Bursal Disease Virus Has a Negative Economic Impact on Broiler Farms in Japan. Avian Pathol. 2021, 50, 6–17. [Google Scholar] [CrossRef] [PubMed]
- Aliyu, H.B.; Hair-Bejo, M.; Omar, A.R.; Ideris, A. Genetic Diversity of Recent Infectious Bursal Disease Viruses Isolated From Vaccinated Poultry Flocks in Malaysia. Front. Vet. Sci. 2021, 8, 643976. [Google Scholar] [CrossRef] [PubMed]
- Legnardi, M.; Poletto, F.; Talaat, S.; Selim, K.; Moawad, M.K.; Franzo, G.; Tucciarone, C.M.; Cecchinato, M.; Sultan, H. First Detection and Molecular Characterization of Novel Variant Infectious Bursal Disease Virus (Genotype A2dB1b) in Egypt. Viruses 2023, 15, 2388. [Google Scholar] [CrossRef]
- Jaton, J.; Lozano, L.C.; Gambini, P.; Ponti, M.; Gómez, E.; König, G.A.; Chimeno Zoth, S. Research Note: Characterization and Phylodynamic Analysis of New Infectious Bursal Disease Virus Variants Circulating in Argentina. Poult. Sci. 2024, 103, 103623. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Fan, L.; Jiang, N.; Gao, L.; Li, K.; Gao, Y.; Liu, C.; Cui, H.; Pan, Q.; Zhang, Y.; et al. An Improved Scheme for Infectious Bursal Disease Virus Genotype Classification Based on Both Genome-Segments A and B. J. Integr. Agric. 2021, 20, 1372–1381. [Google Scholar] [CrossRef]
- Jiang, N.; Wang, Y.; Zhang, W.; Niu, X.; Huang, M.; Gao, Y.; Liu, A.; Gao, L.; Li, K.; Pan, Q.; et al. Genotyping and Molecular Characterization of Infectious Bursal Disease Virus Identified in Important Poultry-Raising Areas of China During 2019 and 2020. Front Vet Sci 2021, 8, 759861. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Kato, A.; Otaki, Y.; Nakamura, T.; Sasmaz, E.; Ueda, S. Sequence Comparisons of a Highly Virulent Infectious Bursal Disease Virus Prevalent in Japan. Avian Dis. 1993, 37, 315. [Google Scholar] [CrossRef] [PubMed]
- Tomás, G.; Hernández, M.; Marandino, A.; Techera, C.; Grecco, S.; Hernández, D.; Banda, A.; Panzera, Y.; Pérez, R. Development of an RT-QPCR Assay for the Specific Detection of a Distinct Genetic Lineage of the Infectious Bursal Disease Virus. Avian Pathol. 2017, 46, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Hou, B.; Wang, C.Y.; Luo, Z.B.; Shao, G.Q. Commercial Vaccines Used in China Do Not Protect against a Novel Infectious Bursal Disease Virus Variant Isolated in Fujian. Vet. Rec. 2022, 191, e1840. [Google Scholar] [CrossRef]
- Fan, L.; Wang, Y.; Jiang, N.; Gao, Y.; Niu, X.; Zhang, W.; Huang, M.; Bao, K.; Liu, A.; Wang, S.; et al. Residues 318 and 323 in Capsid Protein Are Involved in Immune Circumvention of the Atypical Epizootic Infection of Infectious Bursal Disease Virus. Front. Microbiol. 2022, 13, 909252. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Wu, T.; Wang, Y.; Hussain, A.; Jiang, N.; Gao, L.; Li, K.; Gao, Y.; Liu, C.; Cui, H.; et al. Novel Variants of Infectious Bursal Disease Virus Can Severely Damage the Bursa of Fabricius of Immunized Chickens. Vet. Microbiol. 2020, 240, 108507. [Google Scholar] [CrossRef]
- Wang, C.; Hou, B. The Booster Immunization Using Commercial Vaccines Effectively Protect Chickens against Novel Variants of Infectious Bursal Disease Virus (Genotype A2dB1). Poult. Sci. 2024, 103, 103552. [Google Scholar] [CrossRef] [PubMed]
- Tomás, G.; Marandino, A.; Courtillon, C.; Amelot, M.; Keita, A.; Pikula, A.; Hernández, M.; Hernández, D.; Vagnozzi, A.; Panzera, Y.; et al. Antigenicity, Pathogenicity and Immunosuppressive Effect Caused by a South American Isolate of Infectious Bursal Disease Virus Belonging to the “Distinct” Genetic Lineage. Avian Pathol. 2019, 48, 245–254. [Google Scholar] [CrossRef]
- Li, K.; Niu, X.; Jiang, N.; Zhang, W.; Wang, G.; Li, K.; Huang, M.; Gao, Y.; Qi, X.; Wang, X. Comparative Pathogenicity of Three Strains of Infectious Bursal Disease Virus Closely Related to Poultry Industry. Viruses 2023, 15, 1257. [Google Scholar] [CrossRef] [PubMed]
- Nooruzzaman, M.; Hossain, I.; Rahman, M.M.; Uddin, A.J.; Mustari, A.; Parvin, R.; Chowdhury, E.H.; Islam, M.R. Comparative Pathogenicity of Infectious Bursal Disease Viruses of Three Different Genotypes. Microb. Pathog. 2022, 169, 105641. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.S.; Rissi, D.R.; Swayne, D.E. Very Virulent Infectious Bursal Disease Virus Produces More-Severe Disease and Lesions in Specific-Pathogen-Free (SPF) Leghorns Than in SPF Broiler Chickens. Avian Dis. 2015, 60, 63–66. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Wang, Y.; Jiang, N.; Chen, M.; Gao, L.; Li, K.; Gao, Y.; Cui, H.; Pan, Q.; Liu, C.; et al. Novel Variant Infectious Bursal Disease Virus Suppresses Newcastle Disease Vaccination in Broiler and Layer Chickens. Poult. Sci. 2020, 99, 6542–6548. [Google Scholar] [CrossRef] [PubMed]
| Fiscal Year | Total Test Numbers * | Genogroup A1 | Genogroup A2 | Genogroup A3 | Genogroup A4 | Novel AntigenicVariants/Total (%) | |
|---|---|---|---|---|---|---|---|
| Classical | Antigenic Variant | Novel Antigenic Variant | Very Virulent | Distinct | |||
| 2014 | 5 | 2 | 0 | 0 | 2 | 1 | 0.0 |
| 2015 | 5 | 4 | 0 | 0 | 0 | 1 | 0.0 |
| 2016 | 1 | 1 | 0 | 0 | 0 | 0 | 0.0 |
| 2017 | 12 | 9 | 0 | 3 | 0 | 0 | 25.0 |
| 2018 | 16 | 10 | 0 | 6 | 0 | 0 | 37.5 |
| 2019 | 6 | 3 | 0 | 3 | 0 | 0 | 50.0 |
| 2020 | 8 | 6 | 0 | 1 | 0 | 1 | 12.5 |
| 2021 | 23 | 12 | 0 | 6 | 0 | 5 | 26.1 |
| 2022 | 19 | 15 | 0 | 3 | 0 | 1 | 15.8 |
| 2023 | 27 | 9 | 0 | 16 | 0 | 2 | 59.3 |
| 122 | 71 | 0 | 38 | 2 | 11 | ||
| IBDV Infected Chickens Group 1 | Mock Infected Chickens Group 2 | |||||||
|---|---|---|---|---|---|---|---|---|
| BF Atrophy Score * | 3 | 7 | 14 | 21 Days pi | 3 | 7 | 14 | 21 Days pi |
| 0 | 4 | 0 | 0 | 0 | 6 | 6 | 6 | 6 |
| +1 | 2 | 4 | 0 | 0 | 0 | 0 | 0 | 0 |
| +2 | 0 | 2 | 3 | 0 | 0 | 0 | 0 | 0 |
| +3 | 0 | 0 | 3 | 6 | 0 | 0 | 0 | 0 |
| Mean score # | 0.33 | 1.33 | 2.50 | 3.00 | 0.00 | 0.00 | 0.00 | 0.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takahashi, M.; Oguro, S.; Kato, A.; Ito, S.; Tsutsumi, N. Novel Antigenic Variant Infectious Bursal Disease Virus Outbreaks in Japan from 2014 to 2023 and Characterization of an Isolate from Chicken. Pathogens 2024, 13, 1141. https://doi.org/10.3390/pathogens13121141
Takahashi M, Oguro S, Kato A, Ito S, Tsutsumi N. Novel Antigenic Variant Infectious Bursal Disease Virus Outbreaks in Japan from 2014 to 2023 and Characterization of an Isolate from Chicken. Pathogens. 2024; 13(12):1141. https://doi.org/10.3390/pathogens13121141
Chicago/Turabian StyleTakahashi, Mari, Shiori Oguro, Atsushi Kato, Soma Ito, and Nobuyuki Tsutsumi. 2024. "Novel Antigenic Variant Infectious Bursal Disease Virus Outbreaks in Japan from 2014 to 2023 and Characterization of an Isolate from Chicken" Pathogens 13, no. 12: 1141. https://doi.org/10.3390/pathogens13121141
APA StyleTakahashi, M., Oguro, S., Kato, A., Ito, S., & Tsutsumi, N. (2024). Novel Antigenic Variant Infectious Bursal Disease Virus Outbreaks in Japan from 2014 to 2023 and Characterization of an Isolate from Chicken. Pathogens, 13(12), 1141. https://doi.org/10.3390/pathogens13121141







