The Anti-Listeria Activity of Pseudomonas fluorescens Isolated from the Horticultural Environment in New Zealand
Abstract
1. Introduction
2. Materials and Methods
2.1. Screening for Protective Bacteria in Horticultural Produce
2.2. Screening for Resident Biocontrol Bacteria Using Listeria Pour Plates
2.3. Selection of Presumptive Biocontrol Colonies
2.4. Zone of Inhibition Using Agar Gel Diffusion Test
2.5. Culture Supernatant Susceptibility and Liquid Coculture Tests
2.6. Species Identification Using 16SrRNA Sequence Analysis
2.7. Fluorescence Activity and Zone of Inhibition against Listeria spp. of the Sequenced Test BCA Cultures
2.8. Testing for Antibacterial Activity of P. fluorescens in Liquid Media Coculture
2.9. Proteome Analysis of P. fluorescens
2.10. Statistical Analysis
3. Results and Discussion
3.1. BCA Bacterial Culture Screening, 16SrRNA Identification and Fluorescence Testing
3.2. Zone of Inhibition
3.3. Anti-Listeria Activity in Liquid Media in Coculture
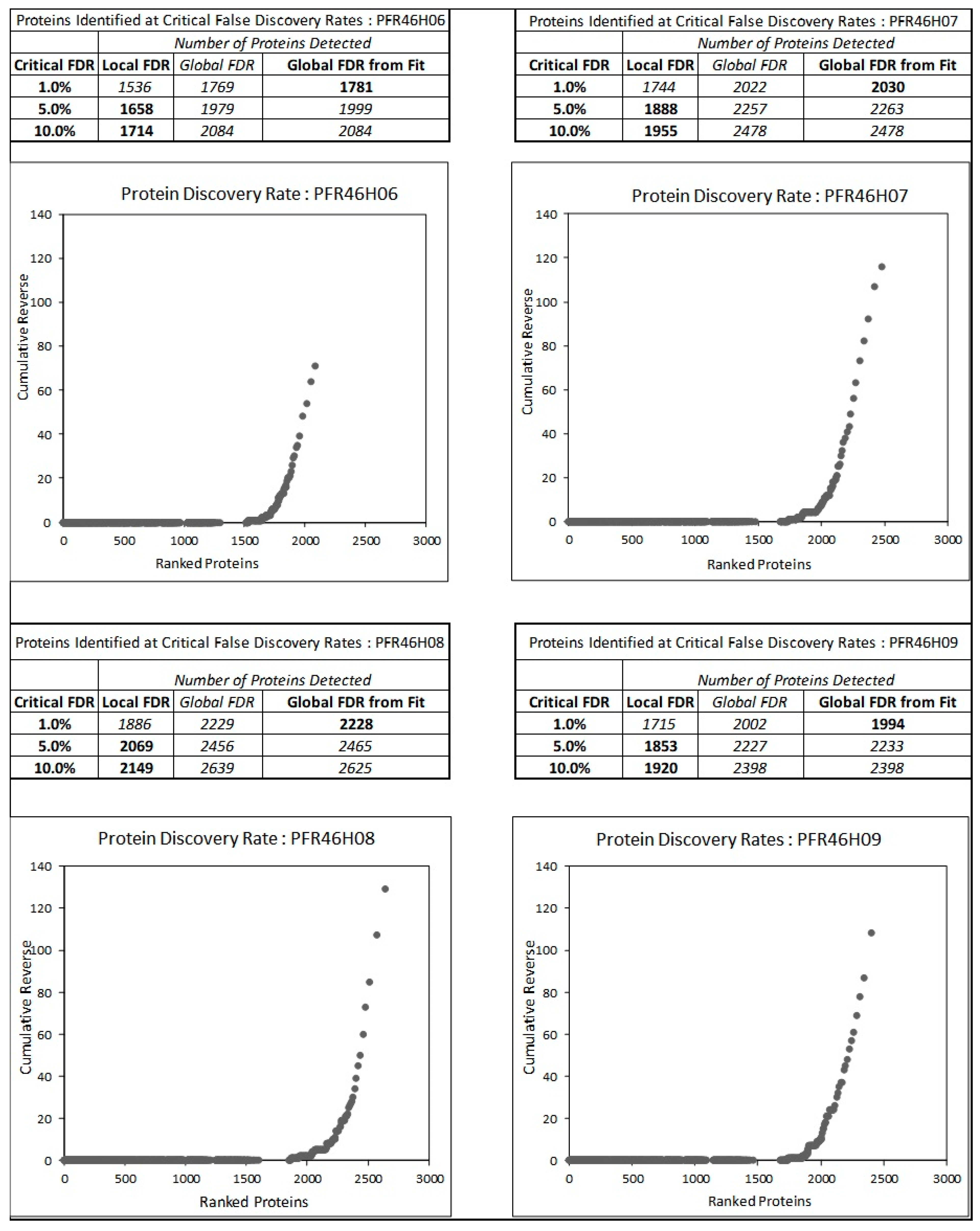
3.4. Proteome Analysis of P. fluorescens Strains
4. Conclusions and Future Directions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Van Haute, S.; Sampers, I.; Holvoet, K.; Uyttendaele, M. Physicochemical quality and chemical safety of chlorine as a reconditioning agent and wash water disinfectant for fresh-cut lettuce washing. Appl. Environ. Microbiol. 2013, 79, 2850–2861. [Google Scholar] [CrossRef] [PubMed]
- Aktar, M.W.; Sengupta, D.; Chowdhury, A. Impact of pesticides use in agriculture: Their benefits and hazards. Interdiscip. Toxicol. 2009, 2, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Weller, D.M. Pseudomonas biocontrol agents of soilborne pathogens: Looking back over 30 years. Phytopathology 2007, 97, 250–256. [Google Scholar] [CrossRef]
- Albano, H.; Oliveira, M.; Aroso, R.; Cubero, N.; Hogg, T.; Teixeira, P. Antilisterial activity of lactic acid bacteria isolated from “Alheiras” (traditional Portuguese fermented sausages): In situ assays. Meat Sci. 2007, 76, 796–800. [Google Scholar] [CrossRef] [PubMed]
- Lakicevic, B.; Nastasijevic, I.; Raseta, M. Sources of Listeria monocytogenes contamination in retail establishments. Procedia Food Sci. 2015, 5, 160–163. [Google Scholar] [CrossRef]
- Buchrieser, C.; Rusniok, C.; Garrido, P.; Hain, T.; Scortti, M.; Lampidis, R.; Kärst, U.; Chakraborty, T.; Cossart, P.; Kreft, J.; et al. Complete genome sequence of the animal pathogen Listeria ivanovii. which provides insights into host specificities and evolution of the genus Listeria. J. Bacteriol. 2011, 193, 6787. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, H.O.; Atwill, E.; Dunbar, L.; Ward, T.; McDonough, P.; Gonzalez, R.; Stipetic, K. The risk of Listeria monocytogenes infection in beef cattle operations. J. Appl. Microbiol. 2010, 108, 349–356. [Google Scholar] [CrossRef] [PubMed]
- Dimitrijevic, M.; Anderson, R.C.; Callaway, T.R.; Jung, Y.S.; Harvey, R.B.; Ricke, S.C.; Nisbet, D.J. Inhibitory effect of select nitrocompounds on growth and survivability of Listeria monocytogenes in vitro. J. Food Prot. 2006, 69, 1061–1065. [Google Scholar] [CrossRef] [PubMed]
- Oevermann, A.; Zurbriggen, A.; Vandevelde, M. Rhombencephalitis caused by Listeria monocytogenes in humans and ruminants: A zoonosis on the rise? Interdiscip. Perspect. Infect. Dis. 2010, 2010, 632513. [Google Scholar] [CrossRef] [PubMed]
- Schuppler, M.; Loessner, M.J. The opportunistic pathogen Listeria monocytogenes: Pathogenicity and interaction with the mucosal immune system. Int. J. Inflamm. 2010, 2010, 704321. [Google Scholar] [CrossRef]
- Zhu, Q.; Gooneratne, R.; Hussain, M.A. Listeria monocytogenes in Fresh Produce: Outbreaks, Prevalence and Contamination Levels. Foods 2017, 6, 21. [Google Scholar] [CrossRef]
- Macarisin, D.; Sheth, I.; Hur, M.; Wooten, A.; Kwon, H.J.; Gao, Z.; De Jesus, A.; Jurick, W.; Chen, Y. Survival of outbreak, food, and environmental strains of Listeria monocytogenes on whole apples as affected by cultivar and wax coating. Sci. Rep. 2019, 9, 12170. [Google Scholar] [CrossRef] [PubMed]
- Gaggia, F.; Di Gioia, D.; Baffoni, L.; Biavati, B. The role of protective and probiotic cultures in food and feed and their impact in food safety. Trends Food Sci. Technol. 2011, 22, S58–S66. [Google Scholar] [CrossRef]
- García, P.; Rodríguez, L.; Rodríguez, A.; Martínez, B. Food biopreservation: Promising strategies using bacteriocins, bacteriophages and endolysins. Trends Food Sci. Technol. 2010, 21, 373–382. [Google Scholar] [CrossRef]
- Leroi, F. Occurrence and role of lactic acid bacteria in seafood products. Food Microbiol. 2010, 27, 698–709. [Google Scholar]
- Sunita, R.; Saleena, L.M.; Vasudevan, P.; Nair, S. Biological suppression of rice diseases by Pseudomonas spp. under saline soil conditions. Plant Soil 2003, 251, 73–82. [Google Scholar] [CrossRef]
- Hoffland, E.; Hakulinen, J.P.; van Pelt, J.A. Comparison of systemic resistance induced by avirulent and nonpathogenic Pseudomonas species. Phytopathology 1996, 86, 757–762. [Google Scholar] [CrossRef]
- Meyer, J.M.; Abdallah, M.A. The fluorescent pigment of Pseudomonas fluorescens: Biosynthesis, purification and physicochemical properties. Microbiology 1978, 107, 319–328. [Google Scholar] [CrossRef]
- Palleroni, N.J. Genus Pseudomonas. In Bergey’s Manual of Systematic Bacteriology; Wilkins, W., Ed.; Springer: Baltimore, MD, USA, 1984; Volume 1. [Google Scholar]
- O’Sullivan, D.J.; O’Gara, F. Traits of fluorescent Pseudomonas spp. involved in suppression of plant root pathogens. Microbiol. Rev. 1992, 56, 662–676. [Google Scholar] [CrossRef]
- Haas, D.; Defago, G. Biological control of soil-borne pathogens by fluorescent Pseudomonads. Nat. Rev. Microbiol. 2005, 3, 307–319. [Google Scholar] [CrossRef]
- Scales, B.S.; Dickson, R.P.; LiPuma, J.J.; Huffnagle, G.B. Microbiology, genomics, and clinical significance of the Pseudomonas fluorescens species complex, an unappreciated colonizer of humans. Clin. Microbiol. Rev. 2014, 27, 927–948. [Google Scholar] [CrossRef]
- Barret, M.; Egan, F.; O’Gara, F. Distribution and diversity of bacterial secretion systems across metagenomic datasets. Environ. Microbiol. Rep. 2013, 5, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, S.; Mande, S.S. Identification and Functional Characterization of Gene Components of Type VI Secretion System in Bacterial Genomes. PLoS ONE 2008, 3, e2955. [Google Scholar] [CrossRef]
- Barret, M.; Egan, F.; Fargier, E.; Morrissey, J.P.; O’Gara, F. Genomic analysis of the type VI secretion systems in Pseudomonas spp.: Novel clusters and putative effectors uncovered. Microbiology 2011, 157, 1726–1739. [Google Scholar] [CrossRef]
- Decoin, V.; Barbey, C.; Bergeau, D.; Latour, X.; Feuilloley, M.G.J.; Orange, N.; Merieau, A. A Type VI secretion system is involved in Pseudomonas fluorescens bacterial competition. PLoS ONE 2014, 9, e89411. [Google Scholar] [CrossRef]
- Gerlach, R.G.; Hensel, M. Protein secretion systems and adhesins: The molecular armory of Gram-negative pathogens. Int. J. Med. Microbiol. 2007, 297, 401–415. [Google Scholar] [CrossRef]
- Miyata, S.T.; Kitaoka, M.; Brooks, T.M.; McAuley, S.B.; Pukatzki, S. Vibrio cholerae requires the type VI secretion system virulence factor VasX to kill Dictyostelium discoideum. Infect. Immun. 2011, 79, 2941–2949. [Google Scholar] [CrossRef]
- Hachani, A.; Lossi, N.S.; Hamilton, A.; Jones, C.; Bleves, S.; Albesa-Jové, D.; Filloux, A. Type VI secretion system in Pseudomonas aeruginosa: Secretion and multimerization of VgrG proteins. J. Biol. Chem. 2011, 286, 12317–12327. [Google Scholar] [CrossRef] [PubMed]
- Hachani, A.; Allsopp, L.P.; Oduko, Y.; Filloux, A. The VgrG proteins are “a la carte” delivery systems for bacterial type VI effectors. J. Biol. Chem. 2014, 289, 17872–17884. [Google Scholar] [CrossRef]
- Cascales, E.; Cambillau, C. Structural biology of type VI secretion systems. Philos. Trans. R. Soc. B Biol. Sci. 2012, 367, 1102–1111. [Google Scholar] [CrossRef] [PubMed]
- Kudryashev, M.; Wang, R.Y.-R.; Brackmann, M.; Scherer, S.; Maier, T.; Baker, D.; DiMaio, F.; Stahlberg, H.; Egelman, E.H.; Basler, M. Structure of the type VI secretion system contractile sheath. Cell 2015, 160, 952–962. [Google Scholar] [CrossRef] [PubMed]
- Shneider, M.M.; Buth, S.A.; Ho, B.T.; Basler, M.; Mekalanos, J.J.; Leiman, P.G. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature 2013, 500, 350–353. [Google Scholar] [CrossRef] [PubMed]
- Whitney, J.C.; Beck, C.M.; Goo, Y.A.; Russell, A.B.; Harding, B.N.; De Leon, J.A.; Cunningham, D.A.; Tran, B.Q.; Low, D.A.; Goodlett, D.R. Genetically distinct pathways guide effector export through the type VI secretion system. Mol. Microbiol. 2014, 92, 529–542. [Google Scholar] [CrossRef] [PubMed]
- Hill, A.M.; Staunton, J. 1.10—Type I Modular PKS. In Comprehensive Natural Products II; Liu, H.-W., Mander, L., Eds.; Elsevier: Oxford, UK, 2010; pp. 385–452. [Google Scholar] [CrossRef]
- Kieser, M.; Wassmer, G. On the Use of the Upper Confidence Limit for the Variance from a Pilot Sample for Sample Size Determination. Biom. J. 1996, 38, 941–949. [Google Scholar] [CrossRef]
- Whitehead, A.L.; Julious, S.A.; Cooper, C.L.; Campbell, M.J. Estimating the sample size for a pilot randomised trial to minimise the overall trial sample size for the external pilot and main trial for a continuous outcome variable. Statiscal Methods Med. Res. 2016, 25, 1057–1073. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, R.; Karaoz, U.; Volegova, M.; MacKichan, J.; Kato-Maeda, M.; Miller, S.; Nadarajan, R.; Brodie, E.L.; Lynch, S.V. Use of 16S rRNA gene for identification of a broad range of clinically relevant bacterial pathogens. PLoS ONE 2015, 10, e0117617. [Google Scholar] [CrossRef]
- Zhou, J.; Davey, M.E.; Figueras, J.B.; Rivkina, E.; Gilichinsky, D.; Tiedje, J.M. Phylogenetic diversity of a bacterial community determined from Siberian tundra soil DNA. Microbiology 1997, 143 Pt 12, 3913–3919. [Google Scholar] [CrossRef]
- Barghouthi, S.A. A universal method for the identification of bacteria based on general PCR primers. Indian J. Microbiol. 2011, 51, 430–444. [Google Scholar] [CrossRef]
- Yu, T.; Jiang, X.; Zhang, Y.; Ji, S.; Gao, W.; Shi, L. Effect of Benzalkonium Chloride Adaptation on Sensitivity to Antimicrobial Agents and Tolerance to Environmental Stresses in Listeria monocytogenes. Front. Microbiol. 2018, 9, 2906. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, G.B.; Favarin, L.; Luchese, R.H.; McIntosh, D. Psychrotrophic bacteria in milk: How much do we really know? Braz. J. Microbiol. 2015, 46, 313–321. [Google Scholar] [CrossRef]
- Ribeiro Júnior, J.C.; de Oliveira, A.M.; Silva, F.d.G.; Tamanini, R.; de Oliveira, A.L.M.; Beloti, V. The main spoilage-related psychrotrophic bacteria in refrigerated raw milk. J. Dairy Sci. 2018, 101, 75–83. [Google Scholar] [CrossRef]
- Miller, A.; Scanlan, R.A.; Lee, J.S.; Libbey, L.M. Volatile compounds produced in sterile fish muscle (Sebastes melanops) by Pseudomonas putrefaciens, Pseudomonas fluorescens, and an Achromobacter species. Appl. Microbiol. 1973, 26, 18–21. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.-M.; Michael, P.D.; Luchanskyi, J.B. Identification of Pseudomonas fluorescens strains Isolated from raw pork and chicken that produce siderophores antagonistic towards foodborne pathogens. J. Food Prot. 1995, 58, 1340–1344. [Google Scholar] [CrossRef]
- Farrag, S.A.; Marth, E.H. Growth of Listeria monocytogenes in the presence of Pseudomonas fluorescens at 7 or 13 °C in skim milk. J. Food Prot. 1989, 52, 852–855. [Google Scholar] [CrossRef]
- Kloepper, J.W.; Lifshitz, R.; Schroth, M.N. Pseudomonas Inoculants to Benefit Plant Production; ISI Atlas of Science, Institute of Information: Philadelphia, PA, USA, 1988. [Google Scholar]
- Douglas, L.M.; Schimdt, R.H. Growth of Listeria monocytogenes at 10 °C in milk preincubated with selected Pseudomonads. J. Food Prot. 1988, 51, 277–282. [Google Scholar]
- Belák, Á.; Maráz, A. Antagonistic Effect of Pseudomonas sp. CMI-1 on Foodborne Pathogenic Listeria monocytogenes. Food Technol. Biotechnol. 2015, 53, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Cascales, E.; Buchanan, S.K.; Duché, D.; Kleanthous, C.; Lloubès, R.; Postle, K.; Riley, M.; Slatin, S.; Cavard, D. Colicin biology. Microbiol. Mol. Biol. Rev. 2007, 71, 158–229. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Kightlinger, W.; Kwon, Y.C.; Hong, S.H. Rapid production and characterization of antimicrobial colicins using Escherichia coli-based cell-free protein synthesis. Synth. Biol. (Oxf) 2018, 3, ysy004. [Google Scholar] [CrossRef]
- Gallique, M.; Bouteiller, M.; Merieau, A. The type VI secretion system: A dynamic system for bacterial communication? Front. Microbiol. 2017, 8, 1454. [Google Scholar] [CrossRef] [PubMed]
- Paul, D.; Dineshkumar, N.; Nair, S. Proteomics of a plant growth-promoting rhizobacterium, Pseudomonas fluorescens MSP-393, subjected to salt shock. World J. Microbiol. Biotechnol. 2006, 22, 369–374. [Google Scholar] [CrossRef]
- Kim, W.; Silby, M.W.; Purvine, S.O.; Nicoll, J.S.; Hixson, K.K.; Monroe, M.; Nicora, C.D.; Lipton, M.S.; Levy, S.B. Proteomic detection of non-annotated protein-coding genes in Pseudomonas fluorescens Pf0-1. PLoS ONE 2009, 4, e8455. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, S.; West, T.E.; Boyer, F.; Chiang, W.-C.; Carl, M.A.; Hood, R.D.; Rohmer, L.; Tolker-Nielsen, T.; Skerrett, S.J.; Mougous, J.D. Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog. 2010, 6, e1001068. [Google Scholar] [CrossRef] [PubMed]
- Costa, T.R.; Felisberto-Rodrigues, C.; Meir, A.; Prevost, M.S.; Redzej, A.; Trokter, M.; Waksman, G. Secretion systems in Gram-negative bacteria: Structural and mechanistic insights. Nat. Rev. Microbiol. 2015, 13, 343–359. [Google Scholar] [CrossRef]
- Filloux, A. The rise of the Type VI secretion system. F1000Prime Rep. 2013, 5, 52. [Google Scholar] [CrossRef]
- Filloux, A. The type VI secretion system: A tubular story. EMBO J. 2009, 28, 309. [Google Scholar] [CrossRef] [PubMed]
- Mougous, J.D.; Cuff, M.E.; Raunser, S.; Shen, A.; Zhou, M.; Gifford, C.A.; Goodman, A.L.; Joachimiak, G.; Ordoñez, C.L.; Lory, S.; et al. A Virulence Locus of Pseudomonas aeruginosa Encodes a Protein Secretion Apparatus. Science 2006, 312, 1526. [Google Scholar] [CrossRef]
- Leiman, P.G.; Basler, M.; Ramagopal, U.A.; Bonanno, J.B.; Sauder, J.M.; Pukatzki, S.; Burley, S.K.; Almo, S.C.; Mekalanos, J.J. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. USA 2009, 106, 4154. [Google Scholar] [CrossRef]
- Pell, L.G.; Kanelis, V.; Donaldson, L.W.; Lynne Howell, P.; Davidson, A.R. The phage λ major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc. Natl. Acad. Sci. USA 2009, 106, 4160–4165. [Google Scholar] [CrossRef]
- Ballister, E.R.; Lai, A.H.; Zuckermann, R.N.; Cheng, Y.; Mougous, J.D. In vitro self-assembly of tailorable nanotubes from a simple protein building block. Proc. Natl. Acad. Sci. USA 2008, 105, 3733–3738. [Google Scholar] [CrossRef] [PubMed]
- Coulthurst, S. The Type VI secretion system: A versatile bacterial weapon. Microbiology 2019, 165, 503–515. [Google Scholar] [CrossRef]
- Nano, F.E.; Schmerk, C. The Francisella pathogenicity island. Ann. N. Y. Acad. Sci. 2007, 1105, 122–137. [Google Scholar] [CrossRef] [PubMed]
- Lossi, N.S.; Dajani, R.; Freemont, P.; Filloux, A. Structure-function analysis of HsiF, a gp25-like component of the type VI secretion system, in Pseudomonas aeruginosa. Microbiology 2011, 157, 3292–3305. [Google Scholar] [CrossRef] [PubMed]
- Bonemann, G.; Pietrosiuk, A.; Diemand, A.; Zentgraf, H.; Mogk, A. Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. Embo J. 2009, 28, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Basler, M.; Pilhofer, M.; Henderson, G.P.; Jensen, G.J.; Mekalanos, J.J. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 2012, 483, 182–186. [Google Scholar] [CrossRef]
- Brunet, Y.R.; Espinosa, L.; Harchouni, S.; Mignot, T.; Cascales, E. Imaging type VI secretion-mediated bacterial killing. Cell Rep. 2013, 3, 36–41. [Google Scholar] [CrossRef]
- Kapitein, N.; Bonemann, G.; Pietrosiuk, A.; Seyffer, F.; Hausser, I.; Locker, J.K.; Mogk, A. ClpV recycles VipA/VipB tubules and prevents non-productive tubule formation to ensure efficient type VI protein secretion. Mol. Microbiol. 2013, 87, 1013–1028. [Google Scholar] [CrossRef]
- Bladergroen, M.R.; Badelt, K.; Spaink, H.P. Infection-blocking genes of a symbiotic Rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol. Plant-Microbe Interact. MPMI 2003, 16, 53–64. [Google Scholar] [CrossRef]
- Jones, C.; Hachani, A.; Manoli, E.; Filloux, A. An rhs gene linked to the second type VI secretion cluster is a feature of the Pseudomonas aeruginosa strain PA14. J. Bacteriol. 2014, 196, 800. [Google Scholar] [CrossRef]
- Alteri, C.J.; Mobley, H.L.T. The versatile type VI secretion system. Microbiol. Spectr. 2016, 4, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Kung, V.L.; Khare, S.; Stehlik, C.; Bacon, E.M.; Hughes, A.J.; Hauser, A.R. An rhs gene of Pseudomonas aeruginosa encodes a virulence protein that activates the inflammasome. Proc. Natl. Acad. Sci. USA 2012, 109, 1275. [Google Scholar] [CrossRef]
- Pukatzki, S.; Ma, A.T.; Sturtevant, D.; Krastins, B.; Sarracino, D.; Nelson, W.C.; Heidelberg, J.F.; Mekalanos, J.J. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. USA 2006, 103, 1528–1533. [Google Scholar] [CrossRef] [PubMed]
- Silverman, J.M.; Austin, L.S.; Hsu, F.; Hicks, K.G.; Hood, R.D.; Mougous, J.D. Separate inputs modulate phosphorylation-dependent and -independent type VI secretion activation. Mol. Microbiol. 2011, 82, 1277–1290. [Google Scholar] [CrossRef] [PubMed]
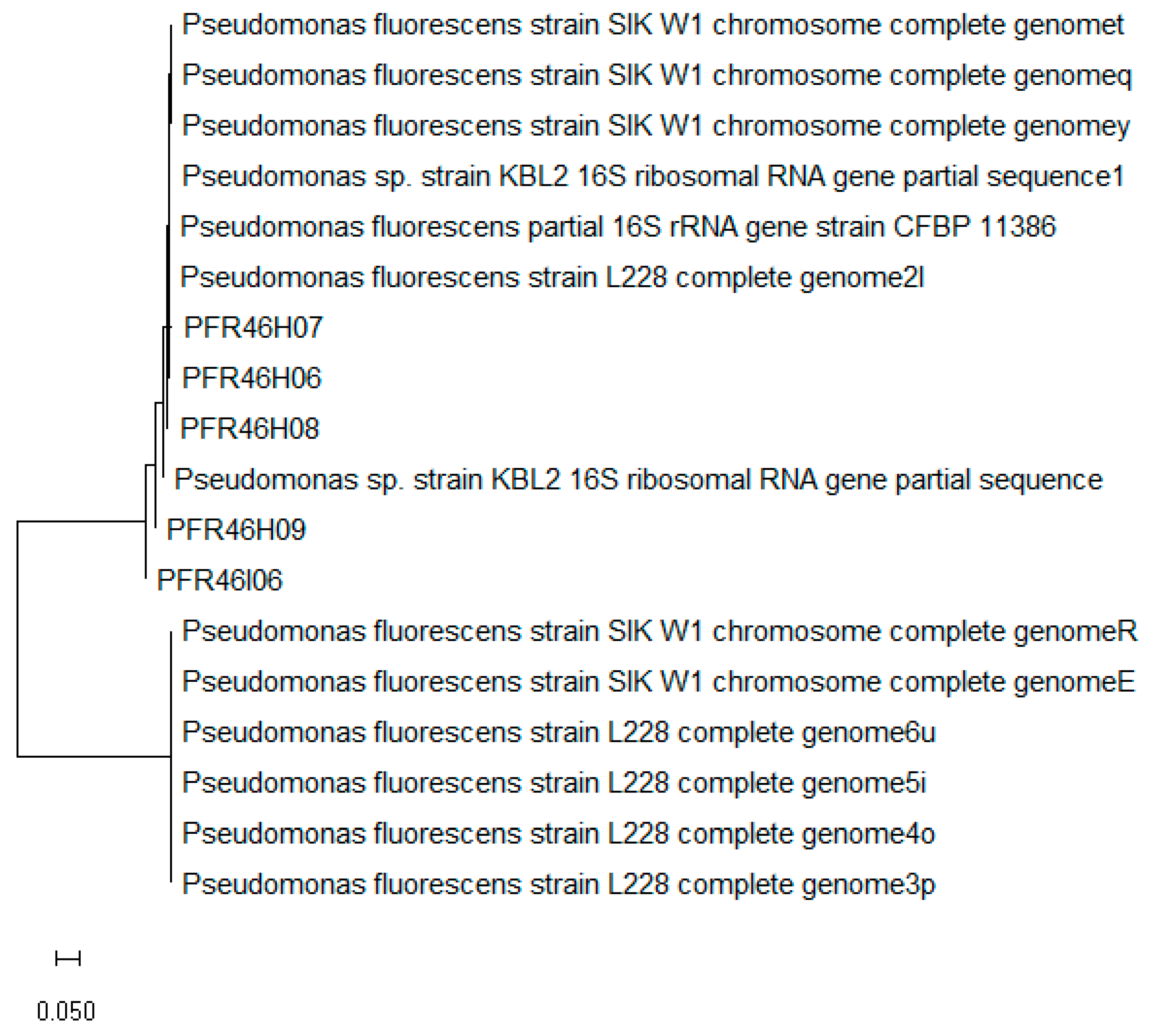

| No. | Strain | Organism | Source |
|---|---|---|---|
| 1 | PFR05A12 | L. seeligeri | Vegetable |
| 2 | PFR12C05 | L. monocytogenes Scott A | Clinical isolate |
| 3 | PFR16B03 | L. monocytogenes ATCC strain 49594 | Clinical isolate |
| 4 | PFR18B09 | L. monocytogenes | Seafood processing environment |
| 5 | PFR18C05 | L. monocytogenes | Seafood processing environment |
| 6 | PFR18C07 | L. monocytogenes | Seafood processing environment |
| 7 | PFR18D01 | L. monocytogenes | Seafood processing environment |
| 8 | PFR18D05 | L. monocytogenes | Seafood processing environment |
| 9 | PFR33F02 | L. monocytogenes | Seafood processing environment |
| 10 | PFR33F03 | L. monocytogenes | Seafood processing environment |
| 11 | PFR33H03 | L. monocytogenes | Seafood processing environment |
| 12 | PFR33H04 | L. monocytogenes | Seafood processing environment |
| 13 | PFR33I04 | L. monocytogenes | Seafood processing environment |
| 14 | PFR40I05 | L. monocytogenes | Horticultural source |
| 15 | PFR40I07 | L. monocytogenes | Horticultural environment |
| 16 | PFR41E01 | L. monocytogenes | Horticultural environment |
| 17 | PFR41E02 | L. monocytogenes | Horticultural environment |
| 18 | PFR41E03 | L. monocytogenes | Horticultural environment |
| 19 | PFR41E05 | L. monocytogenes | Horticultural environment |
| 20 | PFR41F08 | L. monocytogenes | Horticultural environment |
| 21 | PFR41G01 | L. monocytogenes | Horticultural environment |
| 22 | PFR41G02 | L. monocytogenes | Horticultural environment |
| 23 | PFR41H07 | L. monocytogenes | Horticultural environment |
| 24 | PFR41J05 | L. monocytogenes | Horticultural environment |
| 25 | PFR41J08 | L. monocytogenes | Horticultural environment |
| 26 | PFR41J09 | L. monocytogenes | Horticultural environment |
| 27 | PFR42G03 | L. monocytogenes | Horticultural environment |
| 28 | PFR42I05 | L. monocytogenes | Seafood processing environment |
| 29 | PFR42I06 | L. monocytogenes | Seafood processing environment |
| 30 | PFR42I07 | L. monocytogenes | Seafood processing environment |
| 31 | PFR42I08 | L. monocytogenes | Seafood processing environment |
| 32 | PFR42I09 | L. monocytogenes | Seafood processing environment |
| 33 | PFR42I10 | L. monocytogenes | Seafood processing environment |
| 34 | PFR42J03 | L. monocytogenes | Seafood processing environment |
| 35 | PFR05A10 | L. innocua | Processed vegetable |
| Listeria Strains | |||||
|---|---|---|---|---|---|
| Estimate | Std. Error | z Value | Pr(>|z|) | ||
| PFR05A10 (Intercept) | 2.32 | 0.58 | 4.00 | 0.00 | *** |
| PFR05A12 | 0.13 | 0.78 | 0.17 | 0.86 | |
| PFR12C05 | −0.18 | 0.73 | −0.25 | 0.80 | |
| PFR16B03 | −0.34 | 0.70 | −0.48 | 0.63 | |
| PFR18B09 | −1.06 | 0.64 | −1.66 | 0.10 | . |
| PFR18C05 | −1.13 | 0.63 | −1.80 | 0.07 | . |
| PFR18C07 | −1.48 | 0.61 | −2.43 | 0.02 | * |
| PFR18D01 | −1.16 | 0.63 | −1.85 | 0.06 | . |
| PFR18D05 | −1.41 | 0.62 | −2.28 | 0.02 | * |
| PFR33F02 | −0.49 | 0.69 | −0.71 | 0.48 | |
| PFR33F03 | −0.37 | 0.70 | −0.52 | 0.60 | |
| PFR33G10 | −0.47 | 0.69 | −0.68 | 0.50 | |
| PFR33H03 | −0.43 | 0.70 | −0.62 | 0.54 | |
| PFR33H04 | −0.44 | 0.70 | −0.64 | 0.53 | |
| PFR40I05 | −0.83 | 0.67 | −1.23 | 0.22 | |
| PFR40I07 | −0.28 | 0.72 | −0.40 | 0.69 | |
| PFR41E01 | 0.05 | 0.76 | 0.07 | 0.95 | |
| PFR41E02 | −0.13 | 0.73 | −0.18 | 0.86 | |
| PFR41E03 | −0.62 | 0.68 | −0.91 | 0.37 | |
| PFR41E05 | −0.91 | 0.66 | −1.38 | 0.17 | |
| PFR41F08 | −0.35 | 0.71 | −0.49 | 0.62 | |
| PFR41G01 | 0.01 | 0.75 | 0.01 | 0.99 | |
| PFR41G02 | 0.01 | 0.75 | 0.01 | 0.99 | |
| PFR41H08 | −0.08 | 0.74 | −0.11 | 0.91 | |
| PFR41J05 | −0.17 | 0.72 | −0.24 | 0.81 | |
| PFR41J08 | −1.17 | 0.63 | −1.87 | 0.06 | . |
| PFR41J09 | −0.23 | 0.72 | −0.31 | 0.75 | |
| PFR42G03 | −0.36 | 0.70 | −0.51 | 0.61 | |
| PFR42I05 | −0.43 | 0.70 | −0.61 | 0.54 | |
| PFR42I06 | −0.32 | 0.71 | −0.46 | 0.65 | |
| PFR42I07 | −0.26 | 0.71 | −0.37 | 0.72 | |
| PFR42I08 | −0.31 | 0.71 | −0.43 | 0.67 | |
| PFR42I09 | −0.26 | 0.71 | −0.36 | 0.72 | |
| PFR42I10 | −0.33 | 0.71 | −0.46 | 0.65 | |
| PFR42J03 | −0.29 | 0.71 | −0.41 | 0.68 | |
| Secretion Protein | PFR46H06 | PFR46H07 | PFR46H08 | PFR46H09 |
|---|---|---|---|---|
| Type I restriction enzyme R protein | - | - | - | - |
| Type I restriction enzyme R protein | + REVERSED | - | + | - |
| Type I restriction–modification protein subunit M REVERSED | + | - | - | - |
| Type I secretion membrane fusion protein | - | - | + | + |
| Type I secretion outer-membrane protein | - | + | - | + |
| Type I secretion outer-membrane protein tolC | + | - | - | - |
| Type I secretion system ATP-binding protein PrsD | - | - | + | - |
| Type I secretion system membrane fusion protein PrsE | - | - | + | + |
| Type I secretion system permease/ATPase | - | - | + | + |
| Type II secretion pseudopilin HxcU REVERSED | - | - | + | + |
| Type II secretion system protein F | - | + | + REVERSED | + REVERSED |
| Type II secretion system protein GspJ REVERSED | - | - | - | + |
| Type III effector | - | - | + | + |
| Type III pantothenate kinase, coaX | + | - | - | - |
| Type III PLP-dependent enzyme | + | + | + | - |
| Type III restriction system endonuclease REVERSED | + | + | + | + |
| Type III secretion system transcriptional regulator RspS REVERSED | - | - | + | - |
| Type IV pilus response regulator PilH | + | + | + | + |
| Type IV secretion protein Rhs | + | + | + | + |
| Type IVB pilus formation outer-membrane protein, R64 PilN family | - | + | + | - |
| Type VI polysaccharide biosynthesis protein VipB/TviC | + | + | + | + |
| Type VI secretion ATPase, ClpV2 | - | - | - | + |
| Type VI secretion protein | - | + | + | + REVERSED |
| Type VI secretion protein TssK1 REVERSED | - | - | - | + |
| Type VI secretion protein TssL | - | + | - | - |
| Type VI secretion protein VasK REVERSED | + | - | - | - |
| Type VI secretion system baseplate subunit TssK REVERSED | - | - | - | + |
| Type VI secretion system protein ImpK | + | + | + | + |
| Type VI secretion system protein ImpM | + | - | + | + |
| Protein | PFR46H06 | PFR46H07 | PFR46H08 | PFR46H09 |
|---|---|---|---|---|
| Total number of proteins | 1781 | 2030 | 2228 | 1994 |
| Secretion system proteins | 12 | 11 | 18 | 18 |
| Number of tss-T6SS proteins | 4 | 4 | 4 | 9 |
| Phage-related tube, tail and sheath proteins | Present | Present | Present | Present |
| Type I restriction enzyme proteins | 2 | 1 | 5 | 4 |
| Type II secretion proteins | Absent | 1 | 2 | 3 |
| Type III secretion/effector proteins | 3 | 2 | 4 | 2 |
| Type IV pilus response regulator PilH | Present | Present | Present | Present |
| Type IV secretion protein Rhs | Substitutions in positions 37 (T to V) and 427 (S to T) | Substitutions in positions 37 (T to V) and 427 (S to T) | Substitutions in positions 37 (T to V) and 427 (S to T) | Substitutions in positions 37 (T to V) and 427 (S to T). |
| Type IVB pilus formation outer-membrane protein, R64 PilN family | Absent | Present | Present | Absent |
| Type VI polysaccharide biosynthesis protein VipB/TviC | Present | Present | Present | Present |
| Type VI secretion ATPase, ClpV2 | Absent | Absent | Absent | T4 phage polysheath and ClpV substrate protein |
| Type VI secretion protein TssL (Baseplate proteins homologous to T4 phage) | Absent | Baseplate proteins | Absent | Absent |
| Type VI secretion protein VasK (Transposon-mediated virulence protein) | Present | Absent | Absent | Absent |
| Type VI secretion system baseplate subunit TssK | Absent | Absent | Absent | Baseplate proteins |
| Type VI secretion system protein ImpK (Mechanistic contact for inhibition, as well as enhancement) | Substitution of the amino acid from R to N at the 95th position | R-to-N substitution occurred at the 92nd position | At 63, A to G; at 64, N to M; at 67, V to M; at 68, E to D; at 70, V to M; and at 95, R to N | R-to-N substitution at the 95th position |
| Type VI secretion system protein ImpM | Present | Absent | Present | Present |
| VipA (TssB) and VipB proteins that form a tubular polymer | Present | Present | Present | Present |
| Imp family proteins | Present | Present | Present | Present |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohan, V.; Wibisono, R.; Chalke, S.; Fletcher, G.; Leroi, F. The Anti-Listeria Activity of Pseudomonas fluorescens Isolated from the Horticultural Environment in New Zealand. Pathogens 2023, 12, 349. https://doi.org/10.3390/pathogens12020349
Mohan V, Wibisono R, Chalke S, Fletcher G, Leroi F. The Anti-Listeria Activity of Pseudomonas fluorescens Isolated from the Horticultural Environment in New Zealand. Pathogens. 2023; 12(2):349. https://doi.org/10.3390/pathogens12020349
Chicago/Turabian StyleMohan, Vathsala, Reginald Wibisono, Saili Chalke, Graham Fletcher, and Françoise Leroi. 2023. "The Anti-Listeria Activity of Pseudomonas fluorescens Isolated from the Horticultural Environment in New Zealand" Pathogens 12, no. 2: 349. https://doi.org/10.3390/pathogens12020349
APA StyleMohan, V., Wibisono, R., Chalke, S., Fletcher, G., & Leroi, F. (2023). The Anti-Listeria Activity of Pseudomonas fluorescens Isolated from the Horticultural Environment in New Zealand. Pathogens, 12(2), 349. https://doi.org/10.3390/pathogens12020349






