Establishment of an Inactivation Method for Ebola Virus and SARS-CoV-2 Suitable for Downstream Sequencing of Low Cell Numbers
Abstract
1. Introduction
2. Materials and Methods
2.1. Biosafety Statement
2.2. Cell Lines
2.3. Peripheral Blood Mononuclear Cells (PBMCs)
2.4. Coculture of PBMCs with Epithelial Cells
2.5. Virus Propagation
2.6. Amicon Column Testing
2.7. Cytotoxicity Testing
2.8. TCL Buffer Inactivation Testing
2.9. Heat Inactivation Testing
2.10. Immunofluorescence Analysis
2.11. Flow Sorting of PBMCs in Cocultures
2.12. Population Low-Input RNA-seq
2.13. RNA-seq Data Processing and QC
3. Results
3.1. Use of Columns to Reduce Toxicity of TCL Buffer
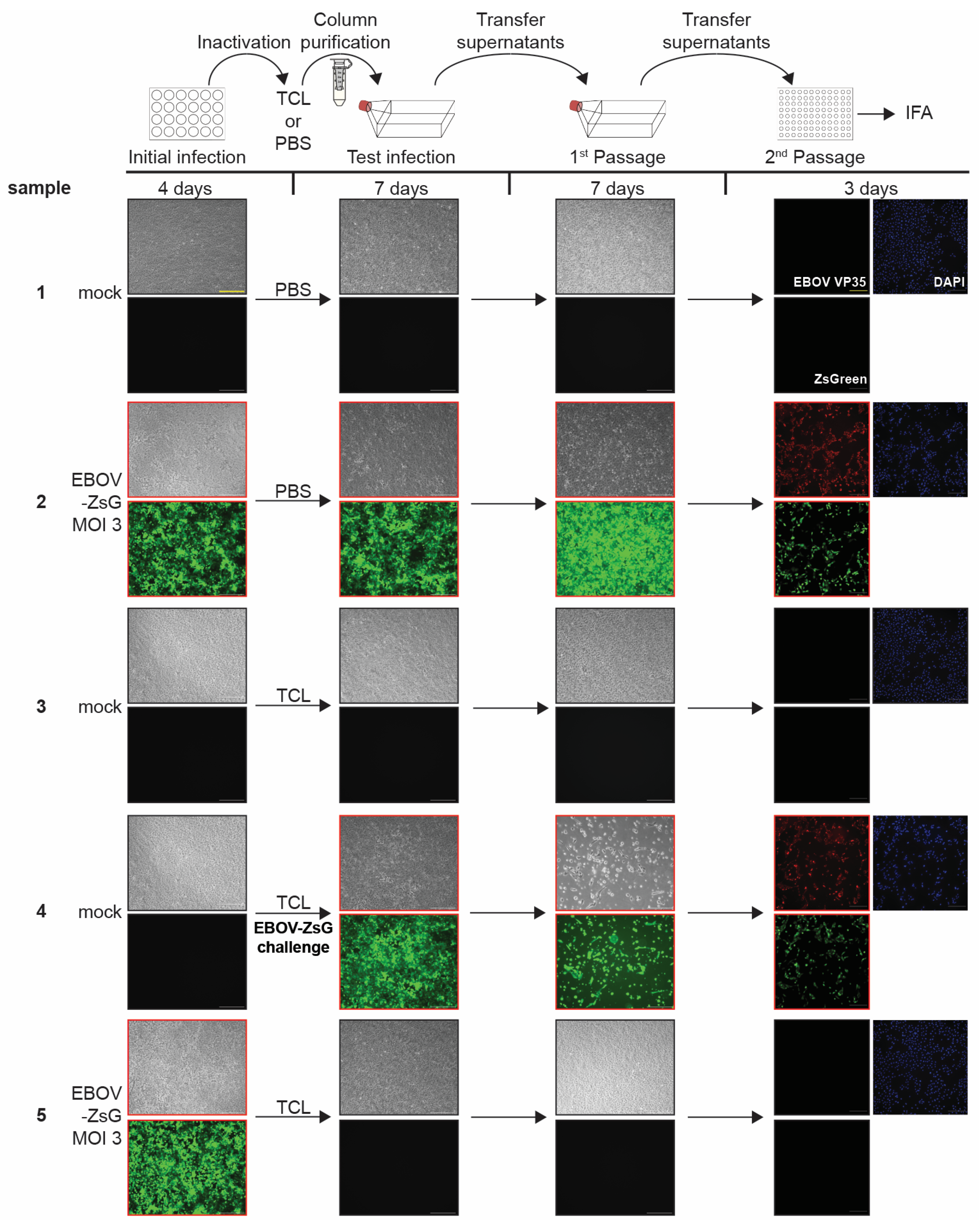
3.2. TCL Buffer Inactivation of Ebola Virus
3.3. Limit of Detection Analysis for EBOV-ZsGreen
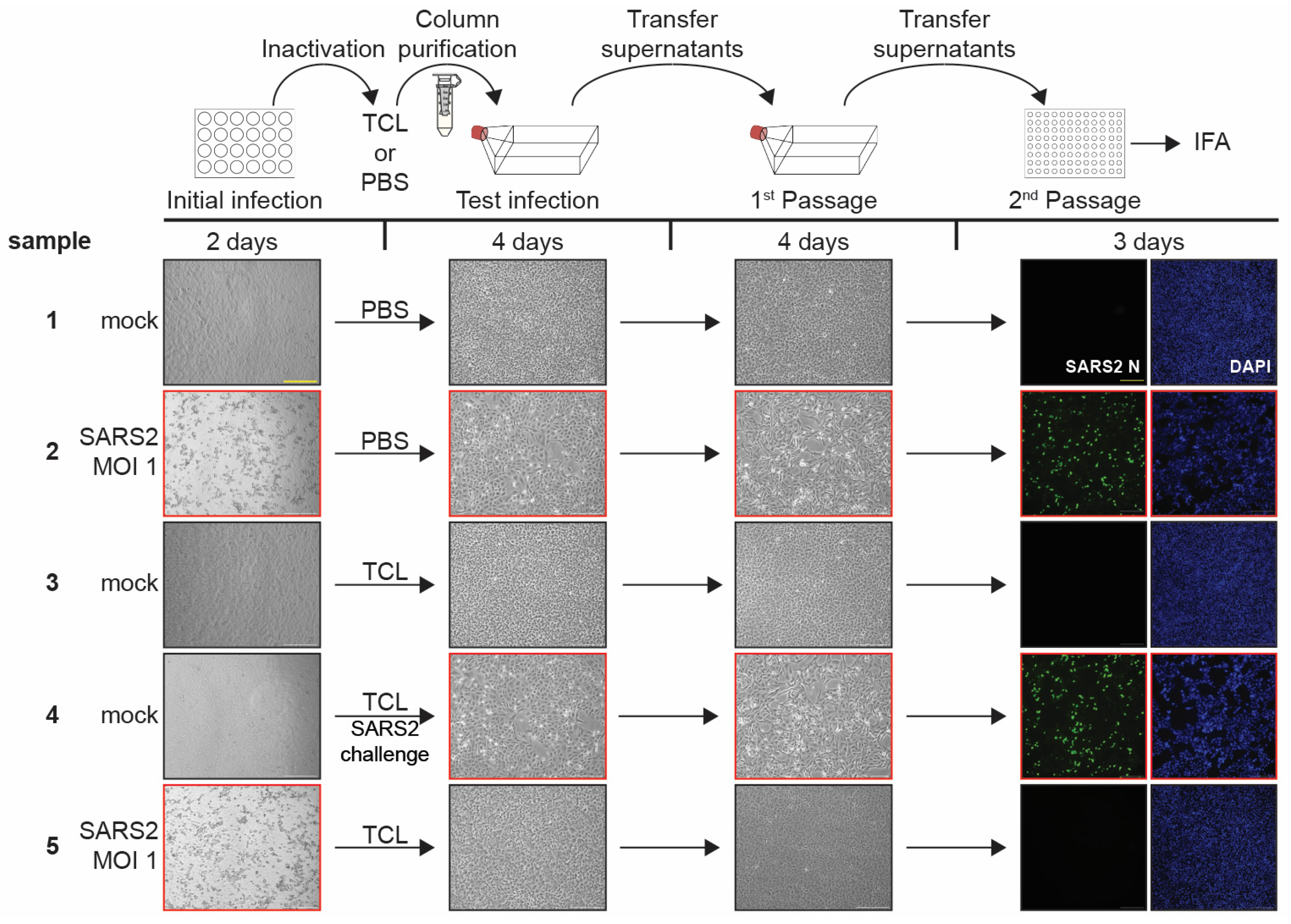
3.4. TCL Buffer Inactivation of SARS-CoV-2 and Limit of Detection Study
3.5. Heat Inactivation of Ebola Virus
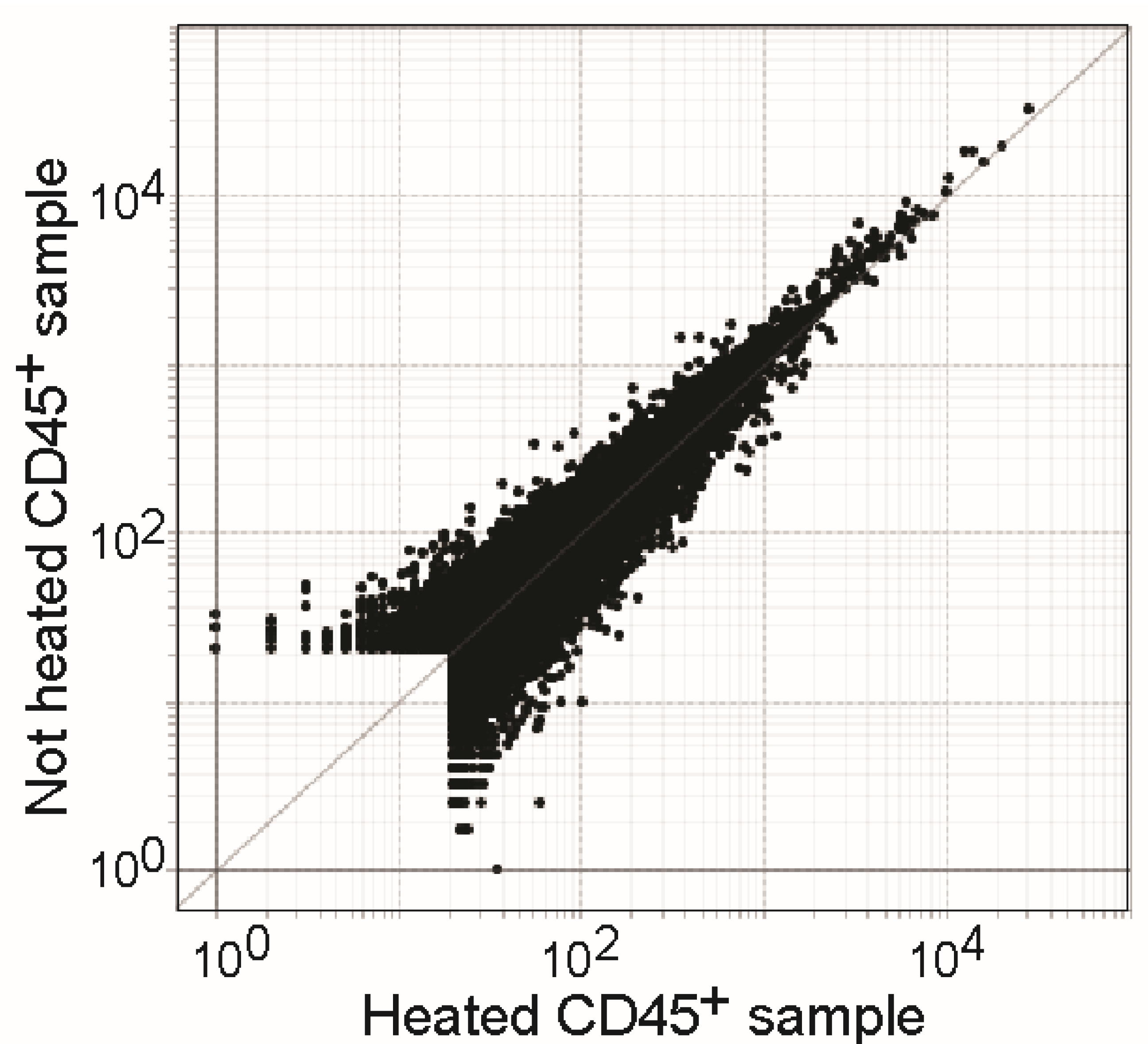
3.6. Heat Treatment Has Minor Effects on RNA Quality of Samples in TCL Buffer
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kanev, K.; Roelli, P.; Wu, M.; Wurmser, C.; Delorenzi, M.; Pfaffl, M.W.; Zehn, D. Tailoring the resolution of single-cell RNA sequencing for primary cytotoxic T cells. Nat. Commun. 2021, 12, 569. [Google Scholar] [CrossRef] [PubMed]
- Picelli, S. Full-Length Single-Cell RNA Sequencing with Smart-seq2. Methods Mol. Biol. 2019, 1979, 25–44. [Google Scholar] [CrossRef] [PubMed]
- Picelli, S.; Faridani, O.R.; Bjorklund, A.K.; Winberg, G.; Sagasser, S.; Sandberg, R. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014, 9, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Trombetta, J.J.; Gennert, D.; Lu, D.; Satija, R.; Shalek, A.K.; Regev, A. Preparation of Single-Cell RNA-Seq Libraries for Next Generation Sequencing. Curr. Protoc. Mol. Biol. 2014, 107, 4–22. [Google Scholar] [CrossRef] [PubMed]
- Immunological Genome Project Ultra-Low-Input RNA-Seq (ULI RNA-Seq). Available online: https://www.immgen.org/Protocols/ImmGenULI_RNAseq_methods.pdf (accessed on 19 November 2022).
- Reyes, M.; Vickers, D.; Billman, K.; Eisenhaure, T.; Hoover, P.; Browne, E.P.; Rao, D.A.; Hacohen, N.; Blainey, P.C. Multiplexed enrichment and genomic profiling of peripheral blood cells reveal subset-specific immune signatures. Sci. Adv. 2019, 5, eaau9223. [Google Scholar] [CrossRef] [PubMed]
- Lam, V.C.; Folkersen, L.; Aguilar, O.A.; Lanier, L.L. KLF12 Regulates Mouse NK Cell Proliferation. J. Immunol. 2019, 203, 981–989. [Google Scholar] [CrossRef]
- Luo, G.; Gao, Q.; Zhang, S.; Yan, B. Probing infectious disease by single-cell RNA sequencing: Progresses and perspectives. Comput. Struct. Biotechnol. J. 2020, 18, 2962–2971. [Google Scholar] [CrossRef]
- Rato, S.; Golumbeanu, M.; Telenti, A.; Ciuffi, A. Exploring viral infection using single-cell sequencing. Virus Res. 2017, 239, 55–68. [Google Scholar] [CrossRef]
- Haddock, E.; Feldmann, F.; Feldmann, H. Effective Chemical Inactivation of Ebola Virus. Emerg. Infect. Dis. 2016, 22, 160233. [Google Scholar] [CrossRef]
- Ngo, K.A.; Jones, S.A.; Church, T.M.; Fuschino, M.E.; George, K.S.; Lamson, D.M.; Maffei, J.; Kramer, L.D.; Ciota, A.T. Unreliable Inactivation of Viruses by Commonly Used Lysis Buffers. Appl. Biosaf. 2017, 22, 56–59. [Google Scholar] [CrossRef]
- Smither, S.J.; Weller, S.A.; Phelps, A.; Eastaugh, L.; Ngugi, S.; O’Brien, L.M.; Steward, J.; Lonsdale, S.G.; Lever, M.S. Buffer AVL Alone Does Not Inactivate Ebola Virus in a Representative Clinical Sample Type. J. Clin. Microbiol. 2015, 53, 3148–3154. [Google Scholar] [CrossRef] [PubMed]
- Hume, A.J.; Heiden, B.; Olejnik, J.; Suder, E.L.; Ross, S.; Scoon, W.A.; Bullitt, E.; Ericsson, M.; White, M.R.; Turcinovic, J.; et al. Recombinant Lloviu virus as a tool to study viral replication and host responses. PLoS Pathog. 2022, 18, e1010268. [Google Scholar] [CrossRef]
- Leon, J.; Michelson, D.A.; Olejnik, J.; Chowdhary, K.; Oh, H.S.; Hume, A.J.; Galvan-Pena, S.; Zhu, Y.; Chen, F.; Vijaykumar, B.; et al. A virus-specific monocyte inflammatory phenotype is induced by SARS-CoV-2 at the immune-epithelial interface. Proc. Natl. Acad. Sci. USA 2022, 119, e2116853118. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Hume, A.J.; Abo, K.M.; Werder, R.B.; Villacorta-Martin, C.; Alysandratos, K.D.; Beermann, M.L.; Simone-Roach, C.; Lindstrom-Vautrin, J.; Olejnik, J.; et al. SARS-CoV-2 Infection of Pluripotent Stem Cell-Derived Human Lung Alveolar Type 2 Cells Elicits a Rapid Epithelial-Intrinsic Inflammatory Response. Cell Stem Cell 2020, 27, 962–973. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Alfson, K.J.; Griffiths, A. Development and Testing of a Method for Validating Chemical Inactivation of Ebola Virus. Viruses 2018, 10, 126. [Google Scholar] [CrossRef] [PubMed]
- Jureka, A.S.; Silvas, J.A.; Basler, C.F. Propagation, Inactivation, and Safety Testing of SARS-CoV-2. Viruses 2020, 12, 622. [Google Scholar] [CrossRef]
- Kochel, T.J.; Kocher, G.A.; Ksiazek, T.G.; Burans, J.P. Evaluation of TRIzol LS Inactivation of Viruses. Appl. Biosaf. 2017, 22, 52–55. [Google Scholar] [CrossRef]
- Auerswald, H.; Yann, S.; Dul, S.; In, S.; Dussart, P.; Martin, N.J.; Karlsson, E.A.; Garcia-Rivera, J.A. Assessment of inactivation procedures for SARS-CoV-2. J. Gen. Virol. 2021, 102, 001539. [Google Scholar] [CrossRef]
- Pastorino, B.; Touret, F.; Gilles, M.; de Lamballerie, X.; Charrel, R.N. Heat Inactivation of Different Types of SARS-CoV-2 Samples: What Protocols for Biosafety, Molecular Detection and Serological Diagnostics? Viruses 2020, 12, 735. [Google Scholar] [CrossRef]
- Avelin, V.; Sissonen, S.; Julkunen, I.; Osterlund, P. Inactivation efficacy of H5N1 avian influenza virus by commonly used sample preparation reagents for safe laboratory practices. J. Virol. Methods 2022, 304, 114527. [Google Scholar] [CrossRef]
- Blow, J.A.; Dohm, D.J.; Negley, D.L.; Mores, C.N. Virus inactivation by nucleic acid extraction reagents. J. Virol. Methods 2004, 119, 195–198. [Google Scholar] [CrossRef] [PubMed]
- Jensen, K.S.; Adams, R.; Bennett, R.S.; Bernbaum, J.; Jahrling, P.B.; Holbrook, M.R. Development of a novel real-time polymerase chain reaction assay for the quantitative detection of Nipah virus replicative viral RNA. PLoS ONE 2018, 13, e0199534. [Google Scholar] [CrossRef] [PubMed]
- Widerspick, L.; Vazquez, C.A.; Niemetz, L.; Heung, M.; Olal, C.; Bencsik, A.; Henkel, C.; Pfister, A.; Brunetti, J.E.; Kucinskaite-Kodze, I.; et al. Inactivation Methods for Experimental Nipah Virus Infection. Viruses 2022, 14, 1052. [Google Scholar] [CrossRef] [PubMed]
- Lo, M.K.; Miller, D.; Aljofan, M.; Mungall, B.A.; Rollin, P.E.; Bellini, W.J.; Rota, P.A. Characterization of the antiviral and inflammatory responses against Nipah virus in endothelial cells and neurons. Virology 2010, 404, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Mire, C.E.; Satterfield, B.A.; Geisbert, J.B.; Agans, K.N.; Borisevich, V.; Yan, L.; Chan, Y.P.; Cross, R.W.; Fenton, K.A.; Broder, C.C.; et al. Pathogenic Differences between Nipah Virus Bangladesh and Malaysia Strains in Primates: Implications for Antibody Therapy. Sci. Rep. 2016, 6, 30916. [Google Scholar] [CrossRef]
- Welch, S.R.; Davies, K.A.; Buczkowski, H.; Hettiarachchi, N.; Green, N.; Arnold, U.; Jones, M.; Hannah, M.J.; Evans, R.; Burton, C.; et al. Analysis of Inactivation of SARS-CoV-2 by Specimen Transport Media, Nucleic Acid Extraction Reagents, Detergents, and Fixatives. J. Clin. Microbiol. 2020, 58, e01713-20. [Google Scholar] [CrossRef]
- Mitchell, S.W.; McCormick, J.B. Physicochemical inactivation of Lassa, Ebola, and Marburg viruses and effect on clinical laboratory analyses. J. Clin. Microbiol. 1984, 20, 486–489. [Google Scholar] [CrossRef]
- Bowen, E.T.; Simpson, D.I.; Bright, W.F.; Zlotnik, I.; Howard, D.M. Vervet monkey disease: Studies on some physical and chemical properties of the causative agent. Br. J. Exp. Pathol. 1969, 50, 400–407. [Google Scholar]
- Patterson, E.I.; Prince, T.; Anderson, E.R.; Casas-Sanchez, A.; Smith, S.L.; Cansado-Utrilla, C.; Solomon, T.; Griffiths, M.J.; Acosta-Serrano, A.; Turtle, L.; et al. Methods of Inactivation of SARS-CoV-2 for Downstream Biological Assays. J. Infect. Dis. 2020, 222, 1462–1467. [Google Scholar] [CrossRef]
- Rabenau, H.F.; Cinatl, J.; Morgenstern, B.; Bauer, G.; Preiser, W.; Doerr, H.W. Stability and inactivation of SARS coronavirus. Med. Microbiol. Immunol. 2005, 194, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Darnell, M.E.; Subbarao, K.; Feinstone, S.M.; Taylor, D.R. Inactivation of the coronavirus that induces severe acute respiratory syndrome, SARS-CoV. J. Virol. Methods 2004, 121, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Behnia, M.; Baer, A.; Skidmore, A.M.; Lehman, C.W.; Bracci, N.; Kehn-Hall, K.; Bradfute, S.B. Inactivation of Venezuelan Equine Encephalitis Virus Genome Using Two Methods. Viruses 2022, 14, 272. [Google Scholar] [CrossRef] [PubMed]
- Thi Nhu Thao, T.; Labroussaa, F.; Ebert, N.; V’Kovski, P.; Stalder, H.; Portmann, J.; Kelly, J.; Steiner, S.; Holwerda, M.; Kratzel, A.; et al. Rapid reconstruction of SARS-CoV-2 using a synthetic genomics platform. Nature 2020, 582, 561–565. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olejnik, J.; Leon, J.; Michelson, D.; Chowdhary, K.; Galvan-Pena, S.; Benoist, C.; Mühlberger, E.; Hume, A.J. Establishment of an Inactivation Method for Ebola Virus and SARS-CoV-2 Suitable for Downstream Sequencing of Low Cell Numbers. Pathogens 2023, 12, 342. https://doi.org/10.3390/pathogens12020342
Olejnik J, Leon J, Michelson D, Chowdhary K, Galvan-Pena S, Benoist C, Mühlberger E, Hume AJ. Establishment of an Inactivation Method for Ebola Virus and SARS-CoV-2 Suitable for Downstream Sequencing of Low Cell Numbers. Pathogens. 2023; 12(2):342. https://doi.org/10.3390/pathogens12020342
Chicago/Turabian StyleOlejnik, Judith, Juliette Leon, Daniel Michelson, Kaitavjeet Chowdhary, Silvia Galvan-Pena, Christophe Benoist, Elke Mühlberger, and Adam J. Hume. 2023. "Establishment of an Inactivation Method for Ebola Virus and SARS-CoV-2 Suitable for Downstream Sequencing of Low Cell Numbers" Pathogens 12, no. 2: 342. https://doi.org/10.3390/pathogens12020342
APA StyleOlejnik, J., Leon, J., Michelson, D., Chowdhary, K., Galvan-Pena, S., Benoist, C., Mühlberger, E., & Hume, A. J. (2023). Establishment of an Inactivation Method for Ebola Virus and SARS-CoV-2 Suitable for Downstream Sequencing of Low Cell Numbers. Pathogens, 12(2), 342. https://doi.org/10.3390/pathogens12020342







