Abstract
Sport facilities represent extreme indoor environments due to intense cleaning and disinfection. The aim of this study was to describe the composition of the cultivated microbiota in dust samples collected in sport facilities during the COVID-19 pandemic. A dust sample is defined as the airborne dust sedimented on 0.02 m2 within 28 d. The results show that the microbial viable counts in samples of airborne dust (n = 9) collected from seven Finnish sport facilities during the pandemic contained a high proportion of pathogenic filamentous fungi and a low proportion of bacteria. The microbial viable counts were between 14 CFU and 189 CFU per dust sample. In seven samples from sport facilities, 20–85% of the microbial viable counts were fungi. Out of 123 fungal colonies, 47 colonies belonged to the potentially pathogenic sections of Aspergillus (Sections Fumigati, Nigri, and Flavi). Representatives of each section were identified as Aspergillus fumigatus, A. flavus, A. niger and A. tubingensis. Six colonies belonged to the genus Paecilomyces. In six samples of dust, a high proportion (50–100%) of the total fungal viable counts consisted of these potentially pathogenic fungi. A total of 70 isolates were considered less likely to be pathogenic, and were identified as Aspergillus section Nidulantes, Chaetomium cochliodes and Penicillium sp. In the rural (n = 2) and urban (n = 7) control dust samples, the microbial viable counts were >2000 CFU and between 44 CFU and 215 CFU, respectively, and consisted mainly of bacteria. The low proportion of bacteria and the high proportion of stress tolerant, potentially pathogenic fungi in the dust samples from sport facilities may reflect the influence of disinfection on microbial communities.
1. Introduction
During the first half of the 20th century, outdoor exercises were the main thrust of health-promoting activities, and outdoor air was considered to be healthy and therapeutic [1,2,3,4]. However, during the latter half of the 20th century, outdoor air pollution caused by vehicle traffic became difficult to avoid in urban environments. In 2013, the WHO declared outdoor urban air as a human carcinogen [1,5,6]. In recent decades, physical activities have moved into the built environment, and indoor athletics have become increasingly popular. Figure 1 illustrates the removal of football playing from outdoor to indoor environments. Nowadays, there are over 10,000 public and private commercial sport facilities, including sport halls, fitness centers and gyms, in Finland [7,8,9,10]. However, after the millennium shift, high hygiene levels and deprivation of exposure to diverse environmental microbiota in outdoor air have been associated with immune-mediated diseases [4]. Outdoor air in green environments has once again been indicated for maintaining health [3,4].

Figure 1.
Football then and now. This figure illustrates the movement of physical activities indoors. Left panel: Depiction of health education in Finland from the first half of the 20th century, which stressed the benefits of outdoor exercises for the prevention of tuberculosis [3]. Right panel: Modern football players inside a built environment, possibly avoiding traffic air pollution in urban air and the effects of the outdoor climate (snow and rain) in Finland (photo: Sanna Sulopuisto).
Physical activities are beneficial for health [8,9,10], but the indoor environment in sport facilities may also contribute to health risks not found in outdoor environments. As in most urban buildings, in sport facilities, mechanical ventilation provides indoor closed spaces with filtered inlet air. Filtering not only removes hazardous outdoor air pollutants, but may also reduce the beneficial components of outdoor air [4]. Specific indoor air quality (IAQ) problems identified in sport facilities are the accumulation of pollutants due to poor ventilation, exposure to high amounts of biocides associated with cleaning chemicals, emissions from plastic and rubber building materials, and contagious bacteria and pathogenic fungi [11,12,13,14]. Moreover, the COVID-19 pandemic has simultaneously increased the use of indoor disinfectants and the occurrence of fungal infections [15,16,17,18,19,20,21].
The risks of fungal and bacterial infections in sport facilities are widely recognized, but the diversity of pathogenic filamentous fungi in sport facilities in Finland is not well documented. The cultivable microbiota in indoor dusts collected during the pandemic have not been investigated. In this study, we show that the airborne dusts collected from Finnish sport facilities during the COVID-19 pandemic contained a high proportion of culturable pathogenic filamentous fungi, and a surprisingly low amount of culturable bacteria. This study is a continuation of the research presented earlier [9,22], but this research focuses on the proportion of cultivable fungi versus bacteria in settled indoor dusts from several Finnish sport facilities.
2. Results
2.1. Culturable Bacteria and Fungi in Samples of Sedimented Airborne Dusts Collected from Sport Facilities during the COVID-19 Pandemic
The culturable microbiota in nine airborne dust samples collected during the COVID-19 pandemic from seven sport facilities were investigated. Nine dust samples collected from urban and rural buildings were used as controls. The dust samples represented the amounts of airborne dust sedimented into three Petri plates within 2–4 weeks, or particles on 0.02 m2 per 14–28 d (Table 1). One of the rural control dust samples from a barn with bad quality hay was used as positive control for cultivation of mesophilic and thermotolerant fungi (able to grow at 37 °C). The other rural control dust sample, collected from an occasionally occupied and hardly cleaned summer house, was used as control for indoor microbiota not exposed to synthetic surfactants and disinfectants. The seven dust samples from urban “complaint-free” educational buildings collected before the pandemic were used as controls for random ordinary urban indoor microbiota.

Table 1.
Cultivable bacteria and fungi grown on malt extract agar (MEA, pH 5.5) and tryptic soy agar (TSA, pH 7.2) at 24 and 37 °C. SF1-SF7 = dust samples from sport facilities, SD1-SD3= samples of school dusts, CD1-CD2 = samples of rural control dusts.
Results in Table 1 visualize the total numbers of colonies identified as fungi or bacteria in each dust sample. In the dust samples from the sport facilities, the fungal and bacterial colony counts varied between no colony detected to 45 CFU, and 5 CFU to 144 CFU per dust sample, respectively. The fungal colony counts in the dust samples from sport facilities were >10 times higher than in the samples of urban indoor dust, but quite similar to the fungal colony counts in the samples of rural control dusts. The number of bacterial CFU in the dust samples from sport facilities was lower than the number of bacterial CFU in all the samples of control dusts, whereas the number of fungal CFUs in dust samples from sport facilities was largely similar to the number of fungal CFU in the control dust samples.
This indicates that the methods used for sampling, sample preparation and cultivation allowed growth of both bacteria and fungi, and enabled estimation of differences in microbial compositions of the dusts. The results also show that bacteria and fungi both grew on tryptic soy agar (TSA). On TSA, fungi able to grow at neutral pH competed successfully with the co-growing microbes. Very few bacterial colonies grew on malt extract agar (MEA).
2.2. Numbers of Dust Particles and Fungi versus Bacterial Colony Counts in the Sedimented Airborne Dusts
The numbers of particles sedimented onto the Petri plates during the sampling period were estimated and shown in Figure 2. The total microbial colony counts (CFU of bacteria + fungi) were related to the number of dust particles (particulate matter, PM), visible in microscope. The results in Figure 2 and Table 2 show that densities of PM and total microbial CFU varied between >100 PM and >100 CFU in dusts from SF1 and SF5 to <10 (PM) and 15 (CFU) in dust from SF3a. Both rural control dust samples contained high amounts of PM (>1000), and a high total microbial CFU (>1000). This indicated that the amounts of airborne dust that settled within the sampling period were very different between the different sport facilities and the rural control buildings.
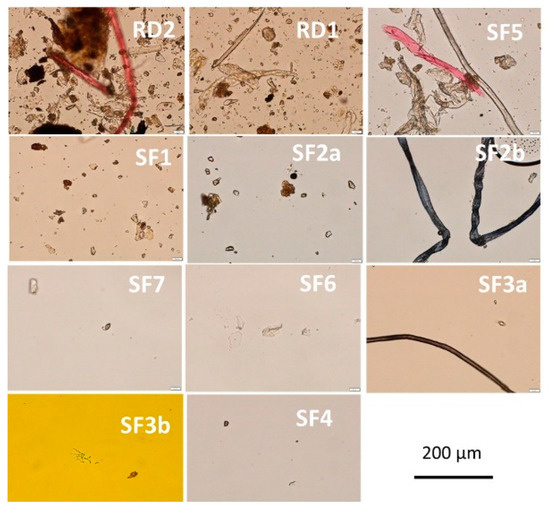
Figure 2.
Densities of particles of 11 settled dusts captured in light microscope (400×). Airborne dust samples SF1-SF7 were collected by sedimentation into Petri plates from sport facilities. The reference dusts RD1 and RD2 were collected from a hay barn and an old summer house. Each dust sample (collected in three fallout plates) was suspended in 2 mL phosphate-buffered saline (PBS). Some 10 µL of the suspension was examined in five microscopic fields. The view shown in each picture is representative of the recorded views in the five examined microscopic fields, thus representing an average view of each dust.

Table 2.
Density of particles visible in microscope (PM) and microbial colony counts (CFU) in sedimented dust samples collected from sport facilities (SFs) in Finland.
The highest number of fungal colonies, 38 CFU and 45 CFU, was detected in dust samples from SF1 and SF5, respectively, which were the samples with the highest densities of PM. Notably, however, fungal colonies of 8 CFU and 15 CFU were also detected in dust samples from SF4 and SF6, respectively, which were samples containing small amounts of PM, <10. These dust samples contained a high proportion of viable fungi compared to the total microbial viable counts. In the rural control dust samples, the amounts of bacteria exceeded the amounts of fungi by >10 times.
The results in Table 2 show that the fungal viable counts in most of the dust samples from sport facilities (8 out of 9 dust samples) were between 13% and 85% of the total microbial viable counts, whereas the fungal colony counts were <1% of the total microbial viable counts in the control samples of rural dusts. In the control samples of urban dusts, the fungal viable counts were between <1% and 5% of the total viable counts. The microbial composition in the dust samples from the sport facilities also differed from each other. The results also revealed a low number of total microorganisms and a high proportion of fungi in two sport facility dusts, SF4 and SF6, exhibiting a low density of PM.
2.3. Tracking Diversity of Fungal Colonies from the Samples of Settled Dusts Collected from Sport Facilities—Identification of the Isolates to Genus, Section and Species Levels
Fungal colonies (n = 123) representing different colony morphology and different microscopic morphology were purely cultured and screened for pathogenic potential by their ability to grow at 37 °C [22]. The 53 potentially pathogenic isolates found were identified to the genus, section and species levels by classical morphological criteria; 11 isolates were identified to species level using molecular methods. A flow chart of the results is presented in Figure 3. Out of the 70 less likely pathogenic isolates (those not growing at 37 °C), only six strains were identified. From the control dust samples collected from a barn and a summer house, 30 out of 217 isolates were identified to section level and considered potentially pathogenic (Figure 4). The results presented in Table 3 show that the fungal colonies from six dust samples collected from the sport facilities consisted mainly of potentially pathogenic isolates belonging to the genera Aspergillus and Paecilomyces, respectively. The majority of the fungal colonies in the rural control dust samples, CD1 and CD2, were the less likely pathogenic Aspergillus and Penicillium isolates not growing at 37 °C. From the urban control dust samples, only five fungal colonies tested positive for growth at 37 °C, and were identified by microscopy as yeasts.
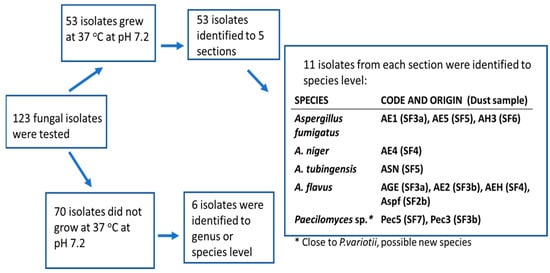
Figure 3.
Identification of the isolates from dust samples collected from sport facilities considered as potentially pathogenic based on their ability to grow at 37 °C and neutral pH. From each section and genus, selected strains from different samples were identified using molecular methods. The codes for the dust samples from the different sport facilities are shown in parentheses (SF2-SF7).
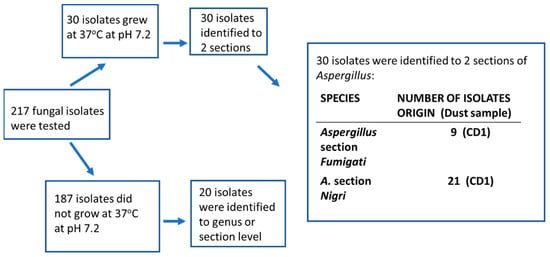
Figure 4.
Identification of the isolates from the rural control dust samples, a hay barn (CD1) and a summer house (CD2). The 30 isolates growing at 37 °C were from the barn; the summer house contained no isolates growing at 37 °C.

Table 3.
Species diversity and pathogenic potential of colonies of filamentous fungi isolated from sedimented dusts collected in sport facilities in Finland. Pathogenic potential was indicated by growth at 37 °C at neutral pH.
Specifically, the results in Table 3 show that out of the nine tested dust samples from sport facilities, six dust samples contained strains of potentially pathogenic fungal species belonging to Aspergillus sections Flavi, Fumigati and Nigri. Four dust samples contained potentially pathogenic strains of Paecilomyces sp. Two dust samples contained a majority of colonies that tested negative for pathogenic potential. Most of these colonies remained unidentified, but some isolates of Penicillium sp., Aspergillus section Nidulantes, and Chaetomium cochliodes were identified. The majority of the isolates from the rural control dust samples represented nonpathogenic species belonging to Aspergillus sections Nidulantes, Circumdati and the genera Trichoderma and Penicillium. The potentially pathogenic colonies from the rural control dust sample CD1 represented Aspergillus sections Fumigati and Nigri. The rural control dust sample CD2 from an uncleaned summer house and one sport facility dust SF2a contained no fungal isolates that tested positive for pathogenic potential. No colonies of potentially pathogenic filamentous fungi were detected from the seven urban control dust samples.
Summarizing the results in Table 1, Table 2 and Table 3, the dust samples collected during the COVID-19 pandemic from the sport facilities contained microbial colony counts that were 100 times smaller than those of the rural control samples from a hay barn and a summer house. However, the dust samples from the sport facilities differed from the control samples by a higher proportion of fungi in the viable counts for total microorganisms, and six of the samples contained a higher proportion of potentially pathogenic fungi, (81%; range: 50–100%), in the viable counts for total fungi. The two control dusts contained 18% and <1%, of potentially pathogenic fungi in the total fungal viable count. The dust samples from the urban control buildings contained no colonies identified as potentially pathogenic fungi.
2.4. Numbers of Persons Visiting the Different Locations, Density of Particles and Microbial Colony Counts in the Settled Dusts
The number of visiting persons was restricted in the sport facilities during the pandemic. The results summarized in Table 4 show that there seemed to be a positive connection between the numbers of visiting persons in the locations, the density of dust particles sedimented into the sampling devices, and the bacterial colony counts of the dusts. However, a high proportion of pathogenic fungi was recorded from facilities with low amounts of visiting persons, low densities of particles in the settled dusts and low bacterial colony counts.

Table 4.
Number of visiting persons in the sport facilities during the sampling period of settled airborne dusts, the density of dust particles visible in microscope (PM) and the microbial colony counts (CFU) in the corresponding dust samples.
2.5. Summary of Results
Summarizing the results in Table 1, Table 2, Table 3 and Table 4, the dust samples from the seven sport facilities contained lower microbial colony counts compared to control dusts from urban educational buildings and those from a barn and a summer house. However, the dust samples from the sport facilities contained a high proportion of fungi in the colony counts for total microorganisms. Six of the samples contained a high proportion of potentially pathogenic species belonging to the Aspergillus sections Flavi, Fumigati and Nigri, and representants of the genus Paecilomyces, comprising 50–100% of the total fungal colony counts. The corresponding percentages for the control dust samples were only 18% (sample of barn dust) and <1% (sample of summer house dust). The proportion of 100% of potentially pathogenic fungi in the total fungal colony counts was recorded in two dusts collected from facilities with low numbers of visiting persons, low numbers of sedimented dust particles and low bacterial colony counts.
3. Discussion
Sport facilities may provide hostile environments for microorganisms because of the effective and regular cleaning and disinfection [23,24]. During the COVID-19 pandemic, disinfection of public buildings was globally intensified [16,17,25,26,27]. This article describes the cultivable microbiota in samples of sedimented dusts collected from sport facilities during the COVID-19 pandemic. The buildings represented ordinary low risk indoor spaces; no serious indoor air quality complaints or water damage were reported. To our knowledge, this is the first report of potentially pathogenic fungi as a large proportion (but not a high number) of the cultivable microbiota of dust samples from sport facilities.
In this study, we used passive collection of settled dust, a sampling method used in several studies characterizing microbial communities in different indoor environments based on analysis of extracted microbial DNA [28,29,30,31]. We hypothesized that this sampling method would favor stress-tolerant and cleaning chemical-resistant microbes as potentially pathogenic Aspergillus spp. [22,32,33]. We chose this dust sampling method for comparing the stress-tolerant culturable microbiota in sedimented dusts from different indoor environments; indoor spaces were subjected to intense cleaning (sport facilities during the pandemic), ordinary cleaning (schools before the pandemic), and an absence of cleaning (a hay barn and a summer house).
The composition of the cultivable microbiota in nine dust samples from sport facilities differed from the compositions in the dust samples from the schools and from the rural buildings: (1) In five dust samples, the fungal colony counts were high compared to the total microbial viable counts (≥20%); (2) in six dust samples, a high proportion (>50%) of the fungal viable counts consisted of potentially pathogenic fungi; and (3) the bacterial viable counts in sport facility dusts were low, and >100 times lower than those in the rural control dusts collected from the buildings never cleaned or disinfected. The low bacterial viable counts coincided with the high proportion of pathogenic fungi. In the urban control dusts (samples of school dust) collected before the pandemic, the proportion of viable fungi was ≤5% of the total microbial viable counts.
The majority of the metabolically active building mycobiota are cultivable and identifiable to the genus level by cultivation-based conventional methods [34]. Non-selective media increase microbial diversity in the obtained viable counts compared to selective media. Complex nonselective culture media without sugars such as TSA (pH 7.2) restrict the growth of rapidly growing microorganisms [35]. The TSA medium also allowed growth of common pathogenic fungi [22].
The virulence of pathogenic fungi is connected to viability and ability to sense and cope with environmental stress in hostile environments. The virulence factors required for invasive fungal infections in humans include the ability to grow at the pH of human tissues, 7.2–7.4, at ≥37 °C [36,37,38]. Strains of the potentially pathogenic species Trichoderma longibrachiatum and the less likely pathogenic Aspergillus westerdijkiae were isolated from viable indoor samples counted on TSA [39,40]. Additional virulence factors of Aspergillus spp., heat resistance and adaptation to extreme pH values, have been shown to be connected to their resistance to disinfectants, cleaning chemicals and antifungal drugs [22,32,33,36,37,38,41]. The explanation for the high proportion of potentially pathogenic fungi in sport facility dusts may be that the intense disinfection in sport facilities irradicated most of the viable microorganisms, but not the stress-tolerant and virulent potentially pathogenic fungi.
Screening for pathogenic potential (growth at 37 °C at neutral pH), and characterization to the genus/section level seemed to be successful methods for the detection and enumeration of the viable potentially pathogenic fungi in the collected dust samples. Identification to the species level for evaluation of risk group and biohazard levels required DNA sequence analysis of marker genes [34,42,43]. Results in Figure 3 and Table 5 show that dust samples from sport facilities contained strains of A. flavus and A. fumigatus classified as risk group 2 (RG-2) and bio-safety level 2 (BSL-2) organisms, species known to cause human infections [22,42]. Recently, the WHO ranked fungal pathogens into three priority groups (critical, high, and medium). A. fumigatus was the only representant of the genus Aspergillus ranked into the critical priority group [44]. A. niger and A. tubingensis are classified as risk group 1 and 2 organisms, respectively, although human infections have been reported [42,43]. Out of the nine control dust samples, the only one which contained strains of Aspergillus section Fumigati and section Nigri was the dust sample from the barn containing moldy hay. This indicates that Aspergillus spp. belonging to RG-2 are not ubiquitous in Finnish buildings. The two potentially pathogenic Paecilomyces sp. were found to be close to, but not identical to Paecilomyces variotii. These strains possibly represent a new, as-yet undescribed species and could not be assigned to any risk group. Nevertheless, P. variotii is classified as an RG-2 organism [42,43].

Table 5.
Selected strains representing the potentially pathogenic genera identified to species level.
The airborne bacteria and fungi measured with an RCS microbial air sampler revealed that the ratios of total bacteria and total fungi were lower indoors than outdoors in all sport facilities [9]. It could be speculated that users might carry fungal spores from outdoor air into sport facilities. The sport facilities were all mechanically ventilated. Uncleaned unsubstituted inlet filters of air conditioning and mechanical ventilation might have served as fungal reservoirs in hospitals [45,46].
In indoor air, impurities are inflated compared to outdoor air wherein the content of every breath differs from the next [4,47]. In forested and green environments, the air contains a diverse microbiota. Environmental microbial exposure, human commensal microbiota, and immunological pathways are generally assumed to be interconnected. A high hygiene level and urban lifestyle may result in microbial imbalance, referred to as dysbiosis, which has been associated with immune-mediated diseases [4,48,49,50,51].
4. Conclusions
As the number of tested dust samples is too low to be able to draw far-reaching conclusions, this study is merely to be considered a pilot work concerning differences in microbial communities in indoor dusts from rural and urban buildings, and from buildings subjected to intense cleaning and disinfection. The viable potentially pathogenic fungi detected meant hardly any health risk for the users. The low bacterial viable counts in the dusts from the sport facilities compared to the control dusts may reflect the use of disinfectants, the low numbers of users and filtration of the inlet air. The microbial composition in the sedimented airborne dust in sport facilities during the COVID-19 pandemic may illustrate some unexpected and unwanted effects of intense cleaning and disinfection of indoor closed spaces.
5. Materials and Methods
5.1. Sampling Design
To investigate the composition of the cultivable microbiota in settled indoor dusts from sport facilities and from urban and rural control buildings, dust collection and cultivation of microbes were designed as shown in Figure 5. Nine dusts from 7 sport facilities were compared to the 9 control dusts, themselves consisting of 7 dusts from 3 urban educational buildings and 2 dusts from 2 rural buildings. The control dusts were collected from urban educational buildings which were complaint-free concerning IAQ, from a barn containing moldy hay, and from a summer house never cleaned and disinfected. The dust samples were investigated for colony counts (CFU) of the following: (a) total microorganisms (bacteria + fungi); (b) total fungi (fungi growing at 24 °C + fungi growing at 37 °C); and (c) fungal isolates with pathogenic potential screened by the ability to grow on TSA (pH 7.2) at 37 °C. Potentially pathogenic isolates were identified to genus, section and species level. The proportion of viable counts of potentially pathogenic fungi in the microbiota was related to the viable counts of total fungi, of total microorganisms, and of bacteria.
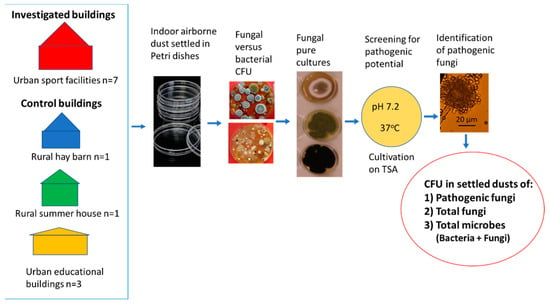
Figure 5.
The experimental design for comparison of the fungal and bacterial colony counts in settled air-borne dusts and for screening for potentially pathogenic fungi. Pathogenic potential was indicated by growth on tryptic soy agar (TSA) at 37 °C.
5.2. The Study Buildings
The seven investigated sport facilities are located in 3 cities in southern Finland. The 5 reference buildings were three educational buildings in the 3 cities, a hay barn in a village in southern Finland, and a summer house in a village in central Finland. Pictures illustrating the three types of buildings are shown in Figure 6. Details of the sampling sites are presented in Table 6.

Figure 6.
The outside (panels A–C) and inside (panels a–c) views of the urban and rural buildings examined in this study. Panels A and a represent views of an urban sport facility in Helsinki. The further panels show the two rural reference buildings, a hay barn (B,b), collection site for dust CD1, and a rural summer house (C,c), collection site for dust CD2. (Panels D,d) shows views of an urban educational building in Espoo.

Table 6.
Locations, activities and occupancy characterizing the sampling sites of urban sport facilities and two rural reference buildings. The sport facilities were built between 1935–2008, had mechanical ventilation and were under modest use during the sampling time [3]. The reference buildings were >100 years old, had natural ventilation and were only sporadically visited by humans.
5.3. Collection of Sedimented Airborne Dust by a Standardised Method
Passive collection of airborne dust settling into Petri plates over a specified time period is a method of detecting differences in amounts of aerosolized dust particles, and also enables the detection of differences in the composition of dust-borne microbiota. Passive collections of settled dust have been used in several studies wherein the microbial communities in different indoor environments have been investigated, characterized and compared [52,53,54,55].
From the sport facilities and the control buildings, dusts were sedimented for 2–4 weeks into sterile polystyrene Petri plates (Berner, 90 × 15 mm, SILEÄ0099/S). The plates were placed 150 cm to 220 cm above floor level, and as far as possible from the inlet and outlet ventilation ducts, doors, windows, and heating sources. A 50 cm minimum air column was ensured above the plates to allow free air movement. After sampling, the plates were sealed with parafilm and stored at room temperature. The dusts from the sport facilities were analyzed at 2–4 months after collection, and the reference building dust samples 2–6 months after collection.
The use of the sport facilities was strongly restricted during the sampling period due to the ongoing COVID-19 pandemic. Largely, only professional athletes and teams and school classes were permitted to use the facilities. Therefore, the occupancy rate was significantly lower than that during normal use, and the occupancy varied depending on the sampling site. The estimated occupancy rate in each sampling site during the sampling period is shown in Table 5.
5.4. Cultivation of Dust Samples, Enumeration of Bacterial and Fungal Colonies, Isolation of Pure Cultures and Testing for Pathogenic Potential
From each site, 3 Petri plates of sedimented dust were collected. The dusts were removed from the plates by suspending in 0.7 mL PBS to flush the bottom surface of the plate. From each sample, 2.1 mL PBS + 0.05% of Tween (Sigma Aldrich, St. Louis, MO, USA) was collected. The bottoms of the plates were then wiped with a moist cotton swab, and the swab was then immersed in the collected PBS. Ten µL from each sample of the visibly cloudy PBS was inspected by microscopy (400× magnification; Olympus CKX41, Tokyo, Japan) and an image recording software (cellSens® standard v. 11.0.06, 2012, Olympus Soft Imaging Solutions GmbH, Münster, Germany) to enumerate the dust particles removed from the plastic surface of the plates. Of the collected PBS, 100 µL was applied and diluted on cultivation plates, and incubated according to the schedule shown in Figure 7.
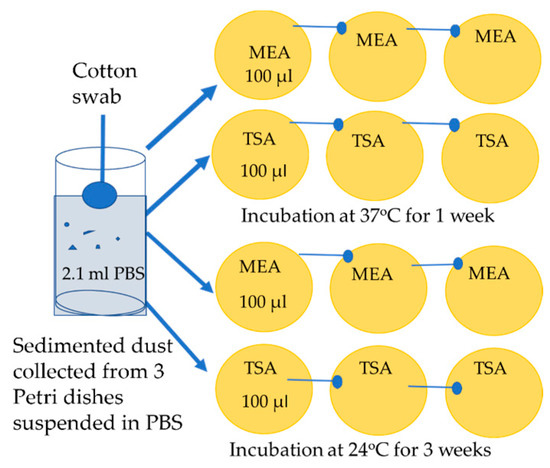
Figure 7.
Cultivation of a sample of sedimented dust. The dust collected from each sample was suspended in 2.1 mL PBS, and 100 µL was applied on the first vertical row of plates. The liquid was spread with the cotton swab on the first plate, the next two plates were wiped with the same swab.
The dusts were cultivated on malt extract agar (MEA), pH 5.4, and tryptic soy agar (TSA), pH 7.2 (Oxoid Ltd., Thermo Fisher, Heysham, Ireland). The plates were sealed with gas-permeable tape before incubation. To detect Trichoderma isolates growing as mycoparasites, the plates incubated at room temperature were inspected for the final time after 6 weeks of incubation.
5.5. Tracking Diversity of Fungal Isolates, Assignation to Genera and Sections, Testing for Pathogenic Potential, and Identification to the Species Level
The fungal colonies cultured pure on MEA at 22 °C were screened for pathogenic potential by their growth at 37 °C on TSA (pH 7.2) [22,30,31,32,33]. Some isolates identified as yeast by the microscope were not further identified. All isolates of filamentous fungi that screened positive for pathogenic potential were identified to genus and section level as Aspergillus sections Flavi, Fumigati, Nigri and Paecilomyces sp. by colony morphology on MEA, size and morphology of conidiophores and conidia in a light microscope (400× magnification), with the equipment described above [22,52]. The identification of the isolates was confirmed by comparison to selected strains isolated from the sport facilities and identified earlier [22,52]. Most of the selected strains listed in Table 6 were identified to the species level in previous studies by sequence analysis of cmdA [53] and/or rbp2 [54]. The identified strains, the gene loci used for identification and the GenBank accession numbers are listed in Table 6. The two Paecilomyces strains in Table 6 could not be assigned to an existing species based on the cmdA locus amplified during this study with primers Cmd5 and Cmd6, according to [55].
The strains that tested negative for pathogenic potential were identified to the species level as Chaetomium cochliodes, as shown in (Table 6). Strains belonging to Aspergillus section Nidulantes and Aspergillus westerdijkiae were identified by comparison to strains SL/3 and PP2, respectively, identified by DSMZ [22] and according to Samson et al. [52]. Trichoderma and Penicillium strains were identified to the genus level based on colony morphology and the morphology of conidiophores, by comparison to reference strains described [56,57] and according to Samson et al. [52]. The morphology of the isolates identified is visualized in Table S1 of the Supplementary Material.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/pathogens12020339/s1, Table S1: Morphology of major genera of fungal strains isolated from settled indoor dusts collected in sport facilities in Finland. Pathogenic potential was estimated based on the ability to grow at 37 °C on Tryptic Soy Agar (TSA), pH 7.2, compared to growth on Malt Extract Agar (MEA), pH 5.5. The strains represent 67% of the total number of colonies.
Author Contributions
Conceptualization, M.A.; data curation, formal analysis, A.V. and M.A.; funding acquisition, H.S.; investigation, A.V., M.A., C.V.-W. and T.K.; methodology, M.A. and A.V.; project administration, H.S., R.M. and C.V.-W.; resources, L.K.; supervision, L.K. and H.S.; validation, L.K. and H.S.; visualization, M.A.; writing—original draft, M.A.; writing—review and editing, M.A., C.V.-W., L.K. and A.V. All authors have read and agreed to the published version of the manuscript.
Funding
The Academy of Finland (grant for CleanSchool, 330150); The Finnish Work Environment Fund (grant for LIIKU, 200068).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Raw data are available upon request to the authors.
Acknowledgments
We wish to thank Emeritus Tari Haahtela for his useful comments and advice.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Martini, M.; Gazzaniga, V.; Behzadifar, M.; Bragazzi, N.L.; Barberis, I. The history of tuberculosis: The social role of sanatoria for the treatment of tuberculosis in Italy between the end of the 19th century and the middle of the 20th. J. Prev. Med. Hyg. 2018, 59, E323–E327. [Google Scholar] [CrossRef] [PubMed]
- De Los Santos, E. Flashback: In the Fight Against Tuberculosis, Open Air Schools in Chicago Took an Unorthodox Approach: Keep Kids Outside, Even in Winter. Chicago Tribune. 1 May 2020. Available online: https://www.chicagotribune.com/history/ct-opinion-flashback-open-air-schools-tuberculosis-20200501-phyphmoe4vhhnelpjj2sck7fmi-story.html (accessed on 15 February 2023).
- Sipinen, J. Terveys on Kallein Aarteesi-Ihannekansalainen 1930-Luvun Suomalaisessa Terveysvalistuksessa. Suomi 2016, 3, 1–43. Available online: https://hybrislehti.net/terveys-on-kallein-aarteesi-ihannekansalainen-1930luvun-suomalaisessa-terveysvalistuksessa (accessed on 15 February 2023).
- Haahtela, T. Biodiversity for resilience—What is needed for allergic children. Pediatr. Allerg. Immunol. 2022, 33, e13779. [Google Scholar] [CrossRef] [PubMed]
- Pascal, M.; Corso, M.; Chanel, O.; Declercq, C.; Badaloni, C.; Cesaroni, G.; Henschel, S.; Meister, K.; Haluza, D.; Martin-Olmedo, P.; et al. Assessing the public health impacts of urban air pollution in 25 European cities: Results of the Aphekom project. Sci. Total Environ. 2013, 449, 390–400. [Google Scholar] [CrossRef] [PubMed]
- IARC. Press Release No. 221. Outdoor Air Pollution a Leading Environmental Cause of Cancer Deaths. 2013. Available online: https://www.iarc.who.int/wp-content/uploads/2018/07/pr221_E.pdf (accessed on 15 February 2023).
- LIPAS Database (LIPAS-Tietokanta). Computer Based Information System of Sports Sites and Facilities. University of Jyväskylä. 2020. Available online: https://www.lipas.fi/liikuntapaikat (accessed on 15 February 2023).
- WHO (World Health Organization). Physical Activity Strategy for the WHO European Region 2016–2025; WHO Regional Office for Europe: Copenhagen, Denmark, 2016; ISBN 978-928-905-147-7. [Google Scholar]
- Koivisto, T. Air Contaminants in Different Indoor Sports Facilities. Master’s Thesis, Aalto University, Aalto, Finland, 2021. Available online: https://aaltodoc.aalto.fi/handle/123456789/110685 (accessed on 15 February 2023).
- Salonen, H.; Salthammer, T.; Morawska, L. Human exposure to air contaminants in sports environments. Indoor Air 2020, 6, 1109–1129. [Google Scholar] [CrossRef]
- Ramos, C.A.; Viegas, C.; Cabo Verde, S.; Wolterbeek, T.H.; Almeida, S.M. Characterizing the fungal and bacterial microflora and concentrations in fitness centers. Indoor Built Environ. 2015, 25, 872–882. [Google Scholar] [CrossRef]
- Ramos, C.A.; Reis, J.F.; Almeida, T.; Wolterbeek, T.H.; Almeida, S.M. Estimating the inhaled dose of pollutants during indoor physical activity. Sci. Total Environ. 2015, 527–528, 111–118. [Google Scholar] [CrossRef]
- Ramos, C.A.; Wolterbeek, T.H.; Almeida, S.M. Exposure to indoor air pollutants during physical activity in fitness centers. Build. Environ. 2014, 82, 349–360. [Google Scholar] [CrossRef]
- Boonrattanakij, N.; Yomchinda, S.; Lin, F.; Luzvisminda, M.; Bellotindos, L.; Lu, M.-C. Investigation and disinfection of bacteria and fungi in sports fitness centers. Environ. Sci. Pollut. Res. 2021, 28, 52576–52586. [Google Scholar] [CrossRef]
- Zheng, G.; Webster, T.; Salamova, A. Quaternary ammonium compounds: Bioaccumulation potentials in humans and levels in blood before and during the COVID-19 pandemic. Environ. Sci. Technol. 2021, 55, 14689–14698. [Google Scholar] [CrossRef]
- Dewey, H.; Jones, J.; Keating, M.; Budhathoki-Upret, J. Increased use of disinfectants during the COVID-19 pandemic and its potential impacts on health and safety. ACS Chem. Health Saf. 2022, 29, 27–38. [Google Scholar] [CrossRef]
- Koronavirus: Uusi Ohje Siivoukseen Suomalaisille Työpaikoille. Available online: https://www.uusisuomi.fi›Uutiset›Työterveys (accessed on 15 February 2023).
- Baddley, J.W.; Thompson, G.R.; Chen, S.C.A.; White, P.L.; Johnson, M.D.; Nguyen, M.H.; Schwartz, I.S.; Spec, A.; Ostrosky-Zeichner, L.; Jackson, B.R.; et al. Coronavirus Disease 2019-associated invasive fungal infection. Open Forum Infect. Dis. 2021, 8, ofab5. [Google Scholar] [CrossRef]
- Fortarezza, F.; Boscolo, A.; Pezzuto, F.; Lunardi, F.; Jesús Acosta, M.; Giraudo, C.; Del Vecchio, C.; Sella, N.; Tiberio, I.; Godi, I.; et al. Proven COVID-19-associated pulmonary aspergillosis in patients with severe respiratory failure. Mycoses 2021, 64, 1223–1229. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Singh, R.; Joshi, S.R.; Misra, A. Mucormycosis in COVID-19: Mucormycosis in COVID-19: A systematic review of cases reported worldwide and in India. Diabetes Metab. Syndr. 2021, 15, 102146. [Google Scholar] [CrossRef] [PubMed]
- Monika, P.; Chandraprabha, M.N. Risks of mucormycosis in the current Covid-19 pandemic: A clinical challenge in both immunocompromised and immunocompetent patients. Mol. Biol. Rep. 2022, 49, 4977–4988. [Google Scholar] [CrossRef] [PubMed]
- Andersson, M.; Varga, A.; Mikkola, R.; Vornanen-Winqvist, C.; Salo, J.; Kredics, L.; Kocsubé, S.; Salonen, H. Aspergillus was the dominant genus found during diversity tracking of cultivable potentially pathogenic indoor fungal isolates. Pathogens 2022, 11, 1171. [Google Scholar] [CrossRef] [PubMed]
- Viegas, C.; Alves, C.; Carolino, E.; Rosado, L.; Santos, C.S. Prevalence of fungi in indoor air with reference to gymnasiums with swimming pools. Indoor Built Environ. 2010, 19, 555–561. [Google Scholar] [CrossRef]
- Liang, Z.; Dong, C.B.; Liang, H.; Zhen, Y.X.; Zhou, R.L.; Han, Y.F.; Liang, Z.Q. A microbiome study reveals the potential relationship between the bacterial diversity of a gymnastics hall and human health. Sci. Rep. 2022, 12, 5663. [Google Scholar] [CrossRef]
- Ekanayake, A.; Rajapaksha, A.U.; Hewawasam, C.; Anand, U.; Bontempi, E.; Kurwadkar, S.; Biswas, J.K.; Vithanage, M. Environmental challenges of COVID-19 pandemic: Resilience and sustainability—A review. Environ. Res. 2022, 216, 114496. [Google Scholar] [CrossRef]
- Guo, J.; Liao, M.; He, B.; Liu, J.; Hu, X.; Yan, D.; Wang, J. Impact of the COVID-19 pandemic on household disinfectant consumption behaviors and related environmental concerns: A questionnaire-based survey in China. J. Environ. Chem. Eng. 2021, 9, 106168. [Google Scholar] [CrossRef]
- Lou, J.; Wang, W.; Lu, H.; Wang, L.; Zhu, L. Increased disinfection byproducts in the air resulting from intensified disinfection during the COVID-19 pandemic. J. Hazard. Mater. 2021, 418, 126249. [Google Scholar] [CrossRef] [PubMed]
- Adams, R.I.; Tian, Y.; Taylor, J.W.; Bruns, T.D.; Hyvarinen, A.; Taubel, M. Passive dust collectors for assessing airborne microbial material. Microbiome 2015, 3, 46. [Google Scholar] [CrossRef] [PubMed]
- Adams, R.I.; Miletto, M.; Taylor, J.W.; Bruns, T.D. Dispersal in microbes: Fungi in indoor air are dominated by outdoor air and show dispersal limitation at short distances. ISME J. 2013, 7, 1262–1273. [Google Scholar] [CrossRef] [PubMed]
- Emerson, J.B.; Keady, P.B.; Brewer, T.E.; Clements, N.; Morgan, E.E.; Awerbuch, J.; Miller, S.L.; Fierer, N. Impacts of flood damage on airborne bacteria and fungi in homes after the 2013 Colorado front range flood. Environ. Sci. Technol. 2015, 49, 2675–2684. [Google Scholar] [CrossRef]
- Madsen, A.M.; Matthiesen, C.B.; Frederiksen, M.W.; Frederiksen, M.; Frankel, M.; Spilak, M.; Gunnarsen, L.; Timm, M. Sampling, extraction and measurement of bacteria, endotoxin, fungi and inflammatory potential of settling indoor dust. J. Environ. Monitor. 2012, 14, 3230–3239. [Google Scholar] [CrossRef]
- Meade, E.; Slattery, M.A.; Garvey, M. Biocidal resistance in clinically relevant microbial species: A major public health risk. Pathogens 2021, 10, 598. [Google Scholar] [CrossRef]
- Barabote, R.D.; Thekkiniath, J.; Strauss, R.E.; Vediyappan, G.; Fralick, J.A.; Francisco, M.J.S. Xenobiotic efflux in bacteria and fungi: A genomics update. Adv. Enzymol. Relat. Areas Mol. Biol. 2011, 77, 237–306. [Google Scholar]
- Andersen, B.; Frisvad, J.C.; Dunn, R.R.; Thrane, U. A pilot study on baseline fungi and moisture indicator fungi in Danish homes. J. Fungi 2021, 7, 71. [Google Scholar] [CrossRef]
- Stewart, S.L.; Grinshpun, S.A.; Willeke, K.; Terzieva, S.; Ulevicius, V.; Donnelly, J. Effect of impact stress on microbial recovery on an agar surface. Appl. Environ. Microbiol. 1995, 61, 1232–1239. [Google Scholar] [CrossRef]
- Cornet, M.; Gaillardin, C. pH signaling in human fungal pathogens: A new target for antifungal strategies. Eukaryot. Cell 2014, 13, 342–352. [Google Scholar] [CrossRef]
- Davis, D. How human pathogenic fungi sense and adapt to pH: The link to virulence. Curr. Opin. Microbiol. 2009, 12, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Dichtl, K.; Samantaray, S.; Wagener, J. Cell wall integrity signalling in human pathogenic fungi. Cell Microbiol. 2016, 18, 1228–1238. [Google Scholar] [CrossRef] [PubMed]
- Mikkola, R.; Andersson, M.A.; Kredics, L.; Grigoriev, P.A.; Sundell, N.; Salkinoja-Salonen, M.S. 20-residue and 11-residue peptaibols from the fungus Trichoderma longibrachiatum are synergistic in forming Na+/K+-permeable channels and adverse action towards mammalian cells. FEBS J. 2012, 279, 4172–4190. [Google Scholar] [CrossRef] [PubMed]
- Mikkola, R.; Andersson, M.A.; Hautaniemi, M.; Salkinoja-Salonen, M.S. Toxic indole alkaloids avrainvillamide and stephacidin B produced by a biocide tolerant indoor mold Aspergillus westerdijkiae. Toxicon 2015, 99, 58–67. [Google Scholar] [CrossRef]
- Raksha; Singh, G.; Urhekar, A.D. Virulence factors detection in Aspergillus isolates from clinical and environmental samples. J. Clin. Diagn. Res. 2017, 11, DC13–DC18. [Google Scholar]
- Góralska, K.; Błaszkowska, J.; Dzikowiec, M. The occurrence of potentially pathogenic filamentous fungi in recreational surface water as a public health risk. J. Water Health 2020, 18, 127–144. [Google Scholar] [CrossRef]
- German Collection of Microorganisms and Cell Cultures GmbH: Catalogue (dsmz.de). Available online: https//dsmz.de (accessed on 15 February 2023).
- WHO Fungal Priority Pathogens List to Guide Research, Development and Public Health Action; Licence: CC BY-NC-SA 3.0 IGO; World Health Organization: Geneva, Switzerland, 2022.
- Ruutu, P.; Valtonen, V.; Tiitanen, L.; Elonen, E.; Volin, L.; Veijalainen, P.; Ruutu, T. An outbreak of invasive aspergillosis in a haematologic unit. Scand J. Infect. Dis. 1987, 19, 347–351. [Google Scholar] [CrossRef]
- Ichai, P.; Saliba, F.; Baune, P.; Daoud, A.; Coilly, A.; Samuel, D. Impact of negative air pressure in ICU rooms on the risk of pulmonary aspergillosis in COVID-19 patients. Crit. Care 2020, 24, 538. [Google Scholar] [CrossRef]
- Dylag, M.; Spychała, K.; Zielinski, J.; Łagowski, D.; Gnat, S. Update on Stachybotrys chartarum—Black mold perceived as toxigenic and potentially pathogenic to humans. Biology 2022, 11, 352. [Google Scholar] [CrossRef]
- Selway, C.A.; Mills, J.G.; Weinstein, P.; Skelly, C.; Yadav, S.; Lowe, A.; Breed, M.F.; Weyrich, L.S. Transfer of environmental microbes to the skin and respiratory tract of humans after urban green space exposure. Environ. Int. 2020, 145, 106084. [Google Scholar] [CrossRef]
- Roslund, M.I.; Puhakka, R.; Grönroos, M.; Nurminen, N.; Oikarinen, S.; Gazali, A.M.; Cinek, O.; Kramná, L.; Siter, N.; Vari, H.K.; et al. Biodiversity intervention enhances immune regulation and health-associated commensal microbiota among daycare children. Sci. Adv. 2020, 6, eaba2578. [Google Scholar] [CrossRef] [PubMed]
- Rook, G.; Bloomfield, S. Microbial exposures that establish immunoregulation are compatible with targeted hygiene. J. Allerg Clin. Immunol. 2021, 148, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Soininen, L.; Roslund, M.I.; Nurminen, N.; Puhakka, R.; Laitinen, O.H.; Hyöty, H.; Sinkkonen, A. Indoor green wall affects health-associated commensal skin microbiota and enhances immune regulation: A randomized trial among urban office workers. Sci Rep. 2022, 12, 6518. [Google Scholar] [CrossRef] [PubMed]
- Samson, R.A.; Hoekstra, E.S.; Frisvad, J.C.; Filtenborg, O. (Eds.) Introduction to Food and Air-Borne Fungi, 6th ed.; Centraalbureau voor Schimmelcultures: Utrecht, The Netherlands, 2002. [Google Scholar]
- Druzhinina, I.S.; Komon-Zelazowsa, M.; Kredics, L.; Hatvani, L.; Antal, Z.; Belayneh, T.; Kubicek, C.P. Alternative reproductive strategies of Hypocrea orientalis and genetically close but clonal Trichoderma longibrachiatum, both capable of causing invasive mycoses of humans. Microbiology 2008, 154, 3447–3459. [Google Scholar] [CrossRef]
- Houbraken, J.; Kocsubé, S.; Visagie, C.M.; Yilmaz, N.; Wang, X.-C.; Meijer, M.; Kraak, B.; Hubka, V.; Bensch, K.; Samson, R.A.; et al. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series and species. Stud. Mycol. 2020, 95, 5–169. [Google Scholar] [CrossRef]
- Hong, S.; Go, S.; Shin, H.; Frisvad, J.; Samson, R. Polyphasic taxonomy of Aspergillus fumigatus and related species. Mycologia 2005, 97, 1316–1329. [Google Scholar] [CrossRef] [PubMed]
- Castagnoli, E.; Marik, T.; Mikkola, R.; Kredics, L.; Andersson, M.A.; Salonen, H.; Kurnitski, J. Indoor Trichoderma strains emitting peptaibols in guttation droplets. J. Appl. Microbiol. 2018, 125, 1408–1422. [Google Scholar] [CrossRef]
- Salo, M.J.; Marik, T.; Mikkola, R.; Andersson, M.A.; Kredics, L.; Salonen, H.; Kurnitski, J. Penicillium expansum strain isolated from indoor building material was able to grow on gypsum board and emitted guttation droplets containing chaetoglobosins and communesins A, B and D. J. Appl. Microbiol. 2019, 127, 1135–1147. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).