Electrostatic Adsorption of Dense AuNPs onto Silica Core as High-Performance SERS Tag for Sensitive Immunochromatographic Detection of Streptococcus pneumoniae
Abstract
1. Introduction
2. Experimental Section
2.1. Chemicals, Materials, and Instruments
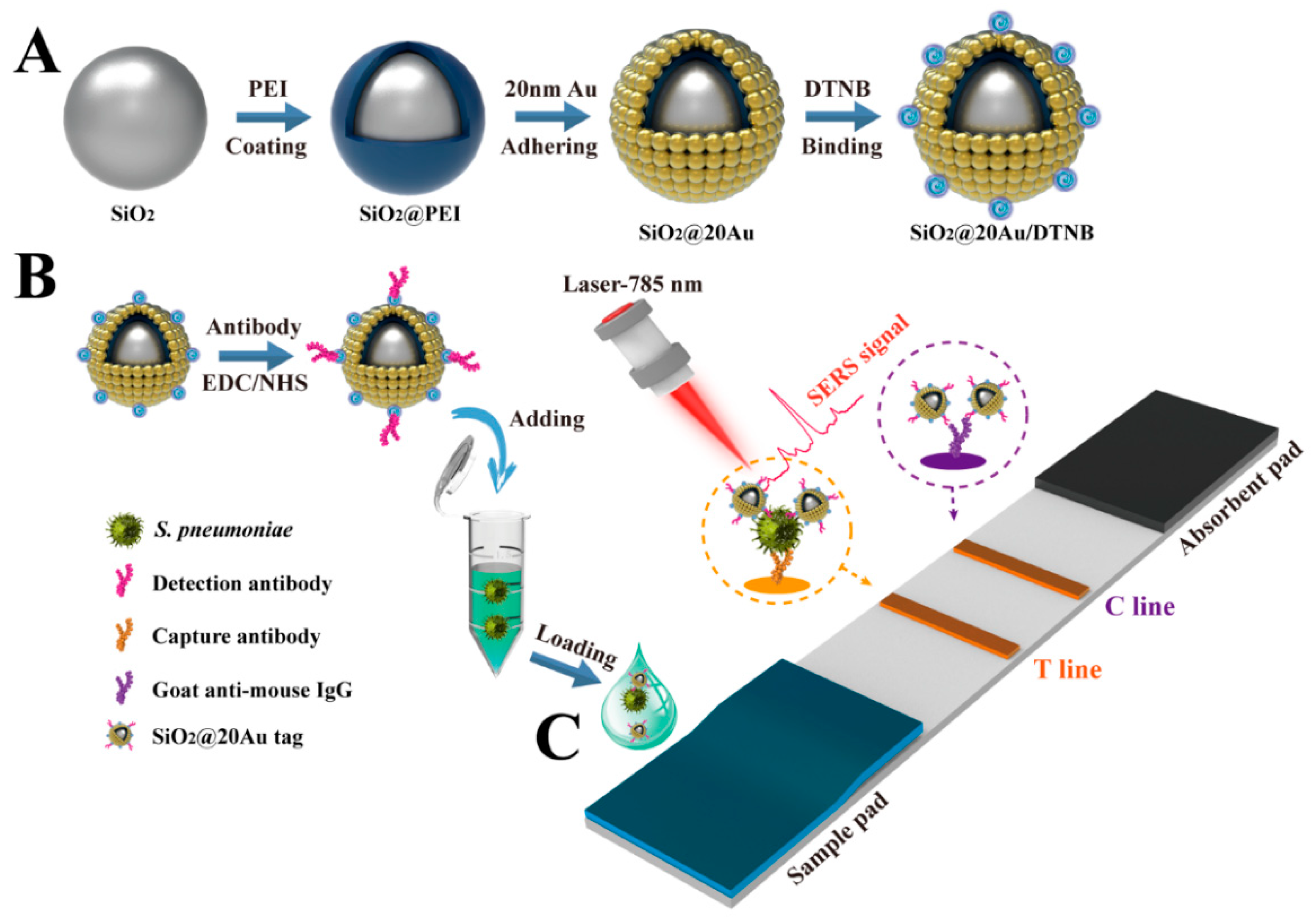
2.2. Preparation of 180 nm SiO2 NPs, 20 nm AuNPs, and SiO2@20Au NPs
2.3. Preparation of SiO2@20Au SERS Tags
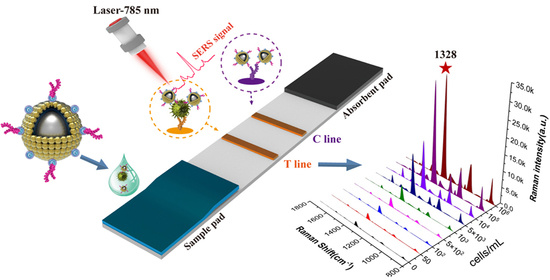
2.4. Preparation of SiO2@20Au-Based SERS-ICA for S. pneumoniae Detection
2.5. Preparation of Bacterial Sample
- Forward primer: 5′-AACTCTTACGCAATCTAGCAGATGAA-3′;
- Reverse primer: 5′-CGTGCAATACTCGTGCGTTTTA-3′
2.6. Bacteria Detection via SiO2@20Au-Based SERS-ICA
3. Results and Discussion
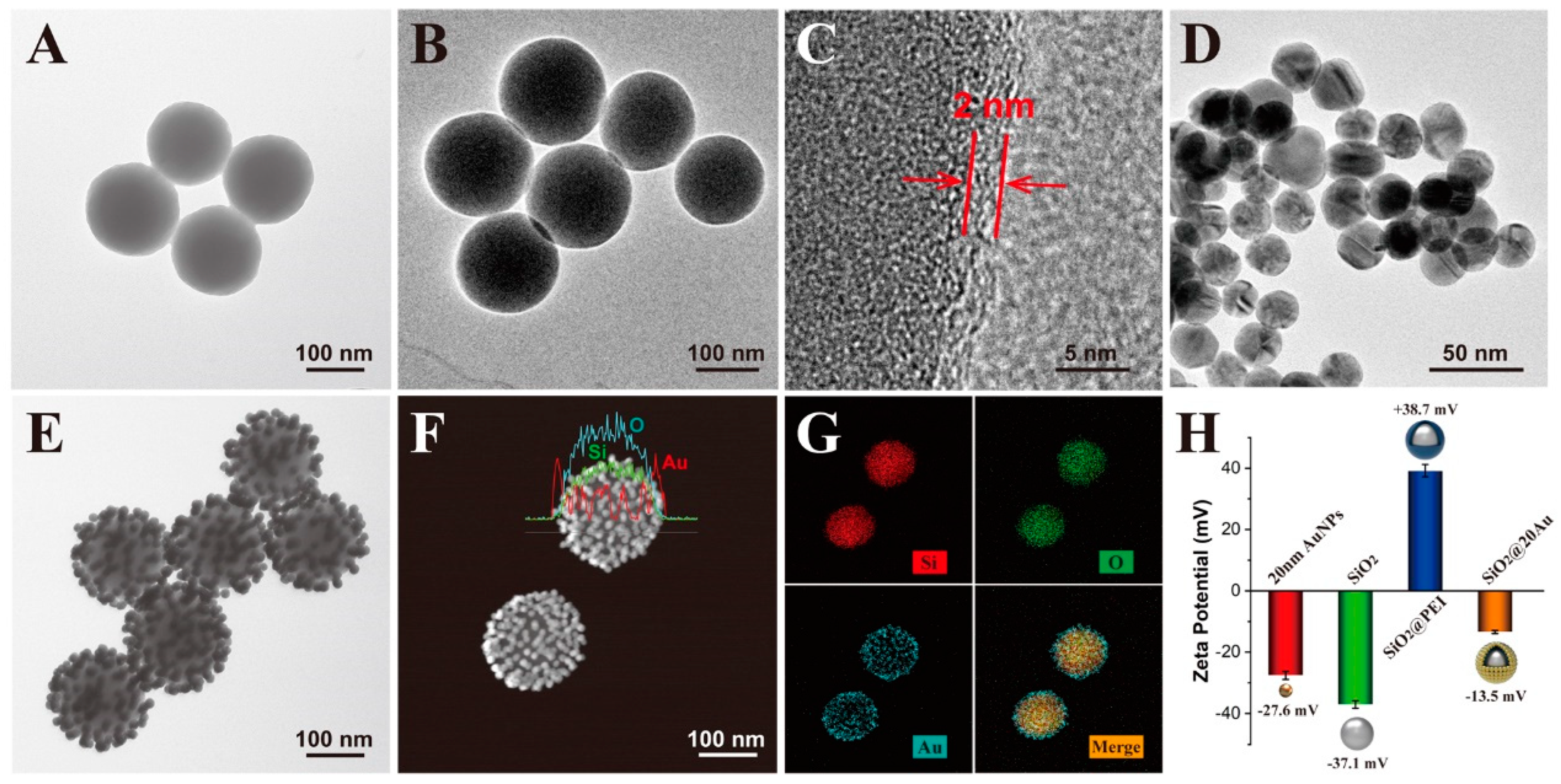
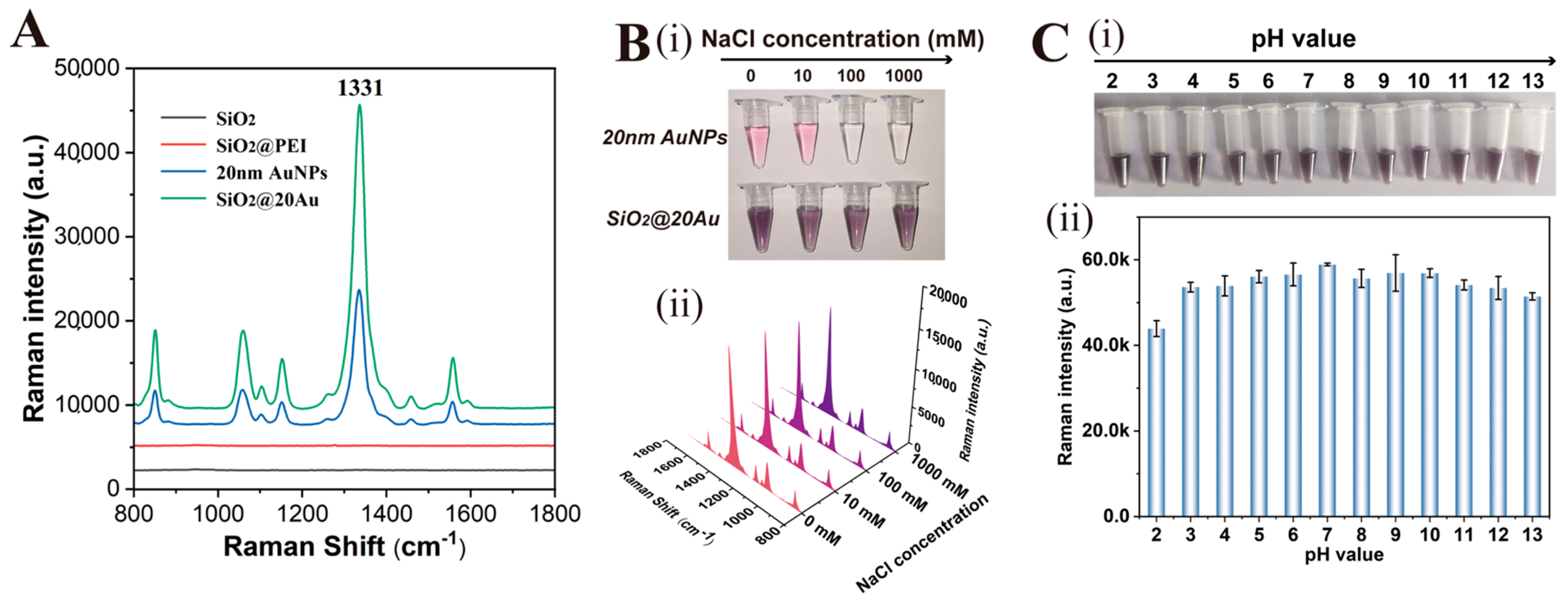
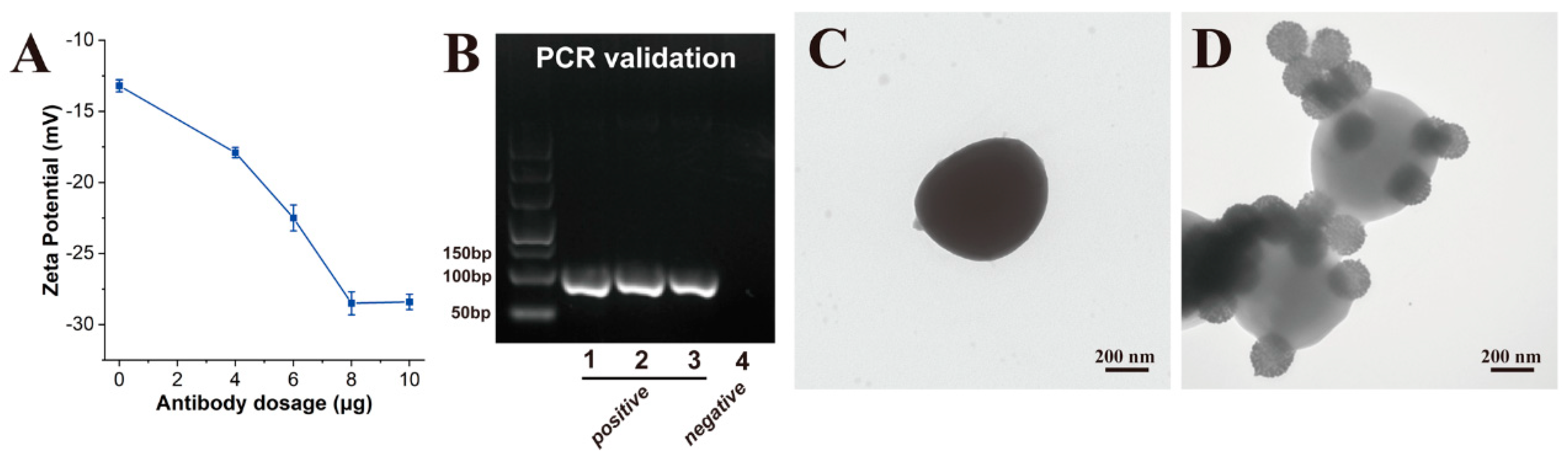
3.1. Preparation and Characterization of SiO2@20Au SERS Tags
3.2. Design and Optimization of SERS-ICA for S. pneumoniae Detection
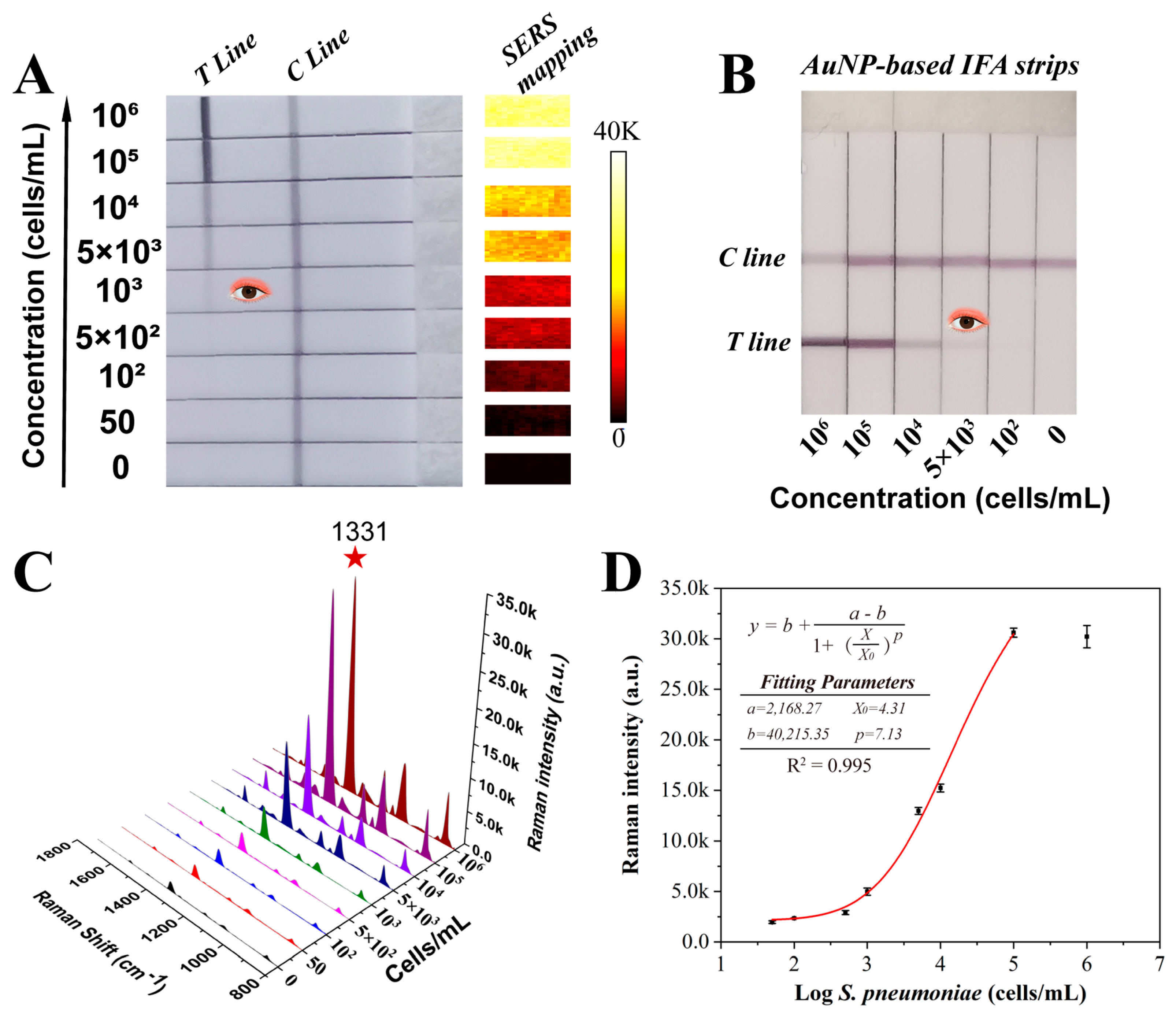
3.3. Detection Performance of SiO2@20Au-SERS-ICA for S. pneumoniae
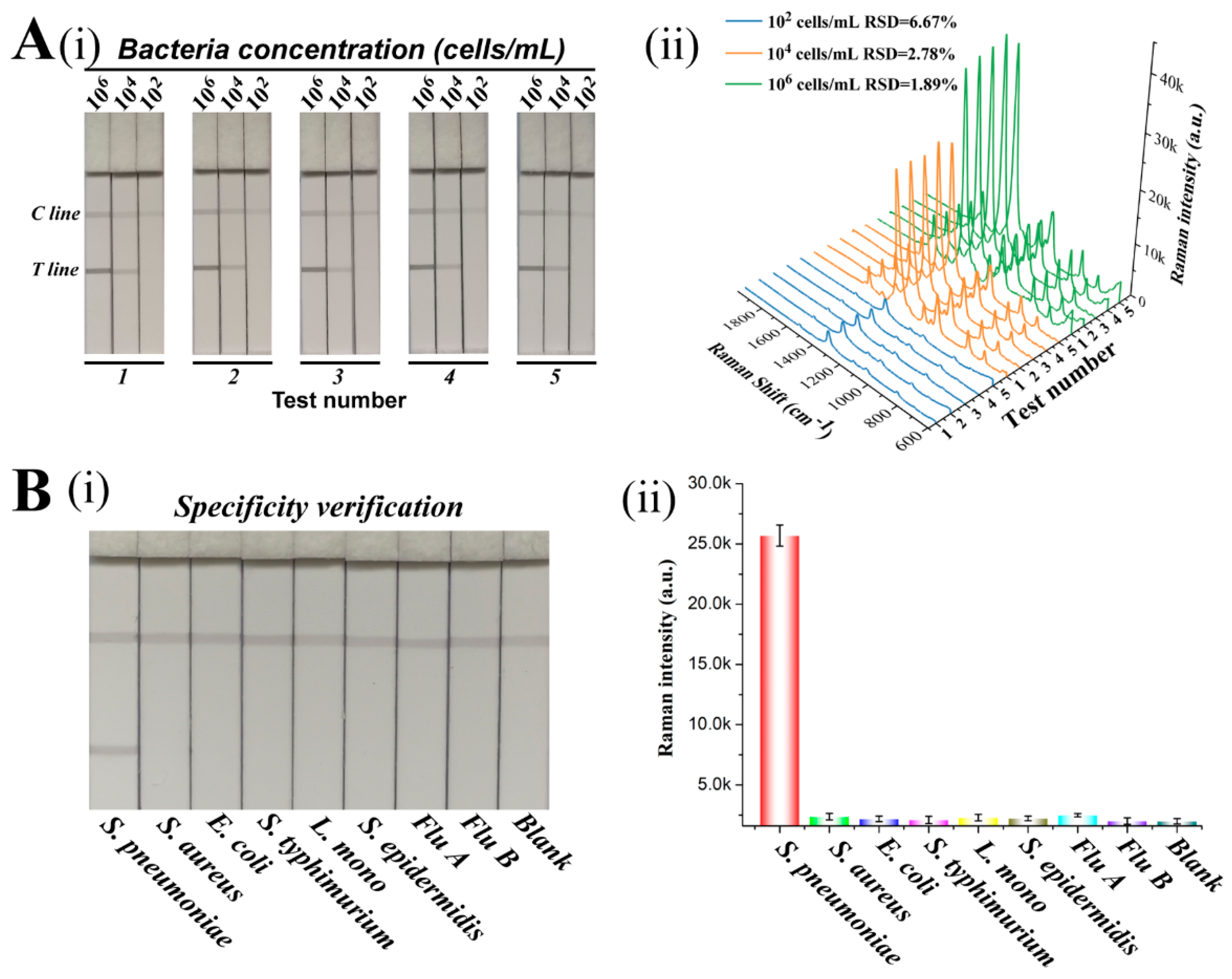
3.4. Reproducibility and Specificity of SiO2@20Au-SERS-ICA
3.5. Detection in Spiked Biological Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Fenton, A.K.; El Mortaji, L.; Lau, D.T.; Rudner, D.Z.; Bernhardt, T.G. CozE is a member of the MreCD complex that directs cell elongation in Streptococcus pneumoniae. Nat. Microbiol. 2016, 2, 16237. [Google Scholar] [CrossRef]
- Troeger, C.; Blacker, B.; Khalil, I.A.; Rao, P.C.; Cao, J.; Zimsen, S.R.M.; Albertson, S.B.; Deshpande, A.; Farag, T.; Abebe, Z.; et al. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory infections in 195 countries, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet. Infect. Dis. 2018, 18, 1191–1210. [Google Scholar] [CrossRef]
- Lella, M.; Oh, M.W.; Kuo, S.H.; Lau, G.W.; Tal-Gan, Y. Attenuating the Streptococcus pneumoniae Competence Regulon Using Urea-Bridged Cyclic Dominant-Negative Competence-Stimulating Peptide Analogs. J. Med. Chem. 2022, 65, 6826–6839. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Zheng, S.; Wang, W.; Han, H.; Yang, X.; Shen, W.; Wang, C.; Wang, S. Synthesis of two-dimensional graphene oxide-fluorescent nanoprobe for ultrasensitive and multiplex immunochromatographic detection of respiratory bacteria. Chem. Eng. J. 2021, 426, 131836. [Google Scholar] [CrossRef]
- Huang, E.; Wang, Y.; Yang, N.; Shu, B.; Zhang, G.; Liu, D. A fully automated microfluidic PCR-array system for rapid detection of multiple respiratory tract infection pathogens. Anal. Bioanal. Chem. 2021, 413, 1787–1798. [Google Scholar] [CrossRef]
- Yang, Y.; Lin, Y.; Qiao, L. Direct MALDI-TOF MS Identification of Bacterial Mixtures. Anal. Chem. 2018, 90, 10400–10408. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Andler, S.M.; Goddard, J.M.; Nugen, S.R.; Rotello, V.M. Integrating recognition elements with nanomaterials for bacteria sensing. Chem. Soc. Rev. 2017, 46, 1272–1283. [Google Scholar] [CrossRef]
- Wang, C.; Wang, C.; Li, J.; Tu, Z.; Gu, B.; Wang, S. Ultrasensitive and multiplex detection of four pathogenic bacteria on a bi-channel lateral flow immunoassay strip with three-dimensional membrane-like SERS nanostickers. Biosens. Bioelectron. 2022, 214, 114525. [Google Scholar] [CrossRef]
- Parolo, C.; Sena-Torralba, A.; Bergua, J.F.; Calucho, E.; Fuentes-Chust, C.; Hu, L.; Rivas, L.; Álvarez-Diduk, R.; Nguyen, E.P.; Cinti, S.; et al. Tutorial: Design and fabrication of nanoparticle-based lateral-flow immunoassays. Nat. Protoc. 2020, 15, 3788–3816. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Zhou, Y.; Ding, L.; Yu, G.; Leng, Y.; Lai, W.; Xiong, Y.; Chen, X. Supramolecular Recognition-Mediated Layer-by-Layer Self-Assembled Gold Nanoparticles for Customized Sensitivity in Paper-Based Strip Nanobiosensors. Small 2019, 15, e1903861. [Google Scholar] [CrossRef]
- Zheng, S.; Wang, C.; Li, J.; Wang, W.; Yu, Q.; Wang, C.; Wang, S. Graphene oxide-based three-dimensional Au nanofilm with high-density and controllable hotspots: A powerful film-type SERS tag for immunochromatographic analysis of multiple mycotoxins in complex samples. Chem. Eng. J. 2022, 448, 137760. [Google Scholar] [CrossRef]
- Guan, T.; Shen, Y.; Jiang, Z.; Wang, J.; Zhang, S.; Koidis, A.; Yao, X.; Yan, Y.; Lei, H. Facile Fabrication of Highly Quantum Dot/AuNP-Loaded Tags for a Dual-Modal Colorimetric/Reversed Ratiometric Fluorescence Immunochromatographic Assay. Anal. Chem. 2022, 94, 13463–13472. [Google Scholar] [CrossRef]
- Wu, T.; Li, J.; Zheng, S.; Yu, Q.; Qi, K.; Shao, Y.; Wang, C.; Tu, J.; Xiao, R. Magnetic Nanotag-Based Colorimetric/SERS Dual-Readout Immunochromatography for Ultrasensitive Detection of Clenbuterol Hydrochloride and Ractopamine in Food Samples. Biosensors 2022, 12, 709. [Google Scholar] [CrossRef]
- Zhou, Y.; Ding, L.; Wu, Y.; Huang, X.; Lai, W.; Xiong, Y. Emerging strategies to develop sensitive AuNP-based ICTS nanosensors. TrAC-Trend. Anal. Chem. 2019, 112, 147–160. [Google Scholar] [CrossRef]
- Wang, J.; Jiang, C.; Jin, J.; Huang, L.; Yu, W.; Su, B.; Hu, J. Ratiometric Fluorescent Lateral Flow Immunoassay for Point-of-Care Testing of Acute Myocardial Infarction. Angew. Chem. Int. Ed. Engl. 2021, 60, 13042–13049. [Google Scholar] [CrossRef]
- Bu, T.; Bai, F.; Zhao, S.; Cao, Y.; He, K.; Sun, X.; Wang, Q.; Jia, P.; Li, M.; Wang, X.; et al. Multifunctional bacteria-derived tags for advancing immunoassay analytical performance with dual-channel switching and antibodies bioactivity sustaining. Biosens. Bioelectron. 2021, 192, 113538. [Google Scholar] [CrossRef]
- Chen, R.; Chen, X.; Zhou, Y.; Lin, T.; Leng, Y.; Huang, X.; Xiong, Y. “Three-in-One” Multifunctional Nanohybrids with Colorimetric Magnetic Catalytic Activities to Enhance Immunochromatographic Diagnosis. ACS Nano 2022, 16, 3351–3361. [Google Scholar] [CrossRef]
- Wang, C.; Cheng, X.; Liu, L.; Zhang, X.; Yang, X.; Zheng, S.; Rong, Z.; Wang, S. Ultrasensitive and Simultaneous Detection of Two Specific SARS-CoV-2 Antigens in Human Specimens Using Direct/Enrichment Dual-Mode Fluorescence Lateral Flow Immunoassay. ACS Appl. Mater. Inter. 2021, 13, 40342–40353. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Jiang, C.; Yuan, J.; Tong, L.; Wang, Y.; Zhuo, D.; Huang, L.; Ni, W.; Zhang, J.; Huang, M.; et al. Hue Recognition Competitive Fluorescent Lateral Flow Immunoassay for Aflatoxin M1 Detection with Improved Visual and Quantitative Performance. Anal. Chem. 2022, 94, 10865–10873. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Yang, X.; Rong, Z.; Tu, Z.; Zhang, X.; Gu, B.; Wang, C.; Wang, S. Introduction of graphene oxide-supported multilayer-quantum dots nanofilm into multiplex lateral flow immunoassay: A rapid and ultrasensitive point-of-care testing technique for multiple respiratory viruses. Nano. Res 2022, 22, 1–11. [Google Scholar] [CrossRef]
- Caro, C.; Gámez, F.; Zaderenko, A.P. Preparation of Surface-Enhanced Raman Scattering Substrates Based on Immobilized Silver-Capped Nanoparticles. J. Spectrosc. 2018, 2018, 4127108. [Google Scholar] [CrossRef]
- Li, J.F.; Huang, Y.F.; Ding, Y.; Yang, Z.L.; Li, S.B.; Zhou, X.S.; Fan, F.R.; Zhang, W.; Zhou, Z.Y.; de Wu, Y.; et al. Shell-isolated nanoparticle-enhanced Raman spectroscopy. Nature 2010, 464, 392–395. [Google Scholar] [CrossRef] [PubMed]
- Tu, J.; Wu, T.; Yu, Q.; Li, J.; Zheng, S.; Qi, K.; Sun, G.; Xiao, R.; Wang, C. Introduction of multilayered magnetic core–dual shell SERS tags into lateral flow immunoassay: A highly stable and sensitive method for the simultaneous detection of multiple veterinary drugs in complex samples. J. Hazard. Mater. 2023, 448, 130912. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, C.; Wang, X.; Wang, K.; Zhu, Y.; Rong, Z.; Wang, W.; Xiao, R.; Wang, S. Magnetic SERS Strip for Sensitive and Simultaneous Detection of Respiratory Viruses. ACS Appl. Mater. Inter. 2019, 11, 19495–19505. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Zheng, P.; Kasani, S.; Wu, S.; Yang, F.; Lewis, S.; Nayeem, S.; Engler-Chiurazzi, E.B.; Wigginton, J.G.; Simpkins, J.W.; et al. Paper-Based Surface-Enhanced Raman Scattering Lateral Flow Strip for Detection of Neuron-Specific Enolase in Blood Plasma. Anal. Chem. 2017, 89, 10104–10110. [Google Scholar] [CrossRef]
- Wang, X.; Choi, N.; Cheng, Z.; Ko, J.; Chen, L.; Choo, J. Simultaneous Detection of Dual Nucleic Acids Using a SERS-Based Lateral Flow Assay Biosensor. Anal. Chem. 2017, 89, 1163–1169. [Google Scholar] [CrossRef]
- Tran, V.; Walkenfort, B.; Konig, M.; Salehi, M.; Schlucker, S. Rapid, Quantitative, and Ultrasensitive Point-of-Care Testing: A Portable SERS Reader for Lateral Flow Assays in Clinical Chemistry. Angew. Chem. Int. Ed. Engl. 2019, 58, 442–446. [Google Scholar] [CrossRef]
- Wang, L.; Wang, X.; Cheng, L.; Ding, S.; Wang, G.; Choo, J.; Chen, L. SERS-based test strips: Principles, designs and applications. Biosens. Bioelectron. 2021, 189, 113360. [Google Scholar] [CrossRef]
- Zhang, D.; Huang, L.; Liu, B.; Ni, H.; Sun, L.; Su, E.; Chen, H.; Gu, Z.; Zhao, X. Quantitative and ultrasensitive detection of multiplex cardiac biomarkers in lateral flow assay with core-shell SERS nanotags. Biosens. Bioelectron. 2018, 106, 204–211. [Google Scholar] [CrossRef]
- Li, J.; Wu, T.; Wang, C.; Tu, J.; Song, X.; Shao, Y.; Wang, C.; Qi, K.; Xiao, R. Nanogapped Fe3O4@Au Surface-Enhanced Raman Scattering Tags for the Multiplex Detection of Bacteria on an Immunochromatographic Strip. ACS Appl. Nano Mater. 2022, 5, 12679–12689. [Google Scholar] [CrossRef]
- Su, L.; Hu, H.; Tian, Y.; Jia, C.; Wang, L.; Zhang, H.; Wang, J.; Zhang, D. Highly Sensitive Colorimetric/Surface-Enhanced Raman Spectroscopy Immunoassay Relying on a Metallic Core-Shell Au/Au Nanostar with Clenbuterol as a Target Analyte. Anal. Chem. 2021, 93, 8362–8369. [Google Scholar] [CrossRef] [PubMed]
- Sheng, E.; Lu, Y.; Xiao, Y.; Li, Z.; Wang, H.; Dai, Z. Simultaneous and ultrasensitive detection of three pesticides using a surface-enhanced Raman scattering-based lateral flow assay test strip. Biosens. Bioelectron. 2021, 181, 113149. [Google Scholar] [CrossRef] [PubMed]
- Tu, D.; Holderby, A.; Guo, H.; Mabbott, S.; Tian, L.; Coté, G.L. Spectrally multiplexed assay using gap enhanced nanoparticle for detection of a myocardial infarction biomarker panel. Anal. Chim. Acta. 2022, 1198, 339562. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Yang, X.; Zheng, S.; Cheng, X.; Xiao, R.; Li, Q.; Wang, W.; Liu, X.; Wang, S. Development of an ultrasensitive fluorescent immunochromatographic assay based on multilayer quantum dot nanobead for simultaneous detection of SARS-CoV-2 antigen and influenza A virus. Sens. Actuators B Chem. 2021, 345, 130372. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, J.; Li, M.; Qu, X.; Zhang, K.; Rong, Z.; Xiao, R.; Wang, S. A rapid SERS method for label-free bacteria detection using polyethylenimine-modified Au-coated magnetic microspheres and Au@Ag nanoparticles. Analyst 2016, 141, 6226–6238. [Google Scholar] [CrossRef]
- Wang, C.; Shen, W.; Rong, Z.; Liu, X.; Gu, B.; Xiao, R.; Wang, S. Layer-by-layer assembly of magnetic-core dual quantum dot-shell nanocomposites for fluorescence lateral flow detection of bacteria. Nanoscale 2020, 12, 795–807. [Google Scholar] [CrossRef]
- Wang, C.; Yang, X.; Gu, B.; Liu, H.; Zhou, Z.; Shi, L.; Cheng, X.; Wang, S. Sensitive and Simultaneous Detection of SARS-CoV-2-Specific IgM/IgG Using Lateral Flow Immunoassay Based on Dual-Mode Quantum Dot Nanobeads. Anal. Chem. 2020, 92, 15542–15549. [Google Scholar] [CrossRef]
- Liu, H.; Dai, E.; Xiao, R.; Zhou, Z.; Zhang, M.; Bai, Z.; Shao, Y.; Qi, K.; Tu, J.; Wang, C.; et al. Development of a SERS-based lateral flow immunoassay for rapid and ultra-sensitive detection of anti-SARS-CoV-2 IgM/IgG in clinical samples. Sens. Actuators B. Chem. 2021, 329, 129196. [Google Scholar] [CrossRef]
- Wang, J.; Chen, Q.; Jin, Y.; Zhang, X.; He, L.; Zhang, W.; Chen, Y. Surface enhanced Raman scattering-based lateral flow immunosensor for sensitive detection of aflatoxin M1 in urine. Anal. Chim. Acta 2020, 1128, 184–192. [Google Scholar] [CrossRef]
- Shen, W.; Wang, C.; Yang, X.; Wang, C.; Zhou, Z.; Liu, X.; Xiao, R.; Gu, B.; Wang, S. Synthesis of raspberry-like nanogapped Fe3O4@Au nanocomposites for SERS-based lateral flow detection of multiple tumor biomarkers. J. Mater. Chem. C. 2020, 8, 12854–12864. [Google Scholar] [CrossRef]
- Zheng, S.; Yang, X.; Zhang, B.; Cheng, S.; Han, H.; Jin, Q.; Wang, C.; Xiao, R. Sensitive detection of Escherichia coli O157:H7 and Salmonella typhimurium in food samples using two-channel fluorescence lateral flow assay with liquid Si@quantum dot. Food Chem. 2021, 363, 130400. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Wang, C.; Zheng, S.; Jiang, B.; Li, J.; Pang, Y.; Wang, C.; Hao, R.; Xiao, R. Ultrasensitive multichannel immunochromatographic assay for rapid detection of foodborne bacteria based on two-dimensional film-like SERS labels. J. Hazard. Mater. 2022, 437, 129347. [Google Scholar] [CrossRef] [PubMed]
- Tu, Z.; Yang, X.; Dong, H.; Yu, Q.; Zheng, S.; Cheng, X.; Wang, C.; Rong, Z.; Wang, S. Ultrasensitive Fluorescence Lateral Flow Assay for Simultaneous Detection of Pseudomonas aeruginosa and Salmonella typhimurium via Wheat Germ Agglutinin-Functionalized Magnetic Quantum Dot Nanoprobe. Biosensors 2022, 12, 942. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, W.; Li, J.; Jiang, B.; Nie, Y.; Pang, Y.; Wang, C.; Xiao, R.; Hao, R. Electrostatic Adsorption of Dense AuNPs onto Silica Core as High-Performance SERS Tag for Sensitive Immunochromatographic Detection of Streptococcus pneumoniae. Pathogens 2023, 12, 327. https://doi.org/10.3390/pathogens12020327
Shen W, Li J, Jiang B, Nie Y, Pang Y, Wang C, Xiao R, Hao R. Electrostatic Adsorption of Dense AuNPs onto Silica Core as High-Performance SERS Tag for Sensitive Immunochromatographic Detection of Streptococcus pneumoniae. Pathogens. 2023; 12(2):327. https://doi.org/10.3390/pathogens12020327
Chicago/Turabian StyleShen, Wanzhu, Jiaxuan Li, Bo Jiang, You Nie, Yuanfeng Pang, Chongwen Wang, Rui Xiao, and Rongzhang Hao. 2023. "Electrostatic Adsorption of Dense AuNPs onto Silica Core as High-Performance SERS Tag for Sensitive Immunochromatographic Detection of Streptococcus pneumoniae" Pathogens 12, no. 2: 327. https://doi.org/10.3390/pathogens12020327
APA StyleShen, W., Li, J., Jiang, B., Nie, Y., Pang, Y., Wang, C., Xiao, R., & Hao, R. (2023). Electrostatic Adsorption of Dense AuNPs onto Silica Core as High-Performance SERS Tag for Sensitive Immunochromatographic Detection of Streptococcus pneumoniae. Pathogens, 12(2), 327. https://doi.org/10.3390/pathogens12020327