Genomic Characteristics and Phylogenetic Analyses of a Multiple Drug-Resistant Klebsiella pneumoniae Harboring Plasmid-Mediated MCR-1 Isolated from Tai’an City, China
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Isolation and Identification
2.2. Antimicrobial Susceptibility Assay
2.3. Conjugation Test
2.4. Bacterial DNA Extraction and Sequencing
2.5. DNA Sequence Analysis
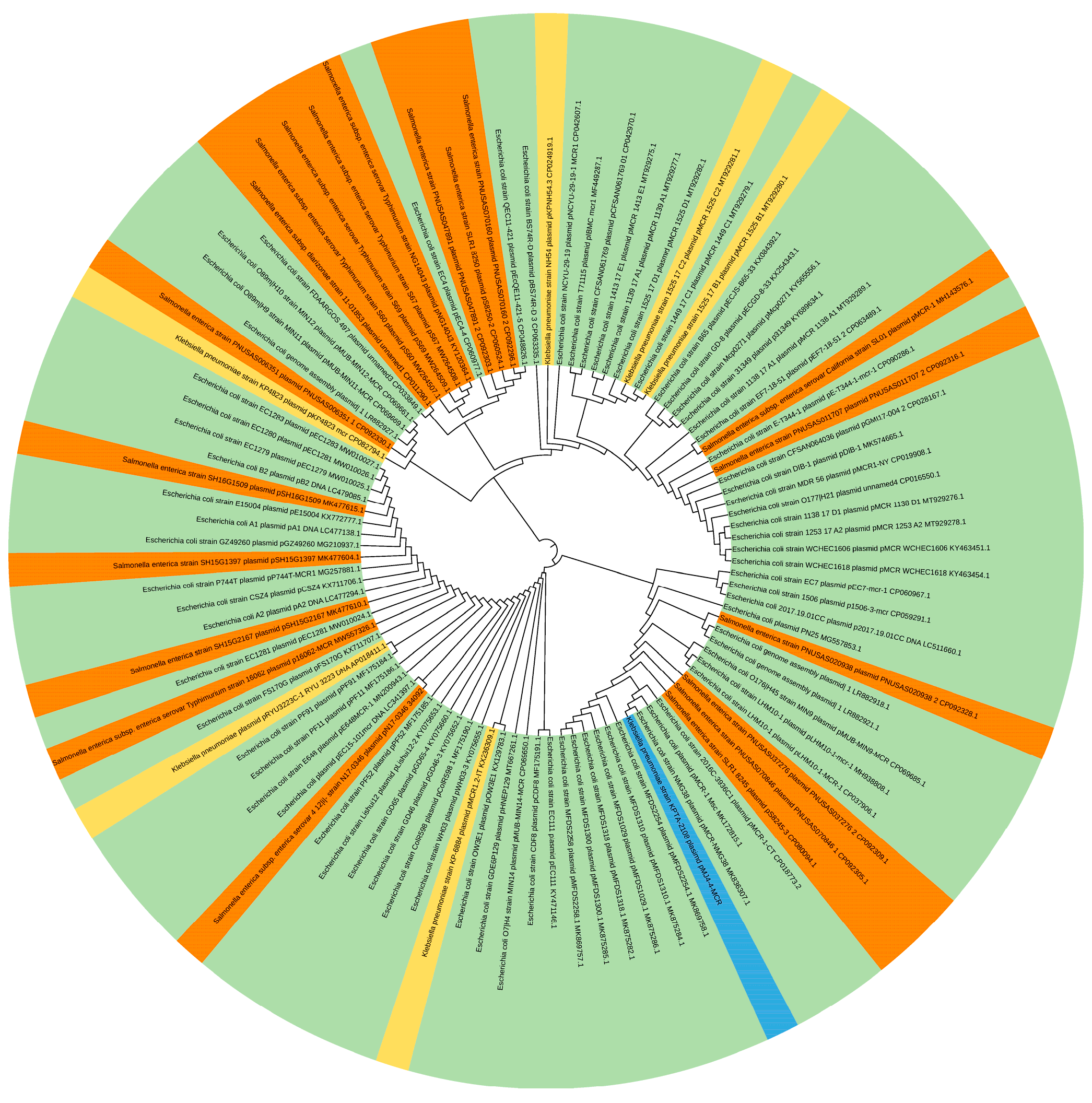
2.6. Bioinformatics Analyses
3. Results
3.1. The Results of the Drug Sensitivity Test
3.2. Genome Characterization and Functional Gene Annotation of KPTA-2108
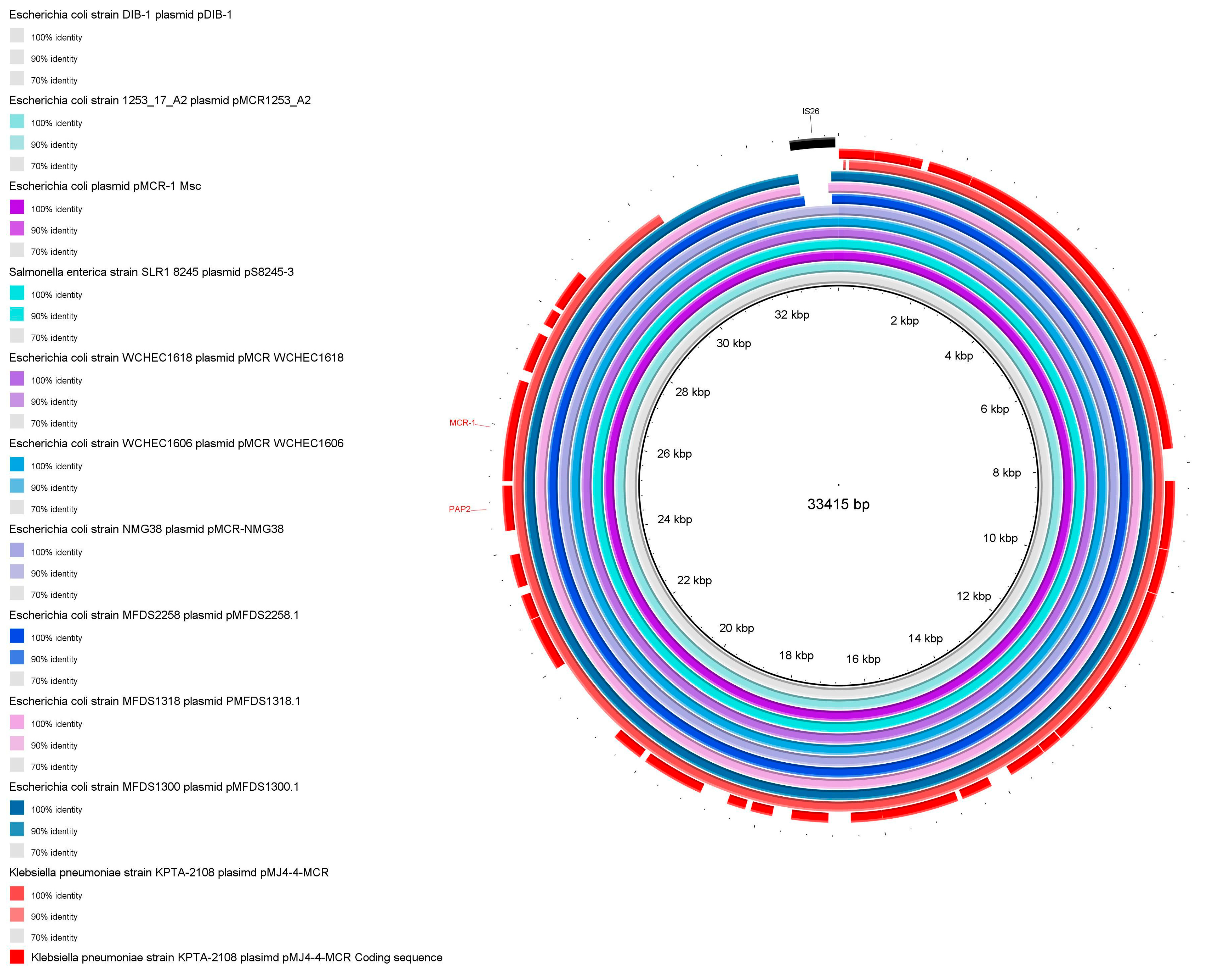
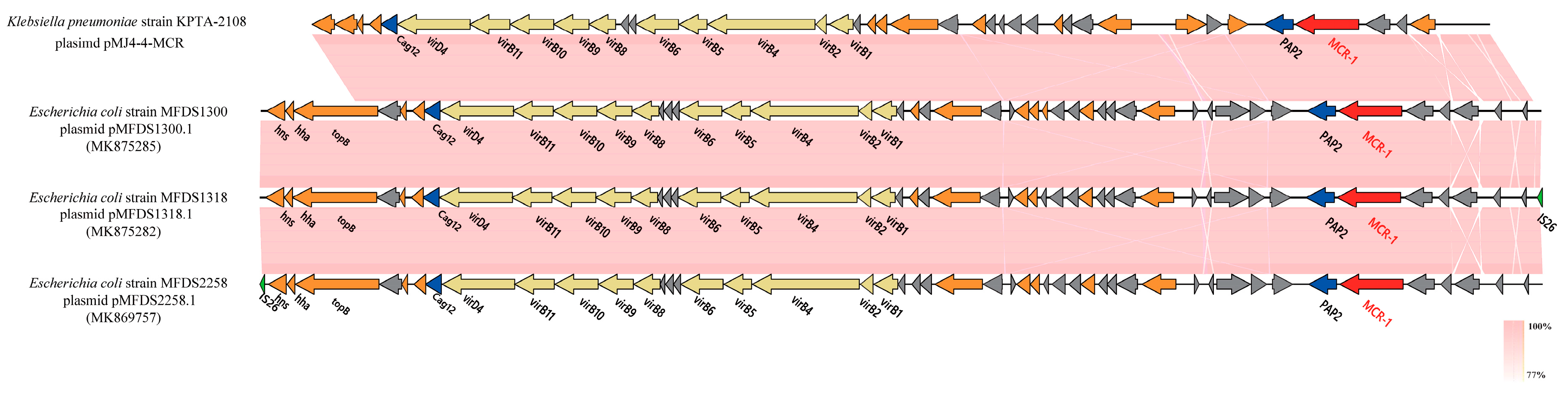
3.3. Characterization of pMJ4-4-MCR
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yang, F.; Deng, B.; Liao, W.; Wang, P.; Chen, P.; Wei, J. High rate of multiresistant Klebsiella pneumoniae from human and animal origin. Infect. Drug Resist. 2019, 12, 2729–2737. [Google Scholar] [CrossRef]
- Sequeira, R.P.; McDonald, J.A.K.; Marchesi, J.R.; Clarke, T.B. Commensal Bacteroidetes protect against Klebsiella pneumoniae coloni-zation and transmission through IL-36 signalling. Nat. Microbiol. 2020, 5, 304–313. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Chen, P.; Deng, B.; Sun, F.H.; Yang, Y.; Yang, Y.; He, R.; Qin, M.; Wu, Y.; Yang, F.; et al. Outcomes and Risk Factors of Bloodstream Infections Caused by Carbapenem-Resistant and Non-Carbapenem-Resistant Klebsiella pneumoniae in China. Infect. Drug Resist. 2022, 15, 3161–3171. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Xiao, M.; Hou, R.; Wang, D.; Yang, M.; Chen, M.; Chen, L. Bundles of care for prevention of ventilator-associated pneumonia caused by carbapenem-resistant Klebsiella pneumoniae in the ICU. Am. J. Transl. Res. 2021, 13, 3561–3572. [Google Scholar]
- Livermore, D.M.; Warner, M.; Mushtaq, S.; Doumith, M.; Zhang, J.; Woodford, N. What remains against carbapenem-resistant En-terobacteriaceae? Evaluation of chloramphenicol, ciprofloxacin, colistin, fosfomycin, minocycline, nitrofurantoin, temocillin and tigecycline. Int. J. Antimicrob. Agents 2011, 7, 415–419. [Google Scholar] [CrossRef]
- Liu, Y.-Y.; Wang, Y.; Walsh, T.R.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Hu, M.; Zhang, Q.; Zhao, C.; Zhang, Y.; Li, L.; Qi, J.; Luo, Y.; Zhou, D.; Liu, Y. Characterization of integrons and antimicrobial resistance in Salmonella from broilers in Shandong, China. Poult. Sci. 2020, 99, 7046–7054. [Google Scholar] [CrossRef] [PubMed]
- Jovcic, B.; Novovic, K.; Dekic, S.; Hrenovic, J. Colistin Resistance in Environmental Isolates of Acinetobacter baumannii. Microb. Drug Resist. 2021, 27, 328–336. [Google Scholar] [CrossRef]
- Hu, Y.-Y.; Wang, Y.-L.; Sun, Q.-L.; Huang, Z.-X.; Wang, H.-Y.; Zhang, R.; Chen, G.-X. Colistin resistance gene mcr-1 in gut flora of children. Int. J. Antimicrob. Agents 2017, 50, 593–597. [Google Scholar] [CrossRef]
- Jian, Z.; Zeng, L.; Xu, T.; Sun, S.; Yan, S.; Yang, L.; Huang, Y.; Jia, J.; Dou, T. Antibiotic resistance genes in bacteria: Occurrence, spread, and control. J. Basic Microbiol. 2021, 61, 1049–1070. [Google Scholar] [CrossRef]
- Shapovalova, V.; Shaidullina, E.; Azizov, I.; Sheck, E.; Martinovich, A.; Dyachkova, M.; Matsvay, A.; Savochkina, Y.; Khafizov, K.; Kozlov, R.; et al. Molecular Epidemiology of mcr-1-Positive Escherichia coli and Klebsiella pneumoniae Isolates: Results from Russian Sentinel Surveillance (2013–2018). Microorganisms 2022, 10, 2034. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Dong, N.; Liu, C.; Zeng, Y.; Sun, Q.; Zhou, H.; Hu, Y.; Chen, S.; Shen, Z.; Zhang, R. Prevalence and molecular epidemiology of mcr-1-positive Klebsiella pneumoniae in healthy adults from China. J. Antimicrob. Chemother. 2020, 75, 2485–2494. [Google Scholar] [CrossRef] [PubMed]
- Ngbede, E.O.; Poudel, A.; Kalalah, A.; Yang, Y.; Adekanmbi, F.; Adikwu, A.A.; Adamu, A.M.; Mamfe, L.M.; Daniel, S.T.; Useh, N.M.; et al. Identification of mobile colistin resistance genes (mcr-1.1, mcr-5 and mcr-8.1) in Enterobacteriaceae and Alcaligenes faecalis of human and animal origin, Nigeria. Int. J. Antimicrob. Agents 2020, 56, 106108. [Google Scholar] [CrossRef] [PubMed]
- Tada, T.; Uechi, K.; Nakasone, I.; Nakamatsu, M.; Satou, K.; Hirano, T.; Kirikae, T.; Fujita, J. Emergence of IncX4 plasmids encoding mcr-1 in a clinical isolate of Klebsiella pneumoniae in Japan. Int. J. Infect. Dis. 2018, 75, 98–100. [Google Scholar] [CrossRef] [PubMed]
- Wyres, K.L.; Holt, K.E. Klebsiella pneumoniae as a key trafficker of drug resistance genes from environmental to clinically im-portant bacteria. Curr. Opin. Microbiol. 2018, 45, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Rossen, J.; Friedrich, A.; Moran-Gilad, J. Practical issues in implementing whole-genome-sequencing in routine diagnostic microbiology. Clin. Microbiol. Infect. 2018, 24, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Balloux, F.; Brynildsrud, O.B.; van Dorp, L.; Shaw, L.P.; Chen, H.; Harris, K.A.; Wang, H.; Eldholm, V. From Theory to Practice: Translating Whole-Genome Sequencing (WGS) into the Clinic. Trends Microbiol. 2018, 26, 1035–1048. [Google Scholar] [CrossRef]
- Pornsukarom, S.; Van Vliet, A.H.M.; Thakur, S. Whole genome sequencing analysis of multiple Salmonella serovars provides insights into phylogenetic relatedness, antimicrobial resistance, and virulence markers across humans, food animals and agriculture environmental sources. BMC Genom. 2018, 19, 801. [Google Scholar] [CrossRef]
- Llarena, A.-K.; Taboada, E.; Rossi, M. Whole-Genome Sequencing in Epidemiology of Campylobacter jejuni Infections. J. Clin. Microbiol. 2017, 55, 1269–1275. [Google Scholar] [CrossRef]
- Prendeville, S.; Sanders, C.; Sherry, J.; Costa, F. Circular Economy: Is It Enough. EcoDesign Centre, Wales. 2014. Available online: http://www.edcw.org/en/resources/circulareconomy-it-enough (accessed on 21 July 2014).
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: CLSI Supplement M100, 31th ed; CLSI: Wayne, PA, USA, 2021. [Google Scholar]
- Figueroa-Bossi, N.; Balbontín, R.; Bossi, L. Preparing Bacterial Genomic DNA. Cold Spring Harb. Protoc. 2022, 2022, Pdb.prot107853. [Google Scholar] [CrossRef]
- Mikhail, K.; Jeffrey, Y.; Yu, L.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar]
- Sergey, K.; Brian, P.W.; Konstantin, B.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. kCanu: Scalable and accurate long-read assembly via adaptive -mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar]
- Hyatt, D.; Chen, G.-L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Staerfeldt, H.H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ri-bosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef] [PubMed]
- Griffiths-Jones, S.; Bateman, A.; Marshall, M.; Khanna, A.; Eddy, S. Rfam: An RNA family database. Nucleic Acids Res. 2003, 31, 439–441. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Kwong, J.C.; da Silva, A.G.; Dyet, K.; Williamson, D.; Stinear, T.; Howden, B.; Seemann, T. NGMASTER: In silico multi-antigen sequence typing for Neisseria gonorrhoeae. Microb. Genom. 2016, 2, e000076. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; Garcìa-Fernandez, A.; Larsen, M.; Lund, O.; Voldby Villa, L.; Møller Aarestrup, F.; Hasman, H. In Silico Detection and Typing of Plasmids. Antimicrob using PlasmidFinder and plasmid multilocus sequence typing. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Chen, L.; Yang, J.; Yu, J.; Yao, Z.; Sun, L.; Shen, Y.; Jin, Q. VFDB: A reference database for bacterial virulence factors. Nucleic Acids Res. 2005, 33, D325–D328. [Google Scholar] [CrossRef]
- Johansson, M.H.K.; Bortolaia, V.; Tansirichaiya, S.; Aarestrup, F.M.; Roberts, A.P.; Petersen, T.N. Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J. Antimicrob. Chemother. 2020, 76, 101–109. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
- Yang, M.; Derbyshire, M.K.; Yamashita, R.A.; Marchler-Bauer, A. NCBI’s Conserved Domain Database and Tools for Protein Domain Analysis. Curr. Protoc. Bioinform. 2020, 69, e90. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2014, 12, 59–60. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, 256–259. [Google Scholar] [CrossRef]
- Russo, T.A.; Olson, R.; MacDonald, U.; Beanan, J.; Davidson, B.A. Aerobactin, but not yersiniabactin, salmochelin, or enterobactin, enables the growth/survival of hypervirulent (hypermucoviscous) Klebsiella pneumoniae ex vivo and in vivo. Infect. Immun. 2015, 83, 3325–3333. [Google Scholar] [CrossRef]
- Cao, X.; Zhong, Q.; Guo, Y.; Hang, Y.; Chen, Y.; Fang, X.; Xiao, Y.; Zhu, H.; Luo, H.; Yu, F.; et al. Emergence of the Coexistence of mcr-1, blaNDM-5, and blaCTX-M-55 in Klebsiella pneumoniae ST485 Clinical Isolates in China. Infect. Drug Resist. 2021, 14, 3449–3458. [Google Scholar] [CrossRef]
- Chen, F.-J.; Lauderdale, T.-L.; Huang, W.-C.; Shiau, Y.-R.; Wang, H.-Y.; Kuo, S.-C. Emergence of mcr-1, mcr-3 and mcr-8 in clinical Klebsiella pneumoniae isolates in Taiwan. Clin. Microbiol. Infect. 2020, 27, 305–307. [Google Scholar] [CrossRef]
- Wu, R.; Yi, L.-X.; Yu, L.-F.; Wang, J.; Liu, Y.; Chen, X.; Lv, L.; Yang, J.; Liu, J.-H. Fitness Advantage of mcr-1–Bearing IncI2 and IncX4 Plasmids in Vitro. Front. Microbiol. 2018, 9, 331. [Google Scholar] [CrossRef]
- Beyrouthy, R.; Robin, F.; Lessene, A.; Lacombat, I.; Dortet, L.; Naas, T.; Ponties, V.; Bonnet, R. MCR-1 and OXA-48 In Vivo Acquisition in KPC-Producing Escherichia coli after Colistin Treatment. Antimicrob. Agents Chemother. 2017, 61, e02540-16. [Google Scholar] [CrossRef]
- Yang, Y.-Q.; Li, Y.-X.; Lei, C.-W.; Zhang, A.; Wang, H.-N. Novel plasmid-mediated colistin resistance gene mcr-7.1 in Klebsiella pneumoniae. J. Antimicrob. Chemother. 2018, 73, 1791–1795. [Google Scholar] [CrossRef]
- Elbediwi, M.; Li, Y.; Paudyal, N.; Pan, H.; Li, X.; Xie, S.; Rajkovic, A.; Feng, Y.; Fang, W.; Rankin, S.; et al. Global Burden of Colistin-Resistant Bacteria: Mobilized Colistin Re-sistance Genes Study (1980–2018). Microorganisms 2019, 7, 461. [Google Scholar] [CrossRef]
- Shafiq, M.; Huang, J.; Shah, J.M.; Wang, X.; Rahman, S.U.; Ali, I.; Chen, L.; Wang, L. Characterization and virulence factors distribution of blaCTX-M and mcr-1carrying Escherichia coli isolates from bovine mastitis. J. Appl. Microbiol. 2021, 131, 634–646. [Google Scholar] [CrossRef]
- Majewski, P.; Gutowska, A.; Smith, D.G.E.; Hauschild, T.; Majewska, P.; Hryszko, T.; Gizycka, D.; Kedra, B.; Kochanowicz, J.; Glowiński, J.; et al. Plasmid Mediated mcr-1.1 Colistin-Resistance in Clinical Extraintestinal Escherichia coli Strains Isolated in Poland. Front. Microbiol. 2021, 12, 547020. [Google Scholar] [CrossRef]
- Nang, S.C.; Li, J.; Velkov, T. The rise and spread of mcr plasmid-mediated polymyxin resistance. Crit. Rev. Microbiol. 2019, 45, 131–161. [Google Scholar] [CrossRef]
- Gogry, F.A.; Siddiqui, M.T.; Sultan, I.; Haq, Q.M.R. Current Update on Intrinsic and Acquired Colistin Resistance Mechanisms in Bacteria. Front. Med. Lausanne 2021, 8, 677720. [Google Scholar] [CrossRef]
- Trimble, M.J.; Mlynárčik, P.; Kolář, M.; Hancock, R.E. Polymyxin: Alternative Mechanisms of Action and Resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a025288. [Google Scholar] [CrossRef]
- Olaitan, A.; Morand, S.; Rolain, J.-M. Mechanisms of polymyxin resistance: Acquired and intrinsic resistance in bacteria. Front. Microbiol. 2014, 5, 643. [Google Scholar] [CrossRef]
- Singh, S.; Pathak, A.; Kumar, A.; Rahman, M.; Singh, A.; Gonzalez-Zorn, B.; Prasad, K.N. Emergence of Chromosome-Borne Colistin Re-sistance Gene mcr-1 in Clinical Isolates of Klebsiella pneumoniae from India. Antimicrob. Agents Chemother. 2018, 62, e01885-17. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Xia, X.; Yuan, T.; Zhu, J.; Shen, Z.; Li, M. Molecular Epidemiology of Antimicrobial Resistance, Virulence and Capsular Serotypes of Carbapenemase-Carrying Klebsiella pneumoniae in China. Antibiotics 2022, 11, 1100. [Google Scholar] [CrossRef]
- Christie, P.J.; Whitaker, N.; González-Rivera, C. Mechanism and structure of the bacterial type IV secretion systems. Biochim. et Biophys. Acta 2014, 1843, 1578–1591. [Google Scholar] [CrossRef] [PubMed]
- Wallden, K.; Rivera-Calzada, A.; Waksman, G. Microreview: Type IV secretion systems: Versatility and diversity in function. Cell. Microbiol. 2010, 12, 1203–1212. [Google Scholar] [CrossRef]
- Stokes, H.W.; Gillings, M. Gene flow, mobile genetic elements and the recruitment of antibiotic resistance genes into Gram-negative pathogens. FEMS Microbiol. Rev. 2011, 35, 790–819. [Google Scholar] [CrossRef]
- Sun, J.; Fang, L.-X.; Wu, Z.; Deng, H.; Yang, R.-S.; Li, X.-P.; Li, S.-M.; Liao, X.-P.; Feng, Y.; Liu, Y.-H. Genetic Analysis of the IncX4 Plasmids: Implications for a Unique Pattern in the mcr-1 Acquisition. Sci. Rep. 2017, 7, 424. [Google Scholar] [CrossRef]
- Johnson, T.J.; Bielak, E.M.; Fortini, D.; Hansen, L.H.; Hasman, H.; Debroy, C.; Nolan, L.K.; Carattoli, A. Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid 2012, 68, 43–50. [Google Scholar] [CrossRef]
- Choi, Y.; Lee, J.-Y.; Lim, S.-K.; Ko, K.S. Intact pap2 downstream of mcr-1 appears to be required for colistin resistance. Diagn. Microbiol. Infect. Dis. 2020, 97, 114997. [Google Scholar] [CrossRef]
- Juhas, M.; Crook, D.W.; Hood, D.W. Type IV secretion systems: Tools of bacterial horizontal gene transfer and virulence. Cell. Microbiol. 2008, 10, 2377–2386. [Google Scholar] [CrossRef]
| Antibiotics | MIC (μg/mL) | KB (mm) | S/R | Antibiotics | MIC (μg/mL) | KB (mm) | S/R | ||
|---|---|---|---|---|---|---|---|---|---|
| Cephalosporins | Cefazolin | >16 | + | Carbapenems | Ertapenem | 0.5 | − | ||
| Cefepime | >16 | + | Imipenem | ≤0.2 | − | ||||
| Ceftriaxone | >32 | + | Meropenem | ≤0.1 | − | ||||
| Cefuroxime | >16 | + | Glycylcyclines | Tigecycline | 8 | + | |||
| Cefoperazone/sulbactam | 32/8 | + | Sulfonamides | Cotrimoxazole | >4/76 | + | |||
| Cefoxitin | >16 | + | 3rd generation cephalosporins | Ceftazidime/avibactam | 25 | − | |||
| Ceftazidime | >32 | + | Polymyxins | Colistin | 4 | + | |||
| Cefotaxime | 6 | + | β-lactamase inhibitors | Piperacillin/tazobactam | 16/4 | − | |||
| Monobactams | Aztreonam | >32 | + | Aminoglycosides | Amikacin | >32 | + | ||
| Fluoroquinolones | Ciprofloxacin | >4 | + | Gentamicin | >8 | + | |||
| Levofloxacin | >8 | + | Tobramycin | >8 | + | ||||
| Penicillins plus β-lactamase inhibitor | Ampicillin/sulbactam | >16/8 | + | Tetracyclines | Minocycline | 16 | + | ||
| Amoxicillin/clavulanic acid | >32/1 | + | Tetracycline | >8 | + | ||||
| Strain/Plasmid | Genome_Size (bp) | Gene_ Num (#) | GC% | rRNA_Num | sRNA_Num | tRNA_Num | DNA Elements | Plasmid Replicon Type | Resistance Genes |
|---|---|---|---|---|---|---|---|---|---|
| Chromosome | 5,306,347 | 4940 | 0.5741 | 25 | 124 | 88 | 9 | − | − |
| pMJ4-1 | 164,378 | 185 | 0.5201 | 0 | 5 | 0 | 0 | IncFIB(K) | bleO, aac, aadA, aph, ARR-3, mph(A), sul, dfrA, tet, qacE, floR |
| pMJ4-2 | 98,026 | 108 | 0.4955 | 0 | 4 | 0 | 2 | IncI1-I | − |
| pMJ4-3 | 72,739 | 88 | 0.524 | 0 | 2 | 0 | 0 | − | blaCTX-M, blaTEM, qnrS1 |
| pMJ4-4-MCR | 30,124 | 47 | 0.4131 | 0 | 0 | 0 | 0 | IncX4 | MCR-1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Q.; Guo, Z.; Zhu, G.; Li, N.; Bai, G.; Jiang, M. Genomic Characteristics and Phylogenetic Analyses of a Multiple Drug-Resistant Klebsiella pneumoniae Harboring Plasmid-Mediated MCR-1 Isolated from Tai’an City, China. Pathogens 2023, 12, 221. https://doi.org/10.3390/pathogens12020221
Liu Q, Guo Z, Zhu G, Li N, Bai G, Jiang M. Genomic Characteristics and Phylogenetic Analyses of a Multiple Drug-Resistant Klebsiella pneumoniae Harboring Plasmid-Mediated MCR-1 Isolated from Tai’an City, China. Pathogens. 2023; 12(2):221. https://doi.org/10.3390/pathogens12020221
Chicago/Turabian StyleLiu, Qinqin, Zhiyun Guo, Gang Zhu, Ning Li, Guanchen Bai, and Meijie Jiang. 2023. "Genomic Characteristics and Phylogenetic Analyses of a Multiple Drug-Resistant Klebsiella pneumoniae Harboring Plasmid-Mediated MCR-1 Isolated from Tai’an City, China" Pathogens 12, no. 2: 221. https://doi.org/10.3390/pathogens12020221
APA StyleLiu, Q., Guo, Z., Zhu, G., Li, N., Bai, G., & Jiang, M. (2023). Genomic Characteristics and Phylogenetic Analyses of a Multiple Drug-Resistant Klebsiella pneumoniae Harboring Plasmid-Mediated MCR-1 Isolated from Tai’an City, China. Pathogens, 12(2), 221. https://doi.org/10.3390/pathogens12020221






