No Changes in the Occurrence of Methicillin-Resistant Staphylococcus aureus (MRSA) in South-East Austria during the COVID-19 Pandemic
Abstract
1. Introduction
2. Material and Methods
2.1. Bacterial Isolates
2.2. Genetic Analyses
2.3. Classification of HA-MRSA, CA-MRSA, LA-MRSA
3. Results
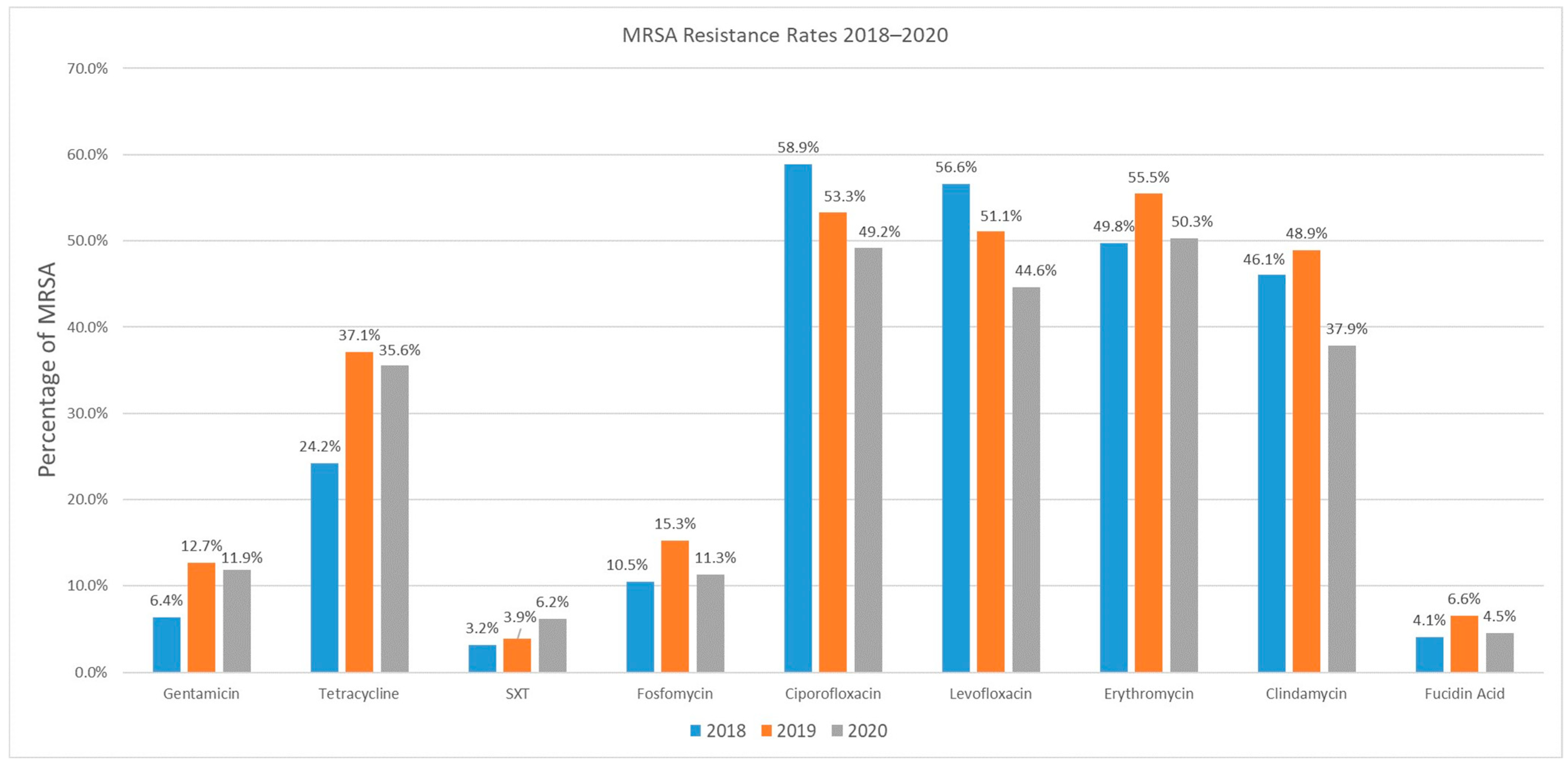
3.1. Antibiotic Resistance
3.2. Spa Typing
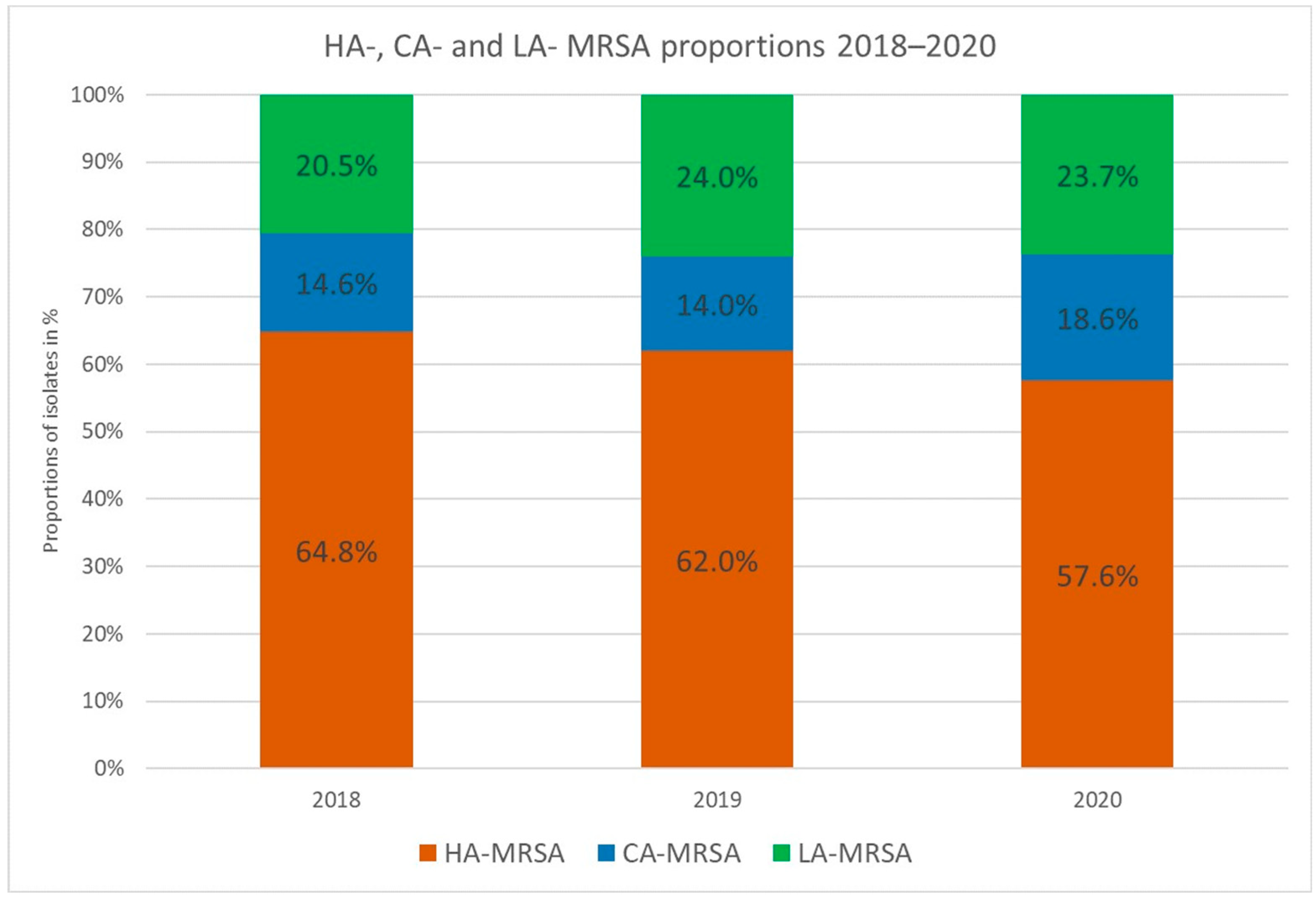
3.3. CA-MRSA
3.4. LA-MRSA
3.5. HA-MRSA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lakhundi, S.; Zhang, K. Methicillin-Resistant Staphylococcus Aureus: Molecular Characterization, Evolution, and Epidemiology. Clin. Microbiol. Rev. 2018, 31, e00020-18. [Google Scholar] [CrossRef] [PubMed]
- Stefani, S.; Chung, D.R.; Lindsay, J.A.; Friedrich, A.W.; Kearns, A.M.; Westh, H.; MacKenzie, F.M. Meticillin-Resistant Staphylococcus aureus (MRSA): Global Epidemiology and Harmonisation of Typing Methods. Int. J. Antimicrob. Agents 2012, 39, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Otto, M. Community-Associated MRSA: What Makes Them Special? Int. J. Med. Microbiol. 2013, 303, 324–330. [Google Scholar] [CrossRef] [PubMed]
- Otter, J.A.; French, G.L. Molecular Epidemiology of Community-Associated Meticillin-Resistant Staphylococcus aureus in Europe. Lancet Infect. Dis. 2010, 10, 227–239. [Google Scholar] [CrossRef]
- Mediavilla, J.R.; Chen, L.; Mathema, B.; Kreiswirth, B.N. Global Epidemiology of Community-Associated Methicillin Resistant Staphylococcus aureus (CA-MRSA). Curr. Opin. Microbiol. 2012, 15, 588–595. [Google Scholar] [CrossRef]
- Aires-de-Sousa, M. Methicillin-Resistant Staphylococcus aureus among Animals: Current Overview. Clin. Microbiol. Infect. 2017, 23, 373–380. [Google Scholar] [CrossRef]
- Witte, W.; Strommenger, B.; Stanek, C.; Cuny, C. Methicillin-Resistant Staphylococcus aureus ST398 in Humans and Animals, Central Europe. Emerg. Infect. Dis. 2007, 13, 255–258. [Google Scholar] [CrossRef]
- Lindsay, J.A. Genomic Variation and Evolution of Staphylococcus aureus. Int. J. Med. Microbiol. 2010, 300, 98–103. [Google Scholar] [CrossRef]
- O’Toole, R.F. The Interface between COVID-19 and Bacterial Healthcare-Associated Infections. Clin. Microbiol. Infect. 2021, 27, 1772–1776. [Google Scholar] [CrossRef]
- Lai, C.-C.; Chen, S.-Y.; Ko, W.-C.; Hsueh, P.-R. Increased Antimicrobial Resistance during the COVID-19 Pandemic. Int. J. Antimicrob. Agents 2021, 57, 106324. [Google Scholar] [CrossRef]
- Ruppitsch, W.; Indra, A.; Stöger, A.; Mayer, B.; Stadlbauer, S.; Wewalka, G.; Allerberger, F. Classifying Spa Types in Complexes Improves Interpretation of Typing Results for Methicillin-Resistant Staphylococcus aureus. J. Clin. Microbiol. 2006, 44, 2442–2448. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lina, G.; Piemont, Y.; Godail-Gamot, F.; Bes, M.; Peter, M.-O.; Gauduchon, V.; Vandenesch, F.; Etienne, J. Involvement of Panton-Valentine Leukocidin--Producing Staphylococcus aureus in Primary Skin Infections and Pneumonia. Clin. Infect. Dis. 1999, 29, 1128–1132. [Google Scholar] [CrossRef] [PubMed]
- Mallah, S.I.; Ghorab, O.K.; Al-Salmi, S.; Abdellatif, O.S.; Tharmaratnam, T.; Iskandar, M.A.; Sefen, J.A.N.; Sidhu, P.; Atallah, B.; El-Lababidi, R.; et al. COVID-19: Breaking down a Global Health Crisis. Ann. Clin. Microbiol. Antimicrob. 2021, 20, 35. [Google Scholar] [CrossRef] [PubMed]
- Talic, S.; Shah, S.; Wild, H.; Gasevic, D.; Maharaj, A.; Ademi, Z.; Li, X.; Xu, W.; Mesa-Eguiagaray, I.; Rostron, J.; et al. Effectiveness of Public Health Measures in Reducing the Incidence of COVID-19, SARS-CoV-2 Transmission, and COVID-19 Mortality: Systematic Review and Meta-Analysis. BMJ 2021, 17, e068302. [Google Scholar] [CrossRef]
- Moore, L.D.; Robbins, G.; Quinn, J.; Arbogast, J.W. The Impact of COVID-19 Pandemic on Hand Hygiene Performance in Hospitals. Am. J. Infect. Control 2021, 49, 30–33. [Google Scholar] [CrossRef]
- Ghosh, S.; Bornman, C.; Zafer, M.M. Antimicrobial Resistance Threats in the Emerging COVID-19 Pandemic: Where Do We Stand? J. Infect. Public Health 2021, 14, 555–560. [Google Scholar] [CrossRef]
- Monnet, D.L.; Harbarth, S. Will Coronavirus Disease (COVID-19) Have an Impact on Antimicrobial Resistance? Eurosurveillance 2020, 25, 2001886. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, Research, and Development of New Antibiotics: The WHO Priority List of Antibiotic-Resistant Bacteria and Tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Abubakar, U.; Al-Anazi, M.; Alanazi, Z.; Rodríguez-Baño, J. Impact of COVID-19 Pandemic on Multidrug Resistant Gram Positive and Gram Negative Pathogens: A Systematic Review. J. Infect. Public Health 2023, 16, 320–331. [Google Scholar] [CrossRef]
- Apfalter, P.; Berning, L.; Eigentler, A.; El Belazi, G.; Fellinger, F.; Fuchs, K.; Fuchs, R.; Hain, C.; Hartl, R.; Igler, G.; et al. Resistenzbericht Österreich AURES 2020; Resistenzbericht Österreich AURES; Federal Ministry of Social Affairs, Health, Care and Consumer Protection: Vienna, Austria, 2021.
- Yang, M.; Feng, Y.; Yuan, L.; Zhao, H.; Gao, S.; Li, Z. High Concentration and Frequent Application of Disinfection Increase the Detection of Methicillin-Resistant Staphylococcus aureus Infections in Psychiatric Hospitals During the COVID-19 Pandemic. Front. Med. 2021, 8, 722219. [Google Scholar] [CrossRef]
- Kourtis, A.P.; Hatfield, K.; Baggs, J.; Mu, Y.; See, I.; Epson, E.; Nadle, J.; Kainer, M.A.; Dumyati, G.; Petit, S.; et al. Vital Signs: Epidemiology and Recent Trends in Methicillin-Resistant and in Methicillin-Susceptible Staphylococcus aureus Bloodstream Infections—United States. MMWR Morb. Mortal. Wkly. Rep. 2019, 68, 214–219. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Liu, J.; Guo, W.; Meng, H.; Huang, Q.; He, L.; Gao, Q.; Lv, H.; Liu, Y.; Wang, Y.; et al. Decreasing Methicillin-Resistant Staphylococcus aureus (MRSA) Infections Is Attributable to the Disappearance of Predominant MRSA ST239 Clones, Shanghai, 2008–2017. Emerg. Microbes Infect. 2019, 8, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Zarfel, G.; Luxner, J.; Folli, B.; Leitner, E.; Feierl, G.; Kittinger, C.; Grisold, A. Increase of Genetic Diversity and Clonal Replacement of Epidemic Methicillin-Resistant Staphylococcus aureus Strains in South-East Austria. FEMS Microbiol. Lett. 2016, 363, fnw137. [Google Scholar] [CrossRef] [PubMed]
- Alba, P.; Feltrin, F.; Cordaro, G.; Porrero, M.C.; Kraushaar, B.; Argudín, M.A.; Nykäsenoja, S.; Monaco, M.; Stegger, M.; Aarestrup, F.M.; et al. Livestock-Associated Methicillin Resistant and Methicillin Susceptible Staphylococcus aureus Sequence Type (CC)1 in European Farmed Animals: High Genetic Relatedness of Isolates from Italian Cattle Herds and Humans. PLoS ONE 2015, 10, e0137143. [Google Scholar] [CrossRef] [PubMed]
| spaCC002 | spaCC008 | spaCC011 | spaCC021 | spaCC032 | spaCC127 | |
|---|---|---|---|---|---|---|
| 2018 | 26.5% (58) | 10.0% (22) | 12.8% (28) | 4.6% (10) | 29.7% (65) | 6.8% (15) |
| 2019 | 24.9% (57) | 9.6% (22) | 14.0% (32) | 2.2% (5) | 19.7% (45) | 14.4% (33) |
| 2020 | 18.6% (33) | 11.9% (21) | 15.3% (27) | 4.0% (7) | 20.3% (36) | 14.1% (25) |
| 2018–2020 | 23.7% (148) | 10.4% (65) | 13.9% (87) | 3.5% (22) | 23.4% (146) | 11.7% (73) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zarfel, G.; Schmidt, J.; Luxner, J.; Grisold, A.J. No Changes in the Occurrence of Methicillin-Resistant Staphylococcus aureus (MRSA) in South-East Austria during the COVID-19 Pandemic. Pathogens 2023, 12, 1308. https://doi.org/10.3390/pathogens12111308
Zarfel G, Schmidt J, Luxner J, Grisold AJ. No Changes in the Occurrence of Methicillin-Resistant Staphylococcus aureus (MRSA) in South-East Austria during the COVID-19 Pandemic. Pathogens. 2023; 12(11):1308. https://doi.org/10.3390/pathogens12111308
Chicago/Turabian StyleZarfel, Gernot, Julia Schmidt, Josefa Luxner, and Andrea J. Grisold. 2023. "No Changes in the Occurrence of Methicillin-Resistant Staphylococcus aureus (MRSA) in South-East Austria during the COVID-19 Pandemic" Pathogens 12, no. 11: 1308. https://doi.org/10.3390/pathogens12111308
APA StyleZarfel, G., Schmidt, J., Luxner, J., & Grisold, A. J. (2023). No Changes in the Occurrence of Methicillin-Resistant Staphylococcus aureus (MRSA) in South-East Austria during the COVID-19 Pandemic. Pathogens, 12(11), 1308. https://doi.org/10.3390/pathogens12111308






