Emerging Roles of Noncoding RNAs in Bovine Mastitis Diseases
Abstract
1. Introduction
2. Mastitis Biology
2.1. Concepts of Mastitis
2.2. Clinical Signs of Mastitis
2.3. Effects of Mastitis
2.4. Causative Agents of Mastitis
| Country | Cow Level Prevalence (%) of Mastitis | Pathogen-Wise Prevalence (%) of Mastitis | References | ||||||
|---|---|---|---|---|---|---|---|---|---|
| CM 1 | SCM 2 | Overall Prevalence | Staphylococcus sp. 3 | Streptococcus sp. 4 | E. coli | Klebsiella sp. | Other 5 | ||
| Argentina | -- | 54 | -- | 72.4 | 8.8 | -- | -- | 5.2 | [43] |
| Australia | -- | -- | 55 | 15 | 7.0 | 4 | -- | -- | [44] |
| Bangladesh | -- | 51.0 | 51.0 | 45.7 | 14.8 | 9.9 | -- | 30.9 | [45] |
| Canada | -- | -- | 36.2 | 20.6 | -- | 2 | -- | 2.9 | [46] |
| China | -- | -- | -- | 39.0 | 11.0 | -- | -- | 18.2 | [47] |
| Kenya | 6.8 | 73.1 | 80.0 | 58.5 | 22.2 | -- | -- | 5.8 | [48] |
| Pakistan | 20 | 53 | -- | 34.0 | 9.0 | 19.4 | 8 | -- | [49] |
| Romania | -- | -- | -- | 43.2 | 22.4 | 13.8 | -- | 20.5 | [50] |
| Slovakia | -- | -- | 82.3 | 48.4 | 20.0 | 14.8 | -- | -- | [51] |
| Zimbabwe | 4.8 | 16.3 | 21.1 | 43.9 | 1.6 | 25.2 | 15.5 | -- | [52] |
2.5. Host Immune Responses to Mastitis
3. MiRNAs and Their Roles in Mastitis Biology
3.1. MiRNA Biosynthesis and Roles
3.2. Occurrence of miRNAs in MG Tissues
3.3. Roles of miRNAs in Mammary Gland Infection and Mastitis
3.3.1. Escherichia coli
3.3.2. Mycoplasma bovis
3.3.3. Staphylococcus aureus
3.3.4. Streptococcus agalactiae
3.3.5. Streptococcus uberis
3.3.6. CMT Tests and Other Mastitis Pathogens
4. LncRNAs in Mastitis Disease
5. Circular RNAs and Other ncRNAs in Mastitis
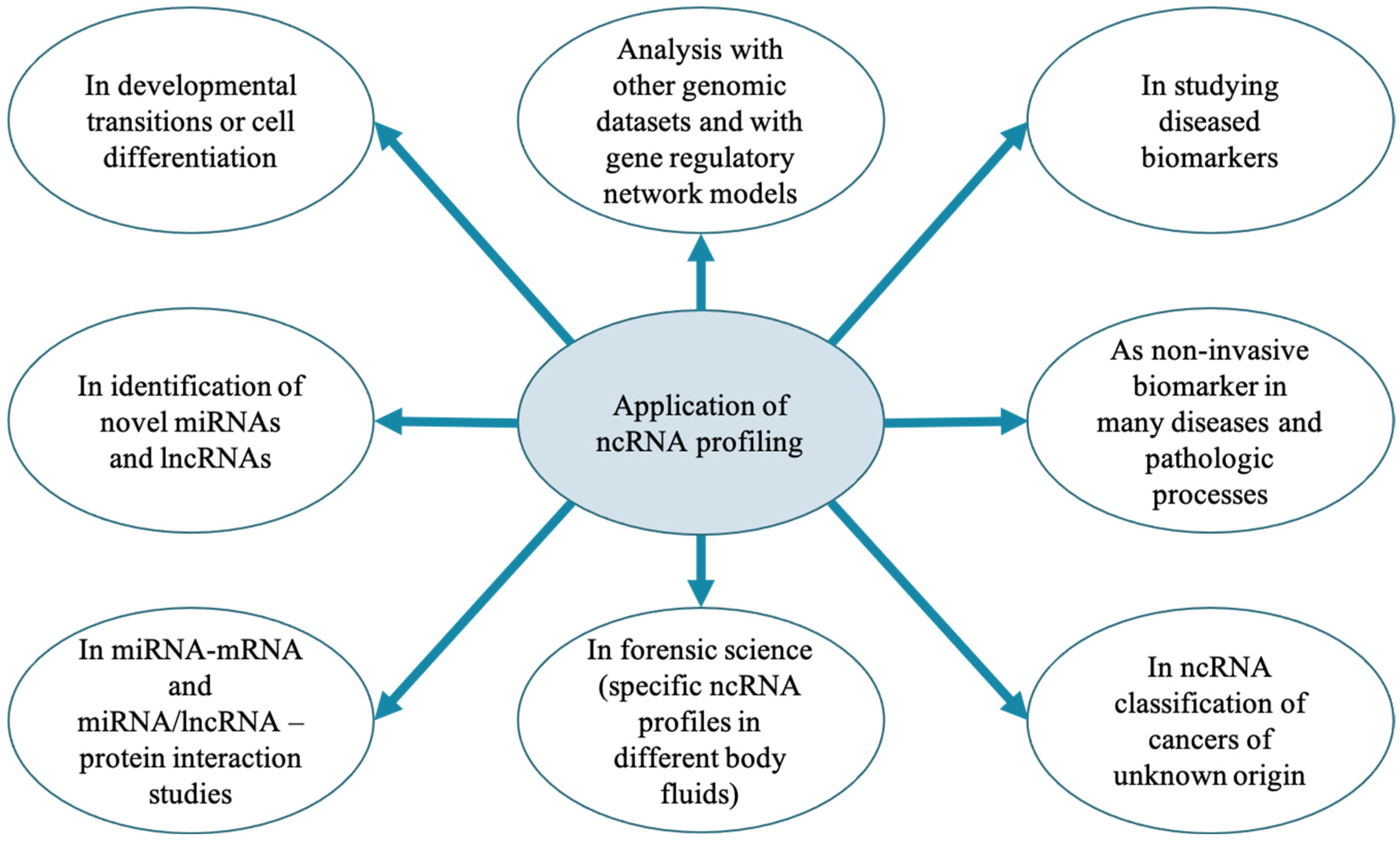
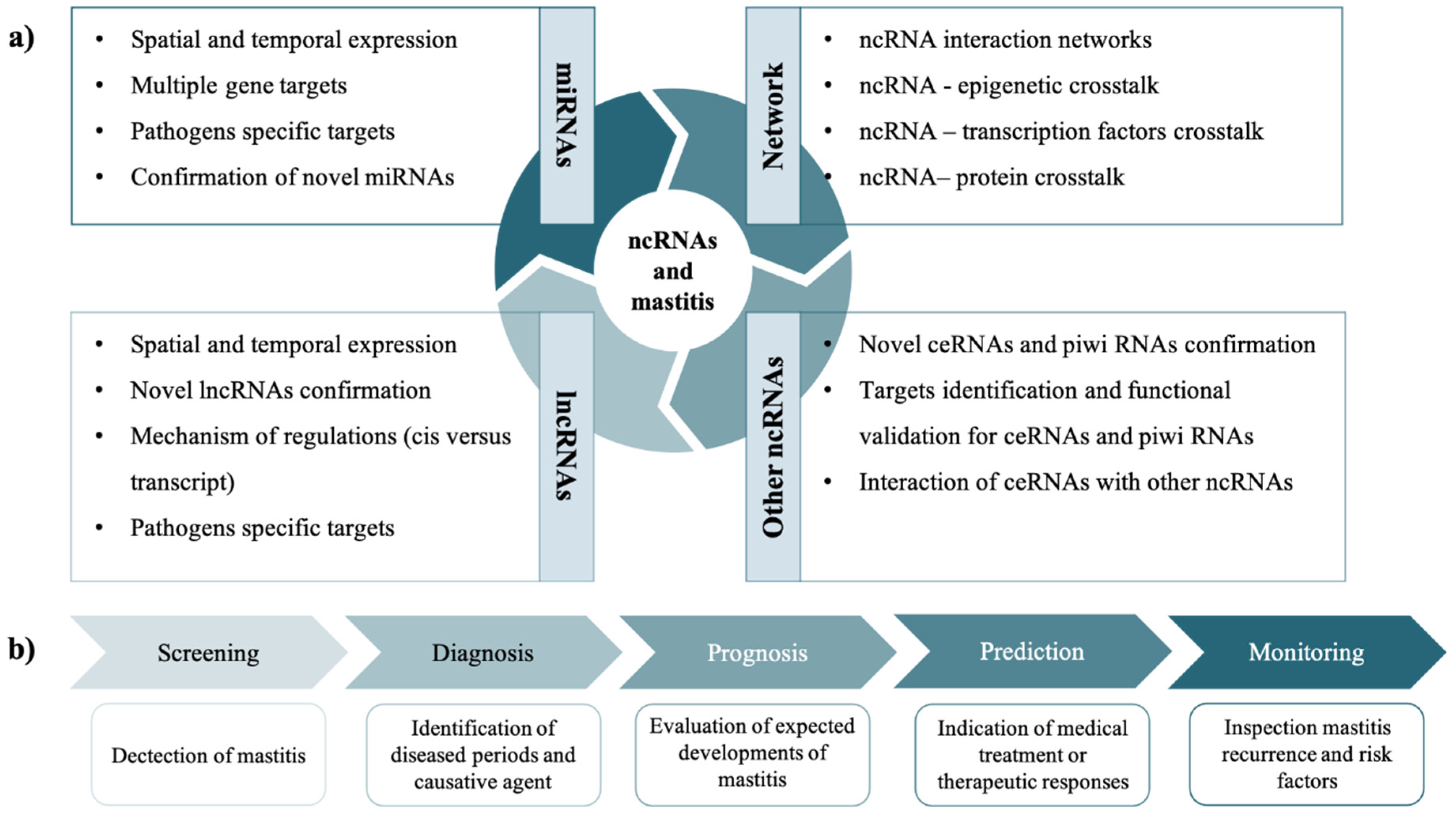
6. Perspectives of ncRNAs Studies in MG Health and Mastitis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- ENCODE Project Consortium; Birney, E.; Stamatoyannopoulos, J.A.; Dutta, A.; Guigo, R.; Gingeras, T.R.; Margulies, E.H.; Weng, Z.; Snyder, M.; Dermitzakis, E.T.; et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007, 447, 799–816. [Google Scholar] [CrossRef] [PubMed]
- Gigli, I.; Maizon, D.O. microRNAs and the mammary gland: A new understanding of gene expression. Genet. Mol. Biol. 2013, 36, 465–474. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Gu, Z.; Jiang, H. MicroRNAs in farm animals. Animal 2013, 7, 1567–1575. [Google Scholar] [CrossRef]
- Do, D.N.; Ibeagha-Awemu, E.M. Non-Coding RNA Roles in Ruminant Mammary Gland Development and Lactation. In Current Topics in Lactation; IntechOpen: London, UK, 2017. [Google Scholar] [CrossRef]
- Dong, H.; Gao, Q.; Peng, X.; Sun, Y.; Han, T.; Zhao, B.; Liu, Y.; Wang, C.; Song, X.; Wu, J.; et al. Circulating MicroRNAs As Potential Biomarkers for Veterinary Infectious Diseases. Front. Vet. Sci. 2017, 4, 186. [Google Scholar] [CrossRef] [PubMed]
- Weikard, R.; Demasius, W.; Kuehn, C. Mining long noncoding RNA in livestock. Anim. Genet. 2016, 48, 3–18. [Google Scholar] [CrossRef]
- Rijnkels, M.; Kabotyanski, E.; Montazer-Torbati, M.B.; Beauvais, C.H.; Vassetzky, Y.; Rosen, J.M.; Devinoy, E. The Epigenetic Landscape of Mammary Gland Development and Functional Differentiation. J. Mammary Gland Biol. Neoplasia 2010, 15, 85–100. [Google Scholar] [CrossRef]
- Piao, H.-L.; Ma, L. Non-Coding RNAs as Regulators of Mammary Development and Breast Cancer. J. Mammary Gland Biol. Neoplasia 2012, 17, 33–42. [Google Scholar] [CrossRef]
- Sandhu, G.K.; Milevskiy, M.J.G.; Wilson, W.; Shewan, A.M.; Brown, M.A. Non-coding RNAs in Mammary Gland Development and Disease. In Non-Coding RNA and the Reproductive System; Springer: Dordrecht, The Netherlands, 2015; Volume 886, pp. 121–153. [Google Scholar] [CrossRef]
- Fang, L.; Sahana, G.; Su, G.; Yu, Y.; Zhang, S.; Lund, M.S.; Sørensen, P. Integrating Sequence-based GWAS and RNA-Seq Provides Novel Insights into the Genetic Basis of Mastitis and Milk Production in Dairy Cattle. Sci. Rep. 2017, 7, srep45560. [Google Scholar] [CrossRef]
- Yang, F.; Chen, F.; Li, L.; Yan, L.; Badri, T.; Lv, C.; Yu, D.; Zhang, M.; Jang, X.; Li, J.; et al. Three Novel Players: PTK2B, SYK, and TNFRSF21 Were Identified to Be Involved in the Regulation of Bovine Mastitis Susceptibility via GWAS and Post-transcriptional Analysis. Front. Immunol. 2019, 10, 1579. [Google Scholar] [CrossRef]
- Wang, X.; Ma, P.; Liu, J.; Zhang, Q.; Zhang, Y.; Ding, X.; Jiang, L.; Wang, Y.; Zhang, Y.; Sun, D.; et al. Genome-wide association study in Chinese Holstein cows reveal two candidate genes for somatic cell score as an indicator for mastitis susceptibility. BMC Genet. 2015, 16, 111. [Google Scholar] [CrossRef]
- Jørgensen, H.B.H.; Buitenhuis, B.; Røntved, C.M.; Jiang, L.; Ingvartsen, K.L.; Sørensen, P. Transcriptional profiling of the bovine hepatic response to experimentally induced E. coli mastitis. Physiol. Genom. 2012, 44, 595–606. [Google Scholar] [CrossRef] [PubMed]
- Rinaldi, M.; Li, R.W.; Capuco, A.V. Mastitis associated transcriptomic disruptions in cattle. Vet. Immunol. Immunopathol. 2010, 138, 267–279. [Google Scholar] [CrossRef] [PubMed]
- Bakhtiarizadeh, M.R.; Mirzaei, S.; Norouzi, M.; Sheybani, N.; Sadi, M.S.V. Identification of Gene Modules and Hub Genes Involved in Mastitis Development Using a Systems Biology Approach. Front. Genet. 2020, 11, 722. [Google Scholar] [CrossRef] [PubMed]
- Boehmer, J.; Bannerman, D.; Shefcheck, K.; Ward, J. Proteomic Analysis of Differentially Expressed Proteins in Bovine Milk During Experimentally Induced Escherichia coli Mastitis. J. Dairy Sci. 2008, 91, 4206–4218. [Google Scholar] [CrossRef]
- Ibeagha-Awemu, E.M.; Ibeagha, A.E.; Messier, S.; Zhao, X. Proteomics, Genomics, and Pathway Analyses of Escherichia coli and Staphylococcus aureus Infected Milk Whey Reveal Molecular Pathways and Networks Involved in Mastitis. J. Proteome Res. 2010, 9, 4604–4619. [Google Scholar] [CrossRef] [PubMed]
- Ibeagha-Awemu, E.M.; Zhao, X. Epigenetic marks: Regulators of livestock phenotypes and conceivable sources of missing variation in livestock improvement programs. Front. Genet. 2015, 6, 302. [Google Scholar] [CrossRef] [PubMed]
- Glynn, C.L. Potential applications of microRNA profiling to forensic investigations. RNA 2019, 26, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Grillone, K.; Riillo, C.; Scionti, F.; Rocca, R.; Tradigo, G.; Guzzi, P.H.; Alcaro, S.; Di Martino, M.T.; Tagliaferri, P.; Tassone, P. Non-coding RNAs in cancer: Platforms and strategies for investigating the genomic “dark matter”. J. Exp. Clin. Cancer Res. 2020, 39, 1–19. [Google Scholar] [CrossRef]
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-Coding RNAs and their Integrated Networks. J. Integr. Bioinform. 2019, 16. [Google Scholar] [CrossRef]
- Santos, M.; Ma, Y.; Barbano, D. Effect of Somatic Cell Count on Proteolysis and Lipolysis in Pasteurized Fluid Milk During Shelf-Life Storage. J. Dairy Sci. 2003, 86, 2491–2503. [Google Scholar] [CrossRef]
- Dervishi, E.; Zhang, G.; Hailemariam, D.; Dunn, S.M.; Ametaj, B.N. Innate immunity and carbohydrate metabolism alterations precede occurrence of subclinical mastitis in transition dairy cows. J. Anim. Sci. Technol. 2015, 57, 46. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N. Alternative Approach to Control Intramammary Infection in Dairy Cows: A Review. Asian J. Anim. Vet. Adv. 2007, 2, 50–62. [Google Scholar] [CrossRef][Green Version]
- Petrovski, K.; Trajcev, M.; Buneski, G. A review of the factors affecting the costs of bovine mastitis: Review article. J. South Afr. Vet. Assoc. 2006, 77, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Halasa, T.; Huijps, K.; Østerås, O.; Hogeveen, H. Economic effects of bovine mastitis and mastitis management: A review. Vet. Q. 2007, 29, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Hogeveen, H. Mastitis Management in an Economic Framework. In Proceedings of the International Dairy Topics; Elsevier: Amsterdam, The Netherlands, 2005; pp. 17–20. [Google Scholar]
- Seegers, H.; Fourichon, C.; Beaudeau, F. Production effects related to mastitis and mastitis economics in dairy cattle herds. Vet. Res. 2003, 34, 475–491. [Google Scholar] [CrossRef]
- Sordillo, L.M.; Streicher, K.L. Mammary Gland Immunity and Mastitis Susceptibility. J. Mammary Gland Biol. Neoplasia 2002, 7, 135–146. [Google Scholar] [CrossRef]
- Novac, C.S.; Andrei, S. The Impact of Mastitis on the Biochemical Parameters, Oxidative and Nitrosative Stress Markers in Goat’s Milk: A Review. Pathogens 2020, 9, 882. [Google Scholar] [CrossRef]
- Izquierdo, A.C.; Liera, J.E.G.; Cervantes, R.E.; Castro, J.F.I.; Mancera, E.A.V.; Crispín, R.H.; Mosqueda, M.D.L.J.; Vazquez, A.G.; Pérez, J.O.; Aparicio, P.S.; et al. Production of Milk and Bovine Mastitis. Adv. Dairy Res. 2017, 5. [Google Scholar] [CrossRef]
- Jones, G.M.; Bailey, T.L. Understanding the Basics of Mastitis. Va. Coop. Ext. 2009. [Google Scholar]
- O’Ryan, M.; Prado, V.; Pickering, L.K. A millennium update on pediatric diarrheal illness in the developing world. Semin. Pediatr. Infect. Dis. 2005, 16, 125–136. [Google Scholar] [CrossRef]
- Podewils, L.J.; Mintz, E.D.; Nataro, J.P.; Parashar, U.D. Acute, infectious diarrhea among children in developing countries. Semin. Pediatr. Infect. Dis. 2004, 15, 155–168. [Google Scholar] [CrossRef] [PubMed]
- Langer, A.J.; Ayers, T.; Grass, J.; Lynch, M.; Angulo, F.J.; Mahon, B.E. Nonpasteurized Dairy Products, Disease Outbreaks, and State Laws—United States, 1993–2006. Emerg. Infect. Dis. 2012, 18, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Mungai, E.A.; Behravesh, C.B.; Gould, L.H. Increased Outbreaks Associated with Nonpasteurized Milk, United States, 2007–2012. Emerg. Infect. Dis. 2015, 21, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Garcia, S.N.; Osburn, B.I.; Cullor, J.S. A one health perspective on dairy production and dairy food safety. One Health 2019, 7, 100086. [Google Scholar] [CrossRef] [PubMed]
- Oviedo-Boyso, J.; Valdez-Alarcón, J.J.; Cajero-Juárez, M.; Ochoa-Zarzosa, A.; López-Meza, J.E.; Bravo-Patiño, A.; Baizabal-Aguirre, V.M. Innate immune response of bovine mammary gland to pathogenic bacteria responsible for mastitis. J. Infect. 2007, 54, 399–409. [Google Scholar] [CrossRef]
- Nickerson, S.; Owens, W.; Boddie, R. Mastitis in Dairy Heifers: Initial Studies on Prevalence and Control. J. Dairy Sci. 1995, 78, 1607–1618. [Google Scholar] [CrossRef]
- Bannerman, D.D.; Paape, M.J.; Lee, J.-W.; Zhao, X.; Hope, J.C.; Rainard, P. Escherichia coli and Staphylococcus aureus Elicit Differential Innate Immune Responses following Intramammary Infection. Clin. Vaccine Immunol. 2004, 11, 463–472. [Google Scholar] [CrossRef]
- Petzl, W.; Zerbe, H.; Günther, J.; Yang, W.; Seyfert, H.-M.; Nürnberg, G.; Schuberth, H.-J. Escherichia coli, but not Staphylococcus aureus triggers an early increased expression of factors contributing to the innate immune defense in the udder of the cow. Vet. Res. 2008, 39, 18–23. [Google Scholar] [CrossRef]
- Fairbrother, J.-H.; Dufour, S.; Fairbrother, J.M.; Francoz, D.; Nadeau, É.; Messier, S. Characterization of persistent and transient Escherichia coli isolates recovered from clinical mastitis episodes in dairy cows. Vet. Microbiol. 2015, 176, 126–133. [Google Scholar] [CrossRef]
- Dieser, S.A.; Vissio, C.; Lasagno, M.C.; Bogni, C.I.; Larriestra, A.J.; Odierno, L.M. Prevalence of Pathogens Causing Subclinical Mastitis in Argentinean Dairy Herds. Pak. Vet. J. 2014, 34, 124–126. [Google Scholar]
- Al-Harbi, H.; Ranjbar, S.; Moore, R.J.; Alawneh, J.I. Bacteria Isolated From Milk of Dairy Cows With and Without Clinical Mastitis in Different Regions of Australia and Their AMR Profiles. Front. Vet. Sci. 2021, 8, 743725. [Google Scholar] [CrossRef] [PubMed]
- Kabir, H.; Ershaduzzaman; Giasuddin; Nazir, K.; Mahmud, M.; Islam, R.; Karim, R.; Yousuf, A.; Rahman, S.; Ali, Y. Prevalence and molecular detection of the causal agents of sub-clinical mastitis in dairy cows in Sirajganj and Pabna districts, Bangladesh. J. Adv. Vet. Anim. Res. 2017, 4, 378. [Google Scholar] [CrossRef]
- Acharya, K.R.; Brankston, G.; Slavic, D.; Greer, A.L. Spatio-Temporal Variation in the Prevalence of Major Mastitis Pathogens Isolated From Bovine Milk Samples Between 2008 and 2017 in Ontario, Canada. Front. Vet. Sci. 2021, 8, 742696. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Huang, X.; Xu, H.; Zhang, C.; Chen, S.; Liu, F.; Guan, S.; Zhang, S.; Zhu, K.; Wu, C. The prevalence of pathogens causing bovine mastitis and their associated risk factors in 15 large dairy farms in China: An observational study. Vet. Microbiol. 2020, 247, 108757. [Google Scholar] [CrossRef]
- Mbindyo, C.M.; Gitao, G.C.; Mulei, C.M. Prevalence, Etiology, and Risk Factors of Mastitis in Dairy Cattle in Embu and Kajiado Counties, Kenya. Vet. Med. Int. 2020, 2020, 8831172. [Google Scholar] [CrossRef] [PubMed]
- Ali, T.; Kamran; Raziq, A.; Wazir, I.; Ullah, R.; Shah, P.; Ali, M.I.; Han, B.; Liu, G. Prevalence of Mastitis Pathogens and Antimicrobial Susceptibility of Isolates From Cattle and Buffaloes in Northwest of Pakistan. Front. Vet. Sci. 2021, 8, 746755. [Google Scholar] [CrossRef]
- Pascu, C.; Herman, V.; Iancu, I.; Costinar, L. Etiology of Mastitis and Antimicrobial Resistance in Dairy Cattle Farms in the Western Part of Romania. Antibiotics 2022, 11, 57. [Google Scholar] [CrossRef]
- Holko, I.; Tančin, V.; Vršková, M.; Tvarožková, K. Prevalence and antimicrobial susceptibility of udder pathogens isolated from dairy cows in Slovakia. J. Dairy Res. 2019, 86, 436–439. [Google Scholar] [CrossRef]
- Katsande, S.; Matope, G.; Ndengu, M.; Pfukenyi, D.M. Prevalence of mastitis in dairy cows from smallholder farms in Zimbabwe. Onderstepoort J. Vet. Res. 2013, 80, 7. [Google Scholar] [CrossRef]
- Sordillo, L.; Shafer-Weaver, K.; DeRosa, D. Immunobiology of the Mammary Gland. J. Dairy Sci. 1997, 80, 1851–1865. [Google Scholar] [CrossRef]
- Paulrud, C. Basic Concepts of the Bovine Teat Canal. Vet. Res. Commun. 2005, 29, 215–245. [Google Scholar] [CrossRef] [PubMed]
- Sudhan, N.A.; Sharma, N. Mastitis-an Important Production Disease of Dairy Animals. In SMVS Dairy Year Book; Sarva Manav Vikash Samiti: Gurgaon, India, 2010; pp. 72–88. [Google Scholar]
- Gray, C.; Strandberg, Y.; Donaldson, L.; Tellam, R.L. Bovine mammary epithelial cells, initiators of innate immune responses to mastitis. Aust. J. Exp. Agric. 2005, 45, 757–761. [Google Scholar] [CrossRef]
- Barber, M.R.; Yang, T.J. Chemotactic Activities in Nonmastitic and Mastitic Mammary Secretions: Presence of Interleukin-8 in Mastitic but Not Nonmastitic Secretions. Clin. Diagn. Lab. Immunol. 1998, 5, 82–86. [Google Scholar] [CrossRef]
- Sordillo, L.M. Factors affecting mammary gland immunity and mastitis susceptibility. Livest. Prod. Sci. 2005, 98, 89–99. [Google Scholar] [CrossRef]
- Stelwagen, K.; Carpenter, E.; Haigh, B.; Hodgkinson, A.; Wheeler, T.T. Immune components of bovine colostrum and milk1. J. Anim. Sci. 2009, 87, 3–9. [Google Scholar] [CrossRef]
- Uthaisangsook, S.; Day, N.K.; Bahna, S.L.; Good, R.A.; Haraguchi, S. Innate immunity and its role against infections. Ann. Allergy, Asthma Immunol. 2002, 88, 253–265. [Google Scholar] [CrossRef]
- Ganz, T. Defensins: Antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 2003, 3, 710–720. [Google Scholar] [CrossRef] [PubMed]
- Wiesner, J.; Vilcinskas, A. Antimicrobial peptides: The ancient arm of the human immune system. Virulence 2010, 1, 440–464. [Google Scholar] [CrossRef] [PubMed]
- Zanetti, M. The Role of Cathelicidins in the Innate Host Defenses of Mammals. Curr. Issues Mol. Biol. 2005, 7, 179–196. [Google Scholar]
- Pecorini, C.; Rebucci, R.; Truchet, S.; Baldi, A. In Vitro Effects of Lactoferrin on Intestinal and Mammary Epithelial Cell Lines. Ital. J. Anim. Sci. 2009, 8, 643–645. [Google Scholar] [CrossRef]
- Enerbäck, C.; A Porter, D.; Seth, P.; Sgroi, D.; Gaudet, J.; Weremowicz, S.; Morton, C.C.; Schnitt, S.; Pitts, R.L.; Stampl, J.; et al. Psoriasin expression in mammary epithelial cells in vitro and in viv. Cancer Res. 2002, 62, 43–47. [Google Scholar] [PubMed]
- Alford, M.A.; Baquir, B.; Santana, F.L.; Haney, E.F.; Hancock, R.E.W. Cathelicidin Host Defense Peptides and Inflammatory Signaling: Striking a Balance. Front. Microbiol. 2020, 11, 1902. [Google Scholar] [CrossRef] [PubMed]
- Paape, M.; Mehrzad, J.; Zhao, X.; Detilleux, J.; Burvenich, C. Defense of the Bovine Mammary Gland by Polymorphonuclear Neutrophil Leukocytes. J. Mammary Gland Biol. Neoplasia 2002, 7, 109–121. [Google Scholar] [CrossRef]
- Harmon, R.J. Physiology of Mastitis and Factors Affecting Somatic Cell Counts. J. Dairy Sci. 1994, 77, 2103–2112. [Google Scholar] [CrossRef]
- Rainard, P.; Riollet, C. Innate immunity of the bovine mammary gland. Vet. Res. 2006, 37, 369–400. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Menon, M.P.; Hua, K.-F. The Long Non-coding RNAs: Paramount Regulators of the NLRP3 Inflammasome. Front. Immunol. 2020, 11, 569524. [Google Scholar] [CrossRef]
- Wang, M.; Jiang, S.; Wu, W.; Yu, F.; Chang, W.; Li, P.; Wang, K. Non-coding RNAs Function as Immune Regulators in Teleost Fish. Front. Immunol. 2018, 9, 2801. [Google Scholar] [CrossRef]
- Tucker, A.R.; Salazar, N.A.; Ayoola, A.O.; Memili, E.; Thomas, B.N.; Morenikeji, O.B. Regulatory network of miRNA, lncRNA, transcription factor and target immune response genes in bovine mastitis. Sci. Rep. 2021, 11, 1–18. [Google Scholar] [CrossRef]
- Almo, M.M.; Sousa, I.G.; Maranhão, A.; Brigido, M.M. The role of long noncoding RNAs in human T CD3+ cells. J. Immunol. Sci. 2018, 2, 32–36. [Google Scholar] [CrossRef][Green Version]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Kim, V.N.; Han, J.; Siomi, M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009, 10, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Borchert, G.M.; Lanier, W.; Davidson, B.L. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006, 13, 1097–1101. [Google Scholar] [CrossRef]
- Brodersen, P.; Voinnet, O. Revisiting the principles of microRNA target recognition and mode of action. Nat. Rev. Mol. Cell Biol. 2009, 10, 141–148. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Pauley, K.M.; Cha, S.; Chan, E.K. MicroRNA in autoimmunity and autoimmune diseases. J. Autoimmun. 2009, 32, 189–194. [Google Scholar] [CrossRef]
- Rottiers, V.; Näär, A.M. MicroRNAs in metabolism and metabolic disorders. Nat. Rev. Mol. Cell Biol. 2012, 13, 239–250. [Google Scholar] [CrossRef]
- Thum, T. MicroRNA therapeutics in cardiovascular medicine. EMBO Mol. Med. 2011, 4, 3–14. [Google Scholar] [CrossRef]
- Alvarez-Garcia, I.; Miska, E.A. MicroRNA functions in animal development and human disease. Development 2005, 132, 4653–4662. [Google Scholar] [CrossRef]
- Esquela-Kerscher, A.; Slack, F. Oncomirs—microRNAs with a role in cancer. Nat. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Croce, C.M. MicroRNA Signatures in Human Cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Paranjape, T.; Slack, F.J.; Weidhaas, J.B. MicroRNAs: Tools for cancer diagnostics. Gut 2009, 58, 1546–1554. [Google Scholar] [CrossRef]
- Wen, P.; Xie, Y.; Wang, L. The Role of microRNA in Pathogenesis, Diagnosis, Different Variants, Treatment and Prognosis of Mycosis Fungoides. Front. Oncol. 2021, 11, 752817. [Google Scholar] [CrossRef]
- Gu, Z.; Eleswarapu, S.; Jiang, H. Identification and characterization of microRNAs from the bovine adipose tissue and mammary gland. FEBS Lett. 2007, 581, 981–988. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Gao, C.; Li, H.; Huang, L.; Sun, Q.; Dong, Y.; Tian, C.; Gao, S.; Dong, H.; Guan, D.; et al. Identification and characterization of microRNAs in raw milk during different periods of lactation, commercial fluid, and powdered milk products. Cell Res. 2010, 20, 1128–1137. [Google Scholar] [CrossRef]
- Izumi, H.; Kosaka, N.; Shimizu, T.; Sekine, K.; Ochiya, T.; Takase, M. Bovine milk contains microRNA and messenger RNA that are stable under degradative conditions. J. Dairy Sci. 2012, 95, 4831–4841. [Google Scholar] [CrossRef]
- Li, R.; Dudemaine, P.-L.; Zhao, X.; Lei, C.; Ibeagha-Awemu, E.M. Comparative Analysis of the miRNome of Bovine Milk Fat, Whey and Cells. PLoS ONE 2016, 11, e0154129. [Google Scholar] [CrossRef]
- Do, D.N.; Li, R.; Dudemaine, P.-L.; Ibeagha-Awemu, E.M. MicroRNA roles in signalling during lactation: An insight from differential expression, time course and pathway analyses of deep sequence data. Sci. Rep. 2017, 7, srep44605. [Google Scholar] [CrossRef]
- Li, Z.; Liu, H.; Jin, X.; Lo, L.; Liu, J. Expression profiles of microRNAs from lactating and non-lactating bovine mammary glands and identification of miRNA related to lactation. BMC Genom. 2012, 13, 731. [Google Scholar] [CrossRef] [PubMed]
- Le Guillou, S.; Marthey, S.; Laloë, D.; Laubier, J.; Mobuchon, L.; Leroux, C.; Le Provost, F. Characterisation and Comparison of Lactating Mouse and Bovine Mammary Gland miRNomes. PLoS ONE 2014, 9, e91938. [Google Scholar] [CrossRef] [PubMed]
- Luoreng, Z.-M.; Wang, X.-P.; Mei, C.-G.; Zan, L.-S. Comparison of microRNA Profiles between Bovine Mammary Glands Infected with Staphylococcus aureus and Escherichia coli. Int. J. Biol. Sci. 2018, 14, 87–99. [Google Scholar] [CrossRef]
- Li, Q.; Yang, C.; Du, J.; Zhang, B.; He, Y.; Hu, Q.; Li, M.; Zhang, Y.; Wang, C.; Zhong, J. Characterization of miRNA profiles in the mammary tissue of dairy cattle in response to heat stress. BMC Genom. 2018, 19, 975. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.-Z.; Chen, Y.; Guan, L.L. MicroRNA expression profiles across blood and different tissues in cattle. Sci. Data 2019, 6, 190013. [Google Scholar] [CrossRef]
- Bourdon, C.; Bardou, P.; Aujean, E.; Le Guillou, S.; Tosser-Klopp, G.; Le Provost, F. RumimiR: A detailed microRNA database focused on ruminant species. Database 2019, 2019, baz099. [Google Scholar] [CrossRef]
- Ethompson-Crispi, K.; Eatalla, H.; Miglior, F.; Mallard, B.A. Bovine Mastitis: Frontiers in Immunogenetics. Front. Immunol. 2014, 5, 493. [Google Scholar] [CrossRef]
- Srikok, S.; Patchanee, P.; Boonyayatra, S.; Chuammitri, P. Potential role of MicroRNA as a diagnostic tool in the detection of bovine mastitis. Prev. Vet. Med. 2020, 182, 105101. [Google Scholar] [CrossRef]
- Lai, Y.; Rahman, M.; Chen, H.; Husna, A.A.; Fujikawa, T.; Ando, T.; Kitahara, G.; Koiwa, M.; Kubota, C.; Miura, N. Bovine milk transcriptome analysis reveals microRNAs and RNU2 involved in mastitis. FEBS J. 2019, 287, 1899–1918. [Google Scholar] [CrossRef]
- Lai, Y.-C.; Fujikawa, T.; Maemura, T.; Ando, T.; Kitahara, G.; Endo, Y.; Yamato, O.; Koiwa, M.; Kubota, C.; Miura, N. Inflammation-related microRNA expression level in the bovine milk is affected by mastitis. PLoS ONE 2017, 12, e0177182. [Google Scholar] [CrossRef]
- Özdemir, S. Expression profiling of microRNAs in the Mycoplasma bovis infected mammary gland tissue in Holstein Friesian cattle. Microb. Pathog. 2020, 147, 104426. [Google Scholar] [CrossRef] [PubMed]
- Özdemir, S. Siyah Alaca ve Doğu Anadolu Kırmızısı Irkına Ait Sığırların Mycoplasma bovis ile Enfekte Sütlerinden Köken Alan Eksozomlardaki Yangı Ile Ilişkili miRNA’ların Ekpresyon Profile. Kahramanmaraş Sütçü İmam Üniversitesi Tarım Ve Doğa Derg. 2020, 23, 762–771. [Google Scholar] [CrossRef]
- Ma, S.; Tong, C.; Ibeagha-Awemu, E.M.; Zhao, X. Identification and characterization of differentially expressed exosomal microRNAs in bovine milk infected with Staphylococcus aureus. BMC Genom. 2019, 20, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ju, Z.; Jiang, Q.; Liu, G.; Wang, X.; Luo, G.; Zhang, Y.; Zhang, J.; Zhong, J.; Huang, J. Solexa sequencing and custom microRNA chip reveal repertoire of microRNAs in mammary gland of bovine suffering from natural infectious mastitis. Anim. Genet. 2018, 49, 3–18. [Google Scholar] [CrossRef]
- Cai, M.; He, H.; Jia, X.; Chen, S.; Wang, J.; Shi, Y.; Liu, B.; Xiao, W.; Lai, S. Genome-wide microRNA profiling of bovine milk-derived exosomes infected with Staphylococcus aureus. Cell Stress Chaperon. 2018, 23, 663–672. [Google Scholar] [CrossRef]
- Ngo, S.; Moloney, S.; Li, X.; McNaughton, L.; Partridge, A.; Sheppard, A.M. Distinct MicroRNA Signatures for Mastitis Measured in Milk Following Natural Exposure in Dairy Herds. Int. J. Anim. Sci. 2017, 1, 1001. [Google Scholar] [CrossRef]
- Ju, Z.; Huang, J.; Jiang, Q.; Wang, C.; Wang, X.; Zhao, S. Identification of bta-miR-15a∼16a cluster expression, localization and regulated target in Holsteins. Mol. Cell. Probes 2018, 40, 8–12. [Google Scholar] [CrossRef]
- Jin, W.; Ibeagha-Awemu, E.M.; Liang, G.; Beaudoin, F.; Zhao, X.; Guan, L.L. Transcriptome microRNA profiling of bovine mammary epithelial cells challenged with Escherichia coli or Staphylococcus aureusbacteria reveals pathogen directed microRNA expression profiles. BMC Genom. 2014, 15, 181. [Google Scholar] [CrossRef]
- Naeem, A.; Zhong, K.; Moisá, S.J.; Drackley, J.K.; Moyes, K.M.; Loor, J.J. Bioinformatics analysis of microRNA and putative target genes in bovine mammary tissue infected with Streptococcus uberis. J. Dairy Sci. 2012, 95, 6397–6408. [Google Scholar] [CrossRef]
- Lawless, N.; Foroushani, A.B.K.; McCabe, M.S.; O’Farrelly, C.; Lynn, D.J. Next Generation Sequencing Reveals the Expression of a Unique miRNA Profile in Response to a Gram-Positive Bacterial Infection. PLoS ONE 2013, 8, e57543. [Google Scholar] [CrossRef]
- Pu, J.; Li, R.; Zhang, C.; Chen, D.; Liao, X.; Zhu, Y.; Geng, X.; Ji, D.; Mao, Y.; Gong, Y.; et al. Expression profiles of miRNAs from bovine mammary glands in response to Streptococcus agalactiae-induced mastitis. J. Dairy Res. 2017, 84, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, H.; Chen, L.; Wang, L.; Liu, X.; Ru, C.; Song, A. Identification and characterization of novel and differentially expressed microRNAs in peripheral blood from healthy and mastitis Holstein cattle by deep sequencing. Anim. Genet. 2013, 45, 20–27. [Google Scholar] [CrossRef]
- Luoreng, Z.-M.; Yang, J.; Wang, X.-P.; Wei, D.-W.; Zan, L.-S. Expression Profiling of microRNA From Peripheral Blood of Dairy Cows in Response to Staphylococcus aureus-Infected Mastitis. Front. Vet. Sci. 2021, 8, 691196. [Google Scholar] [CrossRef]
- Luoreng, Z.-M.; Wang, X.-P.; Mei, C.-G.; Zan, L.-S. Expression profiling of peripheral blood miRNA using RNAseq technology in dairy cows with Escherichia coli-induced mastitis. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Lewandowska-Sabat, A.M.; Hansen, S.F.; Solberg, T.R.; Østerås, O.; Heringstad, B.; Boysen, P.; Olsaker, I. MicroRNA expression profiles of bovine monocyte-derived macrophages infected in vitro with two strains of Streptococcus agalactiae. BMC Genom. 2018, 19, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhou, J.; Wang, X.; Zhang, Y.; Lu, X.; Fan, Y.; Mao, Y.; Loor, J.J.; Yang, Z. Screening candidate microR-15a- IRAK2 regulatory pairs for predicting the response to Staphylococcus aureus-induced mastitis in dairy cows. J. Dairy Res. 2019, 86, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Xu, X.; Tan, T.; Chen, D.; Liang, H.; Sun, K.; Li, M.; Zhang, H.; Mao, Y.; Yang, Z. MicroRNA-145 regulates immune cytokines via targeting FSCN1 in Staphylococcus aureus -induced mastitis in dairy cows. Reprod. Domest. Anim. 2019, 54, 882–891. [Google Scholar] [CrossRef]
- Li, W.; Li, C.; Lu, J.; Zhao, Y. MiR-145 is involved in the proliferation of bovine mammary epithelial cells and regulates bovine insulin receptor substrate 1. Ital. J. Anim. Sci. 2020, 19, 536–543. [Google Scholar] [CrossRef]
- Han, S.; Li, X.; Liu, J.; Zou, Z.; Luo, L.; Wu, R.; Zhao, Z.; Wang, C.; Shen, B. Bta-miR-223 Targeting CBLB Contributes to Resistance to Staphylococcus aureus Mastitis Through the PI3K/AKT/NF-κB Pathway. Front. Vet. Sci. 2020, 7, 529. [Google Scholar] [CrossRef]
- Pu, J.; Chen, D.; Chu, S.; Chen, Z.; Fan, Y.; Zhang, Z.; Loor, J.J.; Mao, Y.; Yang, Z. miR-122 regulates the JAK-STAT signalling pathway by down-regulating EPO in the mammary gland during Streptococcus agalactiae-induced mastitis. Ital. J. Anim. Sci. 2020, 19, 1236–1243. [Google Scholar] [CrossRef]
- Li, Y.; Hu, Q.; Luoreng, Z.; Yang, J.; Wang, X.; Ma, Y.; Wei, D. Differential mRNA Expression Profiling Reveals the Role of MiR-375 in Inflammation of Bovine Mammary Epithelial Cells. Animals 2022, 12, 1431. [Google Scholar] [CrossRef]
- Özdemir, S. Identification and comparison of exosomal microRNAs in the milk and colostrum of two different cow breeds. Gene 2020, 743, 144609. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Zhang, J.; Zhou, Z.; Wang, L.; Liu, Y.; Liu, Y. ALDB: A Domestic-Animal Long Noncoding RNA Database. PLoS ONE 2015, 10, e0124003. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Liu, X.; Li, Z.; Wang, H.; Liu, Y.; He, H.; Yang, J.; Niu, F.; Wang, L.; Guo, J. Expression differences of miRNAs and genes on NF-κB pathway between the healthy and the mastitis Chinese Holstein cows. Gene 2014, 545, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Zhao, Y.; Yu, S.; Zhang, H.; Cheng, M.; Cao, H.; Li, Q.; Min, L. CircRNA as CeRNA mediated by microRNA may be involved in goat lactation. Small Rumin. Res. 2019, 171, 63–72. [Google Scholar] [CrossRef]
- Zhao, X.; Lacasse, P. Mammary tissue damage during bovine mastitis: Causes and control1. J. Anim. Sci. 2008, 86, 57–65. [Google Scholar] [CrossRef]
- Reinoso, E.B.; Lasagno, M.C.; Dieser, S.A.; Odierno, L.M. Distribution of virulence-associated genes in Streptococcus uberis isolated from bovine mastitis. FEMS Microbiol. Lett. 2011, 318, 183–188. [Google Scholar] [CrossRef]
- Li, Z.; Wang, H.; Chen, L.; Zhai, M.; Chen, S.; Li, N.; Liu, X. Identification and expression analysis of miR-144-5p and miR-130b-5p in dairy cattle. Arch. Anim. Breed. 2017, 60, 199–204. [Google Scholar] [CrossRef][Green Version]
- Jiang, Q.; Zhao, H.; Li, R.; Zhang, Y.; Liu, Y.; Wang, J.; Wang, X.; Ju, Z.; Liu, W.; Hou, M.; et al. In silico genome-wide miRNA-QTL-SNPs analyses identify a functional SNP associated with mastitis in Holsteins. BMC Genet. 2019, 20, 46. [Google Scholar] [CrossRef]
- Tzelos, T.; Ho, W.; Charmana, V.I.; Lee, S.; Donadeu, F.X. MiRNAs in milk can be used towards early prediction of mammary gland inflammation in cattle. Sci. Rep. 2022, 12, 1–8. [Google Scholar] [CrossRef]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Mattick, J.S. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 2013, 20, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Vance, K.W.; Ponting, C.P. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 2014, 30, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef]
- Dieci, G.; Fiorino, G.; Castelnuovo, M.; Teichmann, M.; Pagano, A. The expanding RNA polymerase III transcriptome. Trends Genet. 2007, 23, 614–622. [Google Scholar] [CrossRef]
- Kapranov, P.; Cheng, J.; Dike, S.; Nix, D.A.; Duttagupta, R.; Willingham, A.T.; Stadler, P.F.; Hertel, J.; Hackermüller, J.; Hofacker, I.L.; et al. RNA Maps Reveal New RNA Classes and a Possible Function for Pervasive Transcription. Science 2007, 316, 1484–1488. [Google Scholar] [CrossRef]
- Yin, Q.-F.; Yang, L.; Zhang, Y.; Xiang, J.-F.; Wu, Y.-W.; Carmichael, G.G.; Chen, L.-L. Long Noncoding RNAs with snoRNA Ends. Mol. Cell 2012, 48, 219–230. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, H.; Fang, S.; Kang, Y.; Wu, W.; Hao, Y.; Li, Z.; Bu, D.; Sun, N.; Zhang, M.Q.; et al. NONCODE 2016: An informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 2015, 44, D203–D208. [Google Scholar] [CrossRef]
- Weikard, R.; Hadlich, F.; Kuehn, C. Identification of novel transcripts and noncoding RNAs in bovine skin by deep next generation sequencing. BMC Genom. 2013, 14, 789. [Google Scholar] [CrossRef]
- Billerey, C.; Boussaha, M.; Esquerré, D.; Rebours, E.; Djari, A.; Meersseman, C.; Klopp, C.; Gautheret, D.; Rocha, D. Identification of large intergenic non-coding RNAs in bovine muscle using next-generation transcriptomic sequencing. BMC Genom. 2014, 15, 499. [Google Scholar] [CrossRef]
- Koufariotis, L.T.; Chen, Y.-P.P.; Chamberlain, A.; Jagt, C.V.; Hayes, B. A catalogue of novel bovine long noncoding RNA across 18 tissues. PLoS ONE 2015, 10, e0141225. [Google Scholar] [CrossRef] [PubMed]
- Ibeagha-Awemu, E.M.; Li, R.; Dudemaine, P.-L.; Do, D.N.; Bissonnette, N. Transcriptome Analysis of Long Non-Coding RNA in the Bovine Mammary Gland Following Dietary Supplementation with Linseed Oil and Safflower Oil. Int. J. Mol. Sci. 2018, 19, 3610. [Google Scholar] [CrossRef] [PubMed]
- Ibeagha-Awemu, E.M.; Do, D.N.; Dudemaine, P.-L.; Fomenky, B.E.; Bissonnette, N. Integration of lncRNA and mRNA Transcriptome Analyses Reveals Genes and Pathways Potentially Involved in Calf Intestinal Growth and Development during the Early Weeks of Life. Genes 2018, 9, 142. [Google Scholar] [CrossRef] [PubMed]
- Do, D.N.; Dudemaine, P.-L.; Fomenky, B.; Ibeagha-Awemu, E.M. Transcriptome Analysis of Non-Coding RNAs in Livestock Species: Elucidating the Ambiguity. In Applications of RNA-Seq and Omics Strategies—From Microorganisms to Human Health; IntechOpen: London, UK, 2017. [Google Scholar] [CrossRef]
- Tong, C.; Chen, Q.; Zhao, L.; Ma, J.; Ibeagha-Awemu, E.M.; Zhao, X. Identification and characterization of long intergenic noncoding RNAs in bovine mammary glands. BMC Genom. 2017, 18, 1–10. [Google Scholar] [CrossRef]
- Ma, M.; Pei, Y.; Wang, X.; Feng, J.; Zhang, Y.; Gao, M.-Q. LncRNA XIST mediates bovine mammary epithelial cell inflammatory response via NF-κB/NLRP3 inflammasome pathway. Cell Prolif. 2018, 52, e12525. [Google Scholar] [CrossRef]
- Yang, W.; Li, X.; Qi, S.; Li, X.; Zhou, K.; Qing, S.; Zhang, Y.; Gao, M.-Q. lncRNA H19 is involved in TGF-β1-induced epithelial to mesenchymal transition in bovine epithelial cells through PI3K/AKT Signaling Pathway. PeerJ 2017, 5, e3950. [Google Scholar] [CrossRef]
- Wang, H.; Wang, X.; Li, X.; Wang, Q.; Qing, S.; Zhang, Y.; Gao, M. A novel long non-coding RNA regulates the immune response in MAC-T cells and contributes to bovine mastitis. FEBS J. 2019, 286, 1780–1795. [Google Scholar] [CrossRef]
- Bhat, S.A.; Ahmad, S.M.; Mumtaz, P.T.; Malik, A.A.; Dar, M.A.; Urwat, U.; Shah, R.A.; Ganai, N.A. Long non-coding RNAs: Mechanism of action and functional utility. Non-Coding RNA Res. 2016, 1, 43–50. [Google Scholar] [CrossRef]
- Pawar, K.G.; Hanisch, C.; Vera, S.E.P.; Einspanier, R.; Sharbati, S. Down regulated lncRNA MEG3 eliminates mycobacteria in macrophages via autophagy. Sci. Rep. 2016, 6, srep19416. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, L.; Jin, J.; Xia, A.; Wang, C.; Cui, Y.; Qu, B.; Li, Q.; Sheng, C. Comparative transcriptome analysis to investigate the potential role of miRNAs in milk protein/fat quality. Sci. Rep. 2018, 8, 6250. [Google Scholar] [CrossRef]
- Suravajhala, P.; Benso, A. Prioritizing single-nucleotide polymorphisms and variants associated with clinical mastitis. Adv. Appl. Bioinform. Chem. 2017, 10, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Özdemir, S.; Altun, S. Genome-wide analysis of mRNAs and lncRNAs in Mycoplasma bovis infected and non-infected bovine mammary gland tissues. Mol. Cell. Probes 2020, 50, 101512. [Google Scholar] [CrossRef] [PubMed]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Lasda, E.; Parker, R. Circular RNAs: Diversity of form and function. RNA 2014, 20, 1829–1842. [Google Scholar] [CrossRef]
- Hsu, M.-T.; Coca-Prados, M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 1979, 280, 339–340. [Google Scholar] [CrossRef] [PubMed]
- Xuan, L.; Qu, L.; Zhou, H.; Wang, P.; Yu, H.; Wu, T.; Wang, X.; Li, Q.; Tian, L.; Liu, M.; et al. Circular RNA: A novel biomarker for progressive laryngeal cancer. Am. J. Transl. Res. 2016, 8, 932–939. [Google Scholar]
- He, J.; Xie, Q.; Xu, H.; Li, J.; Li, Y. Circular RNAs and cancer. Cancer Lett. 2017, 396, 138–144. [Google Scholar] [CrossRef]
- Wang, Y.; Li, D.; Wang, Y.; Li, M.; Fang, X.; Chen, H.; Zhang, C. The landscape of circular RNAs and mRNAs in bovine milk exosomes. J. Food Compos. Anal. 2019, 76, 33–38. [Google Scholar] [CrossRef]
- Wang, J.-P.; Yang, J.; Jiao, P.; Ren, Q.-Q.; Luoreng, Z.-M.; Wang, X.-P.; Ma, Y.; Wei, D.-W. Differential expression of circRNAs related to lipopolysaccharide-induced inflammation in bovine mammary epithelial cells. Res. Vet. Sci. 2022, 146, 24–27. [Google Scholar] [CrossRef]
- Cheng, J.; Guo, J.-M.; Xiao, B.-X.; Miao, Y.; Jiang, Z.; Zhou, H.; Li, Q. piRNA, the new non-coding RNA, is aberrantly expressed in human cancer cells. Clin. Chim. Acta 2011, 412, 1621–1625. [Google Scholar] [CrossRef]
- Bamezai, S.; Rawat, V.P.; Buske, C. Concise Review: The Piwi-piRNA Axis: Pivotal Beyond Transposon Silencing. Stem Cells 2012, 30, 2603–2611. [Google Scholar] [CrossRef] [PubMed]
- Ernst, C.; Odom, D.T.; Kutter, C. The emergence of piRNAs against transposon invasion to preserve mammalian genome integrity. Nat. Commun. 2017, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef]
- Roovers, E.F.; Rosenkranz, D.; Mahdipour, M.; Han, C.-T.; He, N.; de Sousa Lopes, S.M.C.; van der Westerlaken, L.A.J.; Zischler, H.; Butter, F.; Roelen, B.A.J.; et al. Piwi Proteins and piRNAs in Mammalian Oocytes and Early Embryos. Cell Rep. 2015, 10, 2069–2082. [Google Scholar] [CrossRef] [PubMed]
- Russell, S.J.; Stalker, L.; Gilchrist, G.; Backx, A.; Molledo, G.; Foster, R.A.; LaMarre, J. Identification of PIWIL1 Isoforms and Their Expression in Bovine Testes, Oocytes, and Early Embryos1. Biol. Reprod. 2016, 94, 75. [Google Scholar] [CrossRef]
- Testroet, E.D.; Shome, S.; Reecy, J.M.; Jernigan, R.L.; Zhu, M.; Du, M.; Clark, S.; Beitz, D.C. Profiling of the Exosomal Cargo of Bovine Milk Reveals the Presence of Immune- and Growth-modulatory Non-coding RNAs (ncRNA). Iowa State Univ. Anim. Ind. Rep. 2018, 15. [Google Scholar] [CrossRef][Green Version]
- Laible, G.; Wei, J.; Wagner, S. Improving livestock for agriculture—Technological progress from random transgenesis to precision genome editing heralds a new era. Biotechnol. J. 2014, 10, 109–120. [Google Scholar] [CrossRef]
- Tizard, M.; Hallerman, E.; Fahrenkrug, S.; Newell-McGloughlin, M.; Gibson, J.; De Loos, F.; Wagner, S.; Laible, G.; Han, J.Y.; D’Occhio, M.; et al. Strategies to enable the adoption of animal biotechnology to sustainably improve global food safety and security. Transgenic Res. 2016, 25, 575–595. [Google Scholar] [CrossRef]
- West, J.; Gill, W.W. Genome Editing in Large Animals. J. Equine Vet. Sci. 2016, 41, 1–6. [Google Scholar] [CrossRef]
- Yu, S.; Luo, J.; Song, Z.; Ding, F.; Dai, Y.; Li, N. Highly efficient modification of beta-lactoglobulin (BLG) gene via zinc-finger nucleases in cattle. Cell Res. 2011, 21, 1638–1640. [Google Scholar] [CrossRef]
- Cui, C.; Song, Y.; Liu, J.; Ge, H.; Linyong, H.; Huang, H.; Hu, L.; Zhu, H.; Jin, Y.; Zhang, Y. Gene targeting by TALEN-induced homologous recombination in goats directs production of β-lactoglobulin-free, high-human lactoferrin milk. Sci. Rep. 2015, 5, 10482. [Google Scholar] [CrossRef]
- Liu, X.; Wang, Y.; Guo, W.; Chang, B.; Liu, J.; Guo, Z.; Quan, F.; Zhang, Y. Zinc-finger nickase-mediated insertion of the lysostaphin gene into the beta-casein locus in cloned cows. Nat. Commun. 2013, 4, 2565. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, Y.; Tian, Y.; Yu, Y.; Gao, M.; Hu, G.; Su, F.; Pan, S.; Luo, Y.; Guo, Z.; et al. Generation of mastitis resistance in cows by targeting human lysozyme gene to β-casein locus using zinc-finger nucleases. Proc. R. Soc. B Boil. Sci. 2014, 281, 20133368. [Google Scholar] [CrossRef] [PubMed]
- Jabed, A.; Wagner, S.; McCracken, J.; Wells, D.N.; Laible, G. Targeted microRNA expression in dairy cattle directs production of β-lactoglobulin-free, high-casein milk. Proc. Natl. Acad. Sci. USA 2012, 109, 16811–16816. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, S.; Jonas, E.; Rydhmer, L.; Röcklinsberg, H. Invited review: Breeding and ethical perspectives on genetically modified and genome edited cattle. J. Dairy Sci. 2018, 101, 12962. [Google Scholar] [CrossRef]
- Hu, G.; Do, D.N.; Gray, J.; Miar, Y. Selection for Favorable Health Traits: A Potential Approach to Cope with Diseases in Farm Animals. Animals 2020, 10, 1717. [Google Scholar] [CrossRef]

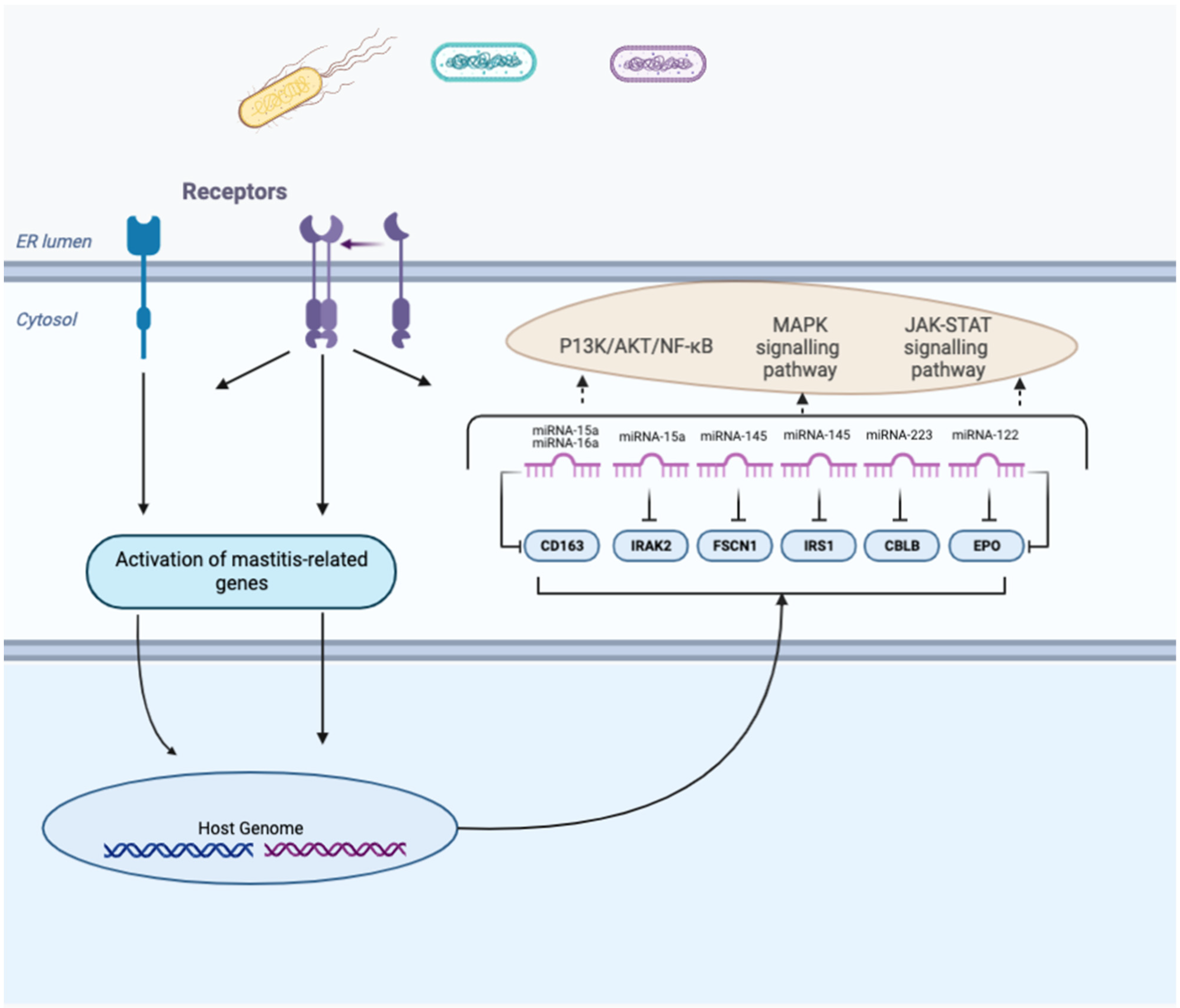
| Pathogens | Phenotypes/Tissues | Upregulated miRNAs | Downregulated miRNAs | References |
|---|---|---|---|---|
| California mastitis test positive (CMT+) | Milk | miR29b-2, miR146A, miR148s, miR155, and miR184. | miR24-2, miR181s1, and miR223 | [102] |
| California mastitis test positive (CMT+) | Milk | miR-221, miR-146a, miR-10a, miR-142-3p, miR-223, miR-21-3p, miR-6529a, miR-338, miR-2284aa, miR-15a, miR-146b, miR-142-5p, miR-30f, miR-1246, miR-147, miR-2285b, miR-2285p, miR-222, miR-2284w, miR-132, miR-130b, miR-301a, miR-505 | miR-23b-3p, miR-874 | [103] |
| California mastitis test positive (CMT+) | Milk | miR-21, miR-122, miR-125b, miR-205, miR-222, and miR-383 | miR-26b and miR-29b | [104] |
| Mycoplasma bovis | Milk | let-7a-5p, miR-100, miR-103, miR-107, miR-10a | miR-125a, miR-126-3p, miR-126-5p, miR-127, miR-1271 | [105] |
| Mycoplasma bovis | Milk | miR-21, miR-146a, miR-155, miR-222, miR-383, miR-200a, miR-205, miR-122, and miR-182 | [106] | |
| Staphylococcus aureus | Milk | miR-1, miR-122, miR-1246, miR-146b, miR-142-5p, miR-146a, miR-154b, miR-184, miR-185, miR-196b, miR-205, miR-2340, miR-2889, miR-2904, miR-378, miR-378c, miR-451, and miR-378. | miR-218, miR-2320-3p, miR-369-3p, miR-582, miR-6525 | [107] |
| Staphylococcus aureus | Milk | miR-1343-5p, miR-2407, miR-296, miR-2360, miR-2374, miR-2328-3p, miR-2412, miR-2904, miR-494, miR-2392, miR-2898 | miR-2373, miR-423-3p, miR-126, miR-19b, miR-148a, miR-21, miR-31, miR-143, miR-26a, miR-145, miR-2881, miR-26b, miR-200b, miR-99a, miR-30a-5p | [108] |
| Staphylococcus aureus | Milk | miR-103, miR-142-3p, miR-142-5p, miR-146a, miR-146b, miR-147, miR-221, miR-223, miR-2284w, miR-2285b, miR-23a | let-7b, miR-1468, miR-423-5p | [109] |
| Streptococcus uberis Coagulase Negative Staphylococcus | Milk Milk | miR-1224, miR-2385-5p, miR-2433 miR-2344 | miR-17-3p, miR-320a, miR-320b miR-1343-3p, miR-345-5p | [110] |
| Escherichia coli | Blood | miR-15a and miR-16a | - | [111] |
| Escherichia coli | MAC-T 1 | miR-365-3p, miR-184 and miR-24-3p (6 hpi 2) miR-21-3p, miR-148a, miR-92a (12 hpi) miR-423-5p and miR-21-3p (24 hpi), miR-486 (48 hpi) | miR-193a-3p, miR-30c and miR-30b-5p (6 hpi) miR-423-5p (12 hpi) let-7a-5p, miR-184, miR-un5 miR-193a-3p (48 hpi) | [112] |
| Streptococcus uberis | BMEC 3 | miR-223, mir-29e and mir-708 (2 hpi) | miR-181a, miR-16a, miR-31, | [113] |
| Streptococcus uberis | BMEC | let-7b, and miR-98 (4 hpi) miR-let-7c and miR-708 (4 hpi in normalized data) let-7b, miR-200c, miR-210, miR-24-2, miR-128-2, let-7d, miR-128-1, let-7e, miR-185, miR-652, miR-494, miR-2342 (6 hpi) | miR-29b-2, miR-193a, and miR-130a (4 hpi) miR-29b-2, miR-29c, miR-29e, and miR-100, miR-130a (6 hpi) miR-15a, miR-17, miR-26a-2, miR-29a, miR-29b-1, and miR-193a (in normalized data) | [114] |
| Streptococcus aureus | MAC-T | miR-2339 (6 hpi), miR-21-3p, miR-92a (12 hpi), miR-23a, miR-21-3p (24 hpi), miR-365-3p (48 hpi) | miR-423-5p and miR-499 (12 hpi) miR-193a-3p, miR-99b, miR-un5 (24 hpi) miR-193a-3p, miR-30c, and miR-30b-5p (48 hpi) | [112] |
| S. agalactiae | BMEC | miR-223, miR-2284k, miR-2484, miR-451, miR-383, miR-486, miR-2332, miR-122, miR-16a, miR-326 | miR-26a, miR-33a, miR-335, miR-3660, miR-146a, miR-206, miR-628, miR-450b, miR-380-p, miR-1388-3p, miR-30e-5p, miR-23b-3p, miR-378b, miR-145, miR-136, miR-135a, miR-126-5p, miR-24, miR-4286, miR-450a, miR-3431, miR-2478, miR-23a, miR-487b, miR-331-5p | [115] |
| Staphylococcus aureus | Blood | miR-486, miR-451, miR-191, miR-342, and miR-30e-5p | miR-339b and miR-25 | [116] |
| Staphylococcus aureus | Blood | miR-1301, miR-30b-5p, miR-193b, miR-320a, miR-19a, and miR-19b | miR-2284r, miR-144, miR-143, miR-205, and miR-24 | [117] |
| Escherichia coli | Blood | miR-200a, miR-205, miR-345-5p, miR-671 (1 hpi) miR-545-3p, miR-190a, let-7a-3p, miR-345-5p, miR-592, miR-324, miR-411b, miR-153, miR-331-3p, miR-144, miR-2299-5p, miR-671, miR-32, miR-30b-5p, miR-29c, miR-1246, miR-142-3p, miR-29d-5p, miR-326, miR-27a-5p, miR-19a (3 hpi) miR-200a, miR-205, miR-182 (5 hpi) miR-200a, miR-205, miR-183, miR-214, miR-182, miR-199a-5p, miR-196a, miR-455-5p, miR-96, miR-143, miR-10b, miR-122, let-7a-3p, miR-126-5p, miR-144, miR-126-3p, miR-2285h, miR-345-5p, miR-3613a, miR-200c (7 hpi) | miR-122 (1 hpi) miR-122, miR-2450a, miR-193a-5p, miR-145, miR-200b, miR-2346 (3 hpi) miR-133a, miR-193b, miR-331-3p (5 hpi) miR-133a, miR-2332, miR-1388-3p, miR-342, miR-1291 (7 hpi) | [118] |
| Streptococcus agalactiae (ST12 and ST103 strain) | Blood | miR-221, miR-628, miR-146b, miR-2285m, miR-2284i, p-miR-3 (both strains) miR-425-5p, miR-425-3p, miR-30b-5p miR-223, miR-155, miR-500, miR-374b, miR-122 miR-2438 (ST12 strain) miR-708, miR-9-5p, miR-222, miR-7858 (ST103 strain) | miR-2427, miR-1306, miR-1249, miR-2898, miR-2478 (both strains) miR-2388-5p, miR-365-3p, miR-92b, miR-2431-3p, miR-197, miR-125a, miR-128, miR-328, miR-484, miR-1343-3p, miR-340, miR-30f, miR-30d, miR-125b, miR-505, miR-2284ab, miR-423-3p, miR-361, miR-92a, miR-1468 miR-669, miR-30c, miR-10a miR-2284w (ST12 strain) miR-2892, miR-1246 (ST103 strain) | [119] |
| Pathogens | Phenotypes /Tissues | miRNAs | Target Genes | Main Consequences | References |
|---|---|---|---|---|---|
| Escherichia coli | Mammary tissues and blood neutrophils | miR-15a and miR-16a | CD163 | Decreases CD163 ability to induce the secretion of anti-inflammatory cytokines | [111] |
| Staphylococcus aureus | Mammary gland tissue | miR-15a | IRAK2 | Might reduce the negative regulatory function of IRAK2 and increase the apoptosis of mast cells | [120] |
| Staphylococcus aureus | Mac-T cells | miR-145 | FSCN1. | Translationally repress FSCN1 function; inhibit the proliferation of Mac-T cells, significantly reduce the secretion of IL-12 and TNF-α, and increase the secretion of IFN-γ | [121] |
| BMEC | miR-145 | IRS1 | Post-transcriptionally regulate IRS1 expression and decrease the proliferation of mammary epithelial cell through the MAPK signaling pathway | [122] | |
| Staphylococcus aureus | Milk | miR-223 | CBLB | Reduce LTA-stimulated inflammation in Mac-T cells by targeting CBLB and the PI3K/AKT/NF-κB downstream pathway | [123] |
| Streptococcus agalactiae | BMEC | miR-122 | EPO | Regulates the JAK-STAT signaling pathway by downregulating EPO in the mammary gland | [124] |
| miR-375 knockdown disease condition | BMEC | miR-375 | NR4A1/ PTPN5 | NR4A1 is an important mediator in early inflammation that upregulates IκBα expression but inhibits NF-κB activation; PTPN5 negatively regulates the activity and localization of MAPK family members | [125] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oyelami, F.O.; Usman, T.; Suravajhala, P.; Ali, N.; Do, D.N. Emerging Roles of Noncoding RNAs in Bovine Mastitis Diseases. Pathogens 2022, 11, 1009. https://doi.org/10.3390/pathogens11091009
Oyelami FO, Usman T, Suravajhala P, Ali N, Do DN. Emerging Roles of Noncoding RNAs in Bovine Mastitis Diseases. Pathogens. 2022; 11(9):1009. https://doi.org/10.3390/pathogens11091009
Chicago/Turabian StyleOyelami, Favour Oluwapelumi, Tahir Usman, Prashanth Suravajhala, Nawab Ali, and Duy N. Do. 2022. "Emerging Roles of Noncoding RNAs in Bovine Mastitis Diseases" Pathogens 11, no. 9: 1009. https://doi.org/10.3390/pathogens11091009
APA StyleOyelami, F. O., Usman, T., Suravajhala, P., Ali, N., & Do, D. N. (2022). Emerging Roles of Noncoding RNAs in Bovine Mastitis Diseases. Pathogens, 11(9), 1009. https://doi.org/10.3390/pathogens11091009