Genome-Wide Association Study for Haemonchus contortus Resistance in Morada Nova Sheep
Abstract
:1. Introduction
2. Results
- -
- Immune response: BTLA (1:175586573–175619634), CD96 (1:174690709–174790572), CD200 (1:175479877–175498685), CFI (6:15708801–15753015), DPP4 (2:146632572–146713094), FER (5:105044035–105461672), LEF1 (6:17197574–17304587), NFE2L2 (2:131754249–131759072), and the TREML1, TREM1, and TREM2 gene cluster (20:15158837–15237353);
- -
- Gastric mucosa and mucins: GALNT10 (5:63016723–63244040), GCNT3 (7:47666417–47667739), and PGC (20:15636993–15649730);
- -
- Hematological parameters: CLEC14A (18:47277443–47278737), DLX1 (2:136599373–136602848), EDNRA (17:10416386–10482226), and SH2B3 (17:54732694–54757389);
- -
- Homeostasis: CCDC80 (1:175729335–175762809), HS3ST5 (8:23301492–23469323), and PLA2G1B (17:62282828–62288877);
- -
- Growth and muscle development: DPPA2 (1:172525876–172536184), HOXD gene cluster (2:132820016–132915560), KLHL41 (2:138842626–138856360), MYF5 (3:116620277–116622399), and PITX2 (6:14934427–14955991);
- -
- Lipid metabolism and fat deposition: MAPK7 (11:33629196–33632861) and SREBF1 (11:34176887–34191779).
3. Discussion
4. Materials and Methods
4.1. Parasitological Tests and Phenotypic Classification of Animals
4.2. Genome-Wide Association Study (GWAS)
4.3. Bioinformatics and Functional Annotation
4.4. GWAS Validation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bueno, M.S.; Cunha, E.A.; Santos, L.E. Morada Nova: Uma Raça Com Potencial Para Produção. 2010. Available online: http://www.iz.sp.gov.br/artigo.php?id=46 (accessed on 8 July 2022).
- McManus, C.; Facó, O.; Shiotsuki, L.; Rolo, J.L.J.P.; Peripolli, V. Pedigree analysis of Brazilian Morada Nova hair sheep. Small Rumin. Res. 2019, 170, 37–42. [Google Scholar] [CrossRef]
- Issakowicz, J.; Issakowicz, A.C.K.S.; Bueno, M.S.; da Costa, R.L.D.; Katiki, L.M.; Geraldo, A.T.; Abdalla, A.L.; McManus, C.; Louvandini, H. Parasitic infection, reproductive and productive performance from Santa Inês and Morada Nova ewes. Small Rumin. Res. 2016, 136, 96–103. [Google Scholar] [CrossRef]
- Amarante, A.F.T.; Bricarello, P.A.; Rocha, R.A.; Gennari, S.M. Resistance of Santa Inês, Suffolk and Ile de France sheep to naturally acquired gastrointestinal nematode infections. Vet. Parasitol. 2004, 120, 91–106. [Google Scholar] [CrossRef]
- Amarante, A.F.T.; Sales, R.O. Control of endoparasitoses of sheep: A revision. Rev. Bras. Hig. San. Anim. 2007, 1, 14–36. [Google Scholar] [CrossRef]
- Oliveira, P.A.; Ruas, J.L.; Riet-Correa, F.; Coelho, A.C.B.; Santos, B.L.; Marcolongo-Pereira, C.; Sallis, E.S.V.; Schild, A.L. Doenças parasitárias em bovinos e ovinos no sul do Brasil: Frequência e estimativa de perdas econômicas. Pesq. Vet. Bras. 2017, 37, 797–801. [Google Scholar] [CrossRef]
- Zvinorova, P.I.; Halimani, T.E.; Muchadeyi, F.C.; Matika, O.; Riggio, V.; Dzama, K. Breeding for resistance to gastrointestinal nematodes–the potential in low-input/output small ruminant production systems. Vet. Parasitol. 2016, 225, 19–28. [Google Scholar] [CrossRef]
- Bishop, S.C.; Stear, M.J. Modeling of host genetics and resistance to infectious diseases: Understanding and controlling nematode infections. Vet. Parasitol. 2003, 115, 147–166. [Google Scholar] [CrossRef]
- Estrada-Reyes, Z.M.; Rae, D.O.; Mateescu, R.G. Genome-wide scan reveals important additive and non-additive genetic effects associated with resistance to Haemonchus contortus in Florida Native sheep. Int. J. Parasitol. 2021, 51, 535–543. [Google Scholar] [CrossRef]
- Haehling, M.B.; Cruvinel, G.G.; Toscano, J.H.B.; Giraldelo, L.A.; Santos, I.B.; Esteves, S.N.; Benavides, M.V.; Barioni Júnior, W.; Niciura, S.C.M.; Chagas, A.C.S. Four single nucleotide polymorphisms (SNPs) are associated with resistance and resilience to Haemonchus contortus in Brazilian Morada Nova sheep. Vet. Parasitol. 2020, 279, 109053. [Google Scholar] [CrossRef]
- Okino, C.H.; Niciura, S.C.M.; Toscano, J.H.B.; Esteves, S.N.; Santos, I.B.; Von Haehling, M.B.; Figueiredo, A.; Oliveira, M.C.S.; Chagas, A.C.S. Ovine β-globin gene: A new qPCR for rapid haplotype identification and association with susceptibility to Haemonchus contortus infection. Vet. Parasitol. 2021, 294, 109434. [Google Scholar] [CrossRef]
- Kalaldeh, M.A.; Gibson, J.; Lee, S.H.; Gondro, C.; Van Der Werf, J.H.J. Detection of genomic regions underlying resistance to gastrointestinal parasites in Australian sheep. Genet. Sel. Evol. 2019, 51, 37. [Google Scholar] [CrossRef] [PubMed]
- Ahbara, A.M.; Rouati, M.; Gharbi, M.; Rekik, M.; Haile, A.; Rischkowsky, B.; Mwacharo, J.M. Genome-wide insights on gastrointestinal nematode resistance in autochthonous Tunisian sheep. Sci. Rep. 2021, 11, 9250. [Google Scholar] [CrossRef] [PubMed]
- Álvarez, I.; Fernández, I.; Soudré, A.; Traoré, A.; Pérez-Pardal, L.; Sanou, M.; Tapsoba, S.A.R.; Menéndez-Arias, N.A.; Goyache, F. Identification of genomic regions and candidate genes of functional importance for gastrointestinal parasite resistance traits in Djallonké sheep of Burkina Faso. Arch. Anim. Breed. 2019, 62, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Goyache, F.; Pérez-Pardal, L.; Fernández, I.; Traoré, A.; Menéndez-Arias, N.A.; Álvarez, I. Ancient autozygous segments subject to positive selection suggest adaptative immune responses in West African cattle. Gene 2021, 803, 145899. [Google Scholar] [CrossRef]
- Arisan, E.D.; Dart, A.; Grant, G.H.; Arisan, S.; Cuhadaroglu, S.; Lange, S.; Uysal-Onganer, P. The prediction of miRNAs in SARS-CoV-2 genomes: Hsa-miR databases identify 7 key miRs linked to host responses and virus pathogenicity-related KEGG pathways significant for comorbidities. Viruses 2020, 12, 614. [Google Scholar] [CrossRef] [PubMed]
- Xiang, H.; Fang, Y.; Tan, Z.; Zhong, R. Haemonchus contortus infection alters gastrointestinal microbial community composition, protein digestion and aminoacid allocations in lambs. Front. Microbiol. 2022, 12, 797746. [Google Scholar] [CrossRef]
- Rainbird, M.A.; Macmillan, D.; Meeusen, E.N. Eosinophil-mediated killing of Haemonchus contortus larvae: Effect of eosinophil activation and role of antibody, complement and interleukin-5. Parasite Immunol. 1998, 20, 93–103. [Google Scholar] [CrossRef]
- Naeem, M.; Iqbal, Z.; Roohi, N. Ovine haemonchosis: A review. Trop. Anim. Health Prod. 2021, 53, 19. [Google Scholar] [CrossRef]
- Bishop, S.C. A consideration of resistance and tolerance for ruminant nematode infections. Front. Genet. 2012, 3, 168. [Google Scholar] [CrossRef]
- Jacobs, J.R.; Middleton, D.; Greiner, S.P.; Bowdridge, S.A. RNA-Sequencing of ovine PBMC after exposure to Haemonchus contortus larval antigen. Parasite Immunol. 2020, 42, e12697. [Google Scholar] [CrossRef]
- Breloer, M.; Hartmann, W.; Blankenhaus, B.; Eschbach, M.L.; Pfeffer, K.; Jacobs, T. Cutting edge: The BTLA-HVEM regulatory pathway interferes with protective immunity to intestinal helminth infection. J. Immunol. 2015, 194, 1413–1416. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; González, J.F.; Hernandez, J.N.; McNeilly, T.N.; Corripio-Miyar, Y.; Frew, D.; Morrison, T.; Yu, P.; Li, R.W. Possible mechanisms of host resistance to Haemonchus contortus infection in sheep breeds native to the Canary Islands. Sci. Rep. 2016, 6, 26200. [Google Scholar] [CrossRef] [PubMed]
- Toscano, J.H.B.; Okino, C.H.; Santos, I.B.; Giraldelo, L.A.; Haehling, M.B.; Esteves, S.N.; Chagas, A.C.S. Innate immune responses associated with resistance against Haemonchus contortus in Morada Nova sheep. J. Immunol. Res. 2019, 2019, 3562672. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Liu, F.; Hunt, P.; Li, C.; Zhang, L.; Ingham, A.; Li, R.W. Transcriptome analysis unrevealed potential mechanisms of resistance to Haemonchus contortus infection in Merino sheep populations bred for parasite resistance. Vet. Res. 2019, 50, 7. [Google Scholar] [CrossRef] [PubMed]
- Estrada-Reyes, Z.M.; Rae, O.; Postley, C.; Medrano, M.B.J.; Gutiérrez, J.D.L.; Mateescu, R.G. Association study reveals Th17, Treg, and Th2 loci related to resistance to Haemonchus contortus in Florida Native sheep. J. Anim. Sci. 2019, 97, 4428–4444. [Google Scholar] [CrossRef]
- Bhuiyan, A.A.; Li, J.; Wu, Z.; Ni, P.; Adetula, A.A.; Wang, H.; Zhang, C.; Tang, X.; Bhuyan, A.A.; Zhao, S.; et al. Exploring the genetic resistance to gastrointestinal nematodes infection in goat using RNA-sequencing. Int. J. Mol. Sci 2017, 18, 751. [Google Scholar] [CrossRef]
- Araujo, R.N.; Padilha, T.; Zarlenga, D.; Sonstegard, T.; Connor, E.E.; Van Tassel, C.; Lima, W.S.; Nascimento, E.; Gasbarre, L.C. Use of a candidate gene array to delineate gene expression patterns in cattle selected for resistance or susceptibility to intestinal nematodes. Vet. Parasitol. 2009, 162, 106–115. [Google Scholar] [CrossRef]
- Rawat, A.K.; Pal, K.; Singh, R.; Anand, A.; Gupta, S.; Kishore, D.; Singh, S.; Singh, R.K. The CD200-CD200R cross-talk helps Leishmania donovani to down regulate macrophage and CD4+ CD44+ T cells effector functions in an NFκB independent manner. Int. J. Biol. Macromol. 2020, 151, 394–401. [Google Scholar] [CrossRef]
- Menezes, J.P.B.; Khouri, R.; Oliveira, C.V.S.; Petersen, A.L.O.A.; Almeida, T.F.; Mendes, F.R.L.; Rebouças, A.A.D.; Lorentz, A.L.; Luz, N.F.; Lima, J.B.; et al. Proteomic analysis reveals a predominant NFE2L2 (NRF2) signature in canonical pathway and upstream regulator analysis of Leishmania-infected macrophages. Front. Immunol. 2019, 10, 1362. [Google Scholar] [CrossRef]
- Otto, P.I.; Guimarães, S.E.F.; Verardo, L.L.; Azevedo, A.L.S.; Vandenplas, J.; Soares, A.C.C.; Sevillano, C.A.; Veroneze, R.; Pires, M.F.A.; Freitas, C.; et al. Genome-wide association studies for tick resistance in Bos taurus × Bos indicus crossbred cattle: A deeper look into this intricate mechanism. J. Dairy Sci. 2018, 101, 11020–11032. [Google Scholar] [CrossRef]
- Jessica, M.O.; Fiorella, R.; Ocatavio, S.; Linnette, R.; Nahomy, L.; Kanth, M.B.; Bismarck, M.; Rondina, M.T.; Valance, W.A. TLT-1-controls early thrombus formation and stability by facilitating AIIBB3 outside-in signaling in mice. Int. J. Adv. Res. 2018, 6, 1143–1149. [Google Scholar] [CrossRef] [PubMed]
- Schmoker, A.M.; Pearson, L.M.P.; Cruz, C.; Flores, L.G.C.; Branfeild, S.; Torres, F.D.P.; Fonseca, K.; Cantres, Y.M.; Ramirez, C.A.S.; Melendez, L.M.; et al. Defining the TLT-1 interactome from resting and activated human platelets. J. Proteomics 2020, 215, 103638. [Google Scholar] [CrossRef]
- Niedziela, D.A.; Naranjo-Lucena, A.; Molina-Hernández, V.; Browne, J.A.; Martínez-Moreno, A.; Pérez, J.; MacHugh, D.E.; Mulcahy, G. Timing of transcriptomic peripheral blood mononuclear cell responses of sheep to Fasciola hepatica infection differs from those of cattle, reflecting different disease phenotypes. Front. Immunol. 2021, 12, 729217. [Google Scholar] [CrossRef] [PubMed]
- Qi, W.; Ebbert, K.V.J.; Craig, A.W.B.; Greer, P.A.; McCafferty, D.M. Absence of Fer protein tyrosine kinase exacerbates endotoxin induced intestinal epithelial barrier dysfunction in vivo. Gut 2005, 54, 1091–1097. [Google Scholar] [CrossRef] [PubMed]
- Vergara-Alert, J.; van den Brand, J.M.; Widagdo, W.; Muñoz, M.; Raj, S.; Schipper, D.; Solanes, D.; Cordón, I.; Bensaid, A.; Haagmans, B.L.; et al. Livestock susceptibility to infection with middle east respiratory syndrome coronavirus. Emerg. Infect. Dis. 2017, 23, 232–240. [Google Scholar] [CrossRef]
- Wei, C.; Wang, H.; Liu, G.; Zhao, F.; Kijas, J.W.; Ma, Y.; Lu, J.; Zhang, L.; Cao, J.; Wu, M.; et al. Genome-wide analysis reveals adaptation to high altitudes in Tibetan sheep. Sci. Rep. 2016, 6, 26770. [Google Scholar] [CrossRef]
- Li, R.W.; Gasbarre, L.C. A temporal shift in regulatory networks and pathways in the bovine small intestine during Cooperia oncophora infection. Int. J. Parasitol. 2009, 39, 813–824. [Google Scholar] [CrossRef]
- Rinaldi, M.; Dreesen, L.; Hoorens, P.R.; Li, R.W.; Claerebout, E.; Goddeeris, B.; Vercruysse, J.; Broek, W.V.D.; Geldhof, P. Infection with the gastrointestinal nematode Ostertagia ostertagi in cattle affects mucus biosynthesis in the abomasum. Vet. Res. 2011, 42, 61. [Google Scholar] [CrossRef]
- Chiba, S.; Takeshita, K.; Imai, Y.; Kumano, K.; Kurokawa, M.; Masuda, S.; Shimizu, K.; Nakamura, S.; Ruddle, F.H.; Hirai, H. Homeoprotein DLX-1 interacts with Smad4 and blocks a signaling pathway from activin A in hematopoietic cells. Proc. Natl. Acad. Sci. USA 2003, 100, 15577–15582. [Google Scholar] [CrossRef]
- Ma, X.; Jia, C.; Fu, D.; Chu, M.; Ding, X.; Wu, X.; Guo, X.; Pei, J.; Bao, P.; Liang, C.; et al. Analysis of hematological traits in polled yak by genome-wide association studies using individual SNPs and haplotypes. Genes 2019, 10, 463. [Google Scholar] [CrossRef]
- Rho, S.S.; Choi, H.J.; Min, J.K.; Lee, H.W.; Park, H.; Park, H.; Kim, Y.M.; Kwon, Y.G. Clec14a is specifically expressed in endothelial cells and mediates cell to cell adhesion. Biochem. Biophys. Res. Commun. 2011, 404, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Guo, T.; Zhao, H.; Qiao, G.; Han, M.; Liu, J.; Yuan, C.; Wang, T.; Li, F.; Yue, Y.; et al. Genome-wide association study using individual single-nucleotide polymorphisms and haplotypes for erythrocyte traits in Alpine Merino sheep. Front. Genet. 2020, 11, 848. [Google Scholar] [CrossRef] [PubMed]
- Entwistle, L.J.; Pelly, V.S.; Coomes, S.M.; Kannan, Y.; Perez-Lloret, J.; Czieso, S.; Santos, M.S.; MacRae, J.I.; Collinson, L.; Sesay, A.; et al. Epithelial-cell-derived phospholipase A2 group 1B is an endogenous anthelmintic. Cell Host Microbe 2017, 22, 484–493. [Google Scholar] [CrossRef] [PubMed]
- Calvet, C.M.; Toma, L.; Souza, F.R.; Meirelles, M.N.S.L.; Pereira, M.C.S. Heparan sulfate proteoglycans mediate the invasion of cardiomyocytes by Trypanosoma cruzi. J. Eukaryot. Microbiol. 2003, 50, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Bishop, J.R.; Crawford, B.E.; Esko, J.D. Cell surface heparin sulfate promotes replication of Toxoplasma gondii. Infect. Immun. 2005, 73, 5395–5401. [Google Scholar] [CrossRef]
- Goldenberg, R.C.S.; Iacobas, D.A.; Iacobas, S.; Rocha, L.L.; Fortes, F.S.A.; Vairo, L.; Nagjyothi, F.; Carvalho, A.C.C.; Tanowitz, H.B.; Spray, D.C. Transcriptomic alterations in Trypanosoma cruzi-infected cardiac myocytes. Microbes Infect. 2009, 11, 1140–1149. [Google Scholar] [CrossRef]
- Doyle, J.L.; Berry, D.P.; Veerkamp, R.F.; Carthy, T.R.; Evans, R.D.; Walsh, S.W.; Purfield, D.C. Genomic regions associated with muscularity in beef cattle differ in five contrasting cattle breeds. Genet. Sel. Evol. 2020, 52, 2. [Google Scholar] [CrossRef]
- Zhao, H.; He, S.; Wang, S.; Zhu, Y.; Xu, H.; Luo, R.; Lan, X.; Cai, Y.; Sun, X. Two new insertion/deletion variants of the PITX2 gene and their effects on growth traits in sheep. Anim. Biotechnol. 2018, 29, 276–282. [Google Scholar] [CrossRef]
- Zhang, S.; Xu, H.; Kang, Z.; Cai, H.; Dang, R.; Lei, C.; Chen, H.; Guo, X.; Lan, X. Bovine pituitary homeobox 2 (PITX2): mRNA expression profiles of different alternatively spliced variants and association analyses with growth traits. Gene 2018, 669, 1–7. [Google Scholar] [CrossRef]
- Wang, L.; Zhao, Y.; Bao, X.; Zhu, X.; Kwok, Y.K.; Sun, K.; Chen, X.; Huang, Y.; Jauch, R.; Esteban, M.A.; et al. LncRNA Dum interacts with Dnmts to regulate Dppa2 expression during myogenic differentiation and muscle regeneration. Cell Res. 2015, 25, 335–350. [Google Scholar] [CrossRef]
- Pak, J.H. KLHL41 in Skeletal Muscle Development. Master’s Thesis, Boston University, Boston, MA, USA, 2019. Available online: https://open.bu.edu/handle/2144/36709 (accessed on 8 July 2022).
- Zhang, Z.; Liu, C.; Hao, W.; Yin, W.; Ai, S.; Zhao, Y.; Duan, Z. Novel single nucleotide polymorphisms and haplotype of MYF5 gene are associated with body measurements and ultrasound traits in Grassland Short-Tailed sheep. Genes 2022, 13, 483. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Wu, Y.; Wang, J.; Chen, J.; Huang, Y.; Rao, J.; Feng, C. MicroRNA-24 promotes 3T3-L2 adipocyte differentiation by directly targeting the MAPK7 signaling. Biochem. Biophys. Res. Commun. 2016, 474, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Qiao, L.; Han, Y.; Liu, J.; Zhang, J.; Liu, W. Regulatory roles of SREBF1 and SRBF2 in lipid metabolism and deposition in two Chinese representative fat-tailed sheep breeds. Animals 2020, 10, 1317. [Google Scholar] [CrossRef] [PubMed]
- Toscano, J.H.B.; Santos, I.B.; Haehling, M.B.; Giraldelo, L.A.; Lopes, L.G.; Silva, M.H.; Figueredo, A.; Esteves, S.N.; Chagas, A.C.S. Morada Nova sheep breed: Resistant or resilient to Haemonchus contortus infection? Vet. Parasitol. X 2019, 2, 100019. [Google Scholar] [CrossRef]
- Ueno, H.; Gonçalves, P.C. Manual para Diagnóstico das Helmintoses de Ruminantes, 4th ed.; Japan International Cooperation Agency: Tokyo, Japan, 1998; pp. 14–45. [Google Scholar]
- Echevarria, F.M.A.; Armour, J.; Duncan, J.L. Efficacy of some anthelmintics on an ivermectin-resistant strain of Haemonchus contortus in sheep. Vet. Parasitol. 1991, 39, 279–284. [Google Scholar] [CrossRef]
- Niciura, S.C.M.; Cruvinel, G.G.; Moraes, C.V.; Bressani, F.A.; Malagó Junior, W.; Benavides, M.V.; Chagas, A.C.S. PCR-based genotyping of SNP markers in sheep. Mol. Biol. Rep. 2018, 45, 651–656. [Google Scholar] [CrossRef]
- Benavides, M.V.; Souza, C.J.H.; Moraes, J.C.F. How efficiently genome-wide association studies (GWAS) identify prolificity-determining genes in sheep. Genet. Mol. Res. 2018, 17, gmr16039909. [Google Scholar] [CrossRef]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef]
- Berton, M.P.; Oliveira Silva, R.M.; Peripolli, E.; Stafuzza, N.B.; Martin, J.F.; Álvarez, M.S.; Gavinã, B.V.; Toro, M.A.; Banchero, G.; Oliveira, P.S.; et al. Genomic regions and pathways associated with gastrointestinal parasites resistance in Santa Inês breed adapted to tropical climate. J. Anim. Sci. Biotechnol. 2017, 8, 73. [Google Scholar] [CrossRef]
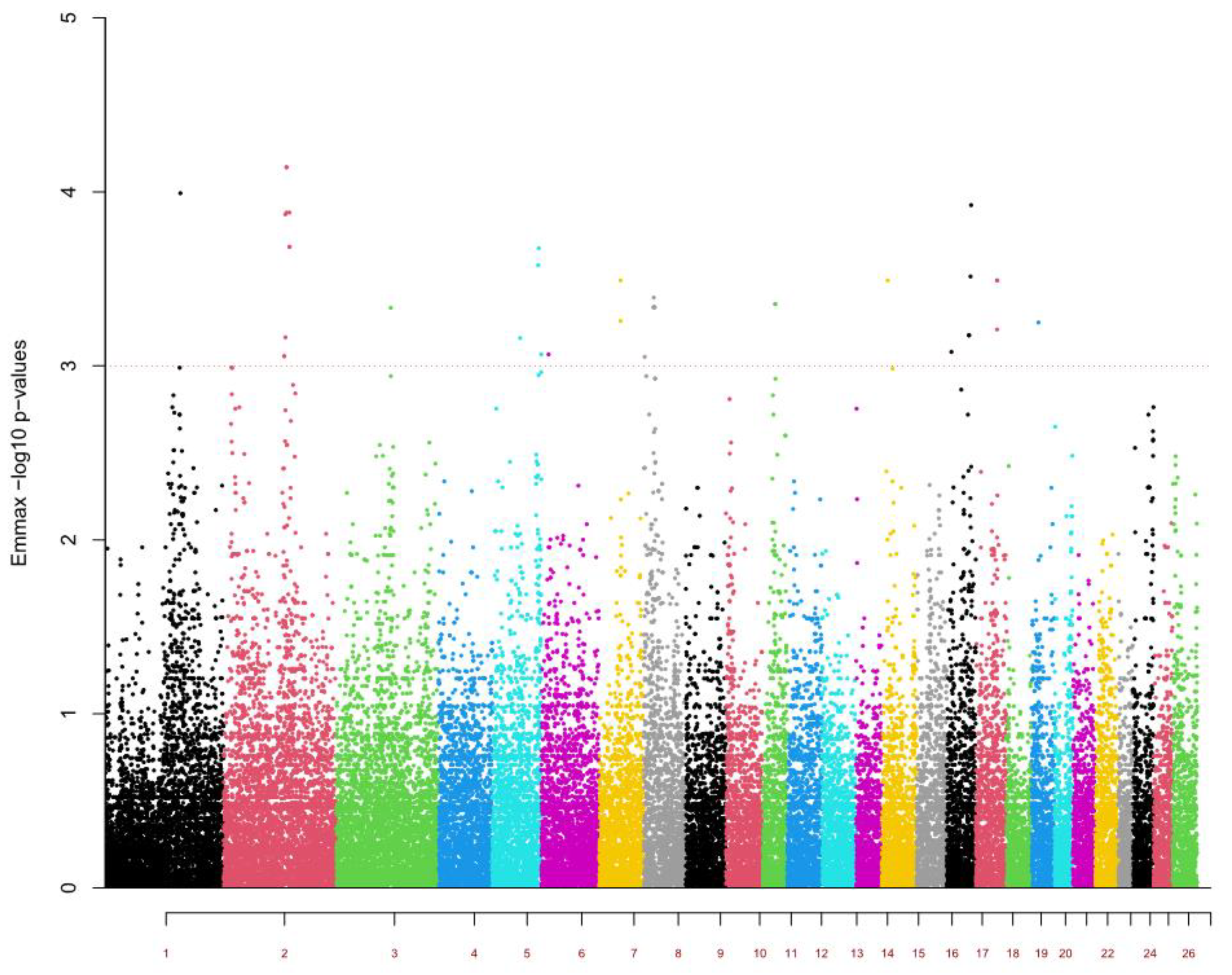

| OAR | Order | p-Value | Illumina SNP ID | SNP ID 1 | Position (bp) | Candidate Protein-Coding Genes 2 | QTL |
|---|---|---|---|---|---|---|---|
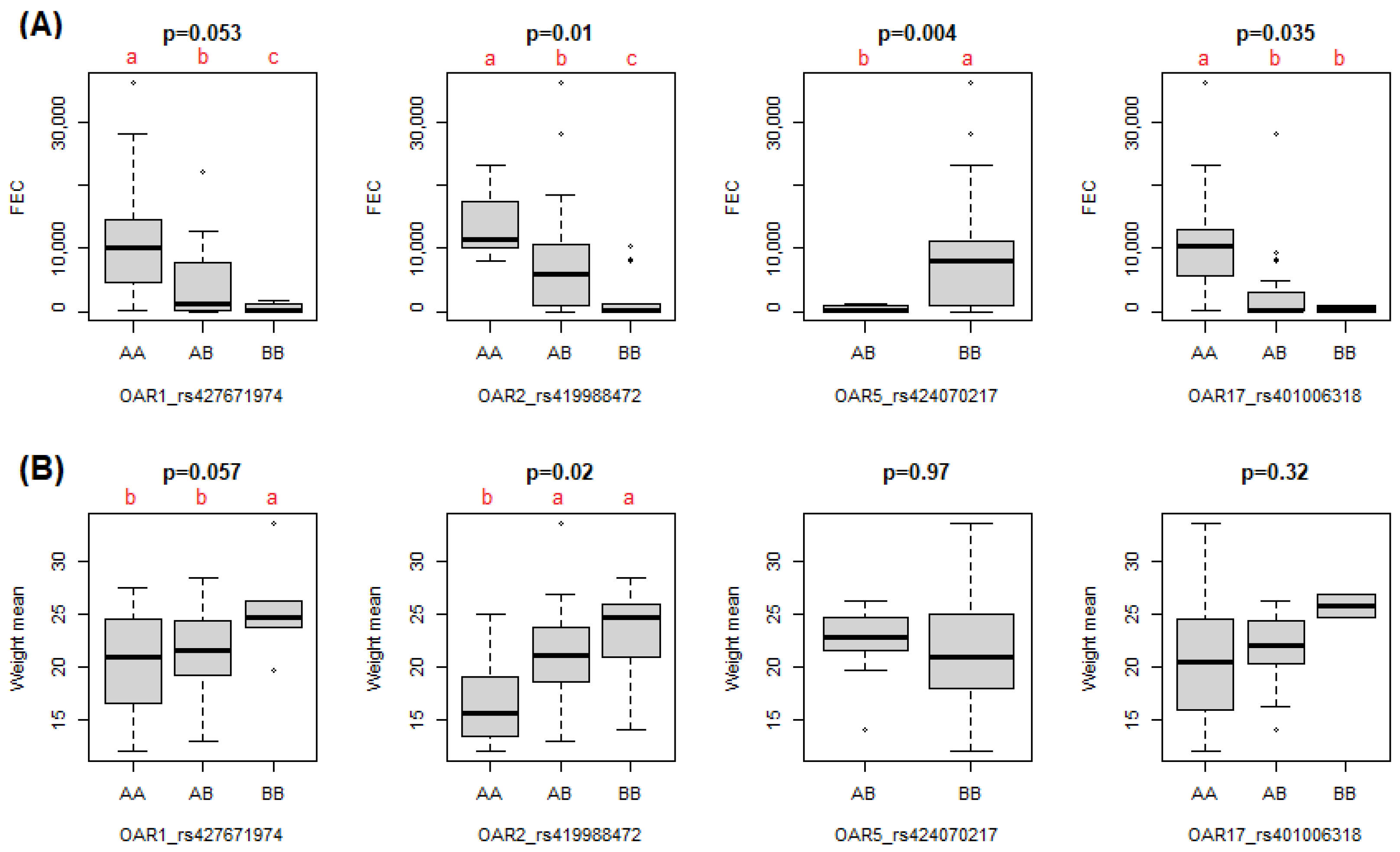
| 1 | 3 | 1.02 × 10−4 | OAR1_187356862.1 | rs427671974 | 173891491 | DPPA2, CD96, CD200, BTLA, CCDC80 | Weight and fat |
| 2 | 1 | 7.22 × 10−5 | s00855.1 | rs409592801 | 139163515 | KLHL41 | Weight |
| 2 | 7.22 × 10−5 | ilmnseq_rs422452493 | rs422452493 | 139188127 | |||
| 5 | 1.32 × 10−4 | ilmnseq_rs419988472 | rs419988472 | 139195167 | |||
| 6 | 1.32 × 10−4 | OAR2_154205756.1 | rs412327523 | 145234800 | DPP4 | ||
| 8 | 2.07 × 10−4 | OAR2_154280201.1 | rs406150872 | 145308492 | |||
| 9 | 2.07 × 10−4 | OAR2_154294235.1 | rs417376212 | 145336020 | |||
| 7 | 1.35 × 10−4 | OAR2_144774839.1 | rs424565808 | 136145355 | NFE2L2, HOXD1, HOXD3, HOXD10, HOXD12, HOXD13, DLX1 | Weight, Trichostrongylus spp. adults and larvae in the abomasum, and Nematodirus FEC | |
| 30 | 6.85 × 10−4 | OAR2_145012995.1 | rs430008551 | 136362050 | |||
| 35 | 8.79 × 10−4 | OAR2_142077352.1 | rs401620358 | 133594324 | |||
| 36 | 8.79 × 10−4 | OAR2_142085758.1 | rs412805133 | 133602325 | |||
| 3 | 24 | 4.63 × 10−4 | OAR3_124041988.1 | rs403393991 | 116331730 | MYF5 | Fat and T. colubriformis FEC |
| 5 | 10 | 2.11 × 10−4 | OAR5_110624576.1 | rs424070217 | 101668310 | - | Weight |
| 11 | 2.64 × 10−4 | OAR5_109379116.1 | rs413371484 | 100428672 | |||
| 31 | 6.92 × 10−4 | s08523.1 | rs411511506 | 63148146 | GALNT10 | ||
| 33 | 8.59 × 10−4 | OAR5_116227818.1 | rs398223820 | 106805224 | FER | - | |
| 6 | 34 | 8.59 × 10−4 | OAR6_19652340.1 | rs421701377 | 16732227 | PITX2, CFI, LEF1 | Weight and fat |
| 7 | 15 | 3.26 × 10−4 | s26745.1 | rs401054470 | 46306835 | GCNT3 | H. contortus FEC |
| 25 | 5.52 × 10−4 | OAR7_50865088.1 | rs413854960 | 46138713 | |||
| 8 | 17 | 4.05 × 10−4 | OAR8_22842614.1 | rs419418467 | 20206310 | HS3ST5 | Fat and Trichostrongylus spp. adults and larvae in the small intestine and abomasum |
| 20 | 4.60 × 10−4 | s58289.1 | rs416090516 | 19586041 | |||
| 21 | 4.60 × 10−4 | OAR8_24000338.1 | rs410048009 | 21552710 | |||
| 22 | 4.60 × 10−4 | OAR8_24026504.1 | rs421189130 | 21578631 | |||
| 23 | 4.60 × 10−4 | OAR8_25075555.1 | rs418914462 | 22667003 | |||
| 37 | 8.87 × 10−4 | OAR8_2336316_X.1 | rs402371066 | 2115027 | - | ||
| 11 | 18 | 4.41 × 10−4 | OAR11_34348656.1 | rs410744616 | 32138419 | MAPK7, SREBF1 | Weight, fat, and Trichostrongylus spp. adults and larvae in the small intestine |
| 19 | 4.41 × 10−4 | OAR11_34401498.1 | rs404901308 | 32191958 | |||
| 15 | 13 | 3.22 × 10−4 | OAR15_15781330_X.1 | rs412682230 | 15625639 | - | - |
| 17 | 4 | 1.19 × 10−4 | s46114.1 | rs401006318 | 60852961 | PLA2G1B | - |
| 12 | 3.06 × 10−4 | OAR17_64517692.1 | rs425080766 | 59101448 | |||
| 28 | 6.67 × 10−4 | OAR17_60253864.1 | rs399621490 | 55225820 | SH2B3 | ||
| 29 | 6.67 × 10−4 | OAR17_60300593.1 | rs410780866 | 55270864 | |||
| 32 | 8.31 × 10−4 | OAR17_12647550.1 | rs422538638 | 11361784 | EDNRA | FEC | |
| 18 | 14 | 3.23 × 10−4 | OAR18_51930638_X.1 | rs416293834 | 48707173 | CLEC14A | Weight and H. contortus FEC |
| 16 | 3.23 × 10−4 | OAR18_52089434.1 | rs428856771 | 48867859 | |||
| 27 | 6.17 × 10−4 | s40949.1 | rs403982333 | 48663303 | |||
| 20 | 26 | 5.63 × 10−4 | s01331.1 | rs406291711 | 14815776 | TREML1, TREM2, TREM1, PGC | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niciura, S.C.M.; Benavides, M.V.; Okino, C.H.; Ibelli, A.M.G.; Minho, A.P.; Esteves, S.N.; Chagas, A.C.d.S. Genome-Wide Association Study for Haemonchus contortus Resistance in Morada Nova Sheep. Pathogens 2022, 11, 939. https://doi.org/10.3390/pathogens11080939
Niciura SCM, Benavides MV, Okino CH, Ibelli AMG, Minho AP, Esteves SN, Chagas ACdS. Genome-Wide Association Study for Haemonchus contortus Resistance in Morada Nova Sheep. Pathogens. 2022; 11(8):939. https://doi.org/10.3390/pathogens11080939
Chicago/Turabian StyleNiciura, Simone Cristina Méo, Magda Vieira Benavides, Cintia Hiromi Okino, Adriana Mercia Guaratini Ibelli, Alessandro Pelegrine Minho, Sergio Novita Esteves, and Ana Carolina de Souza Chagas. 2022. "Genome-Wide Association Study for Haemonchus contortus Resistance in Morada Nova Sheep" Pathogens 11, no. 8: 939. https://doi.org/10.3390/pathogens11080939
APA StyleNiciura, S. C. M., Benavides, M. V., Okino, C. H., Ibelli, A. M. G., Minho, A. P., Esteves, S. N., & Chagas, A. C. d. S. (2022). Genome-Wide Association Study for Haemonchus contortus Resistance in Morada Nova Sheep. Pathogens, 11(8), 939. https://doi.org/10.3390/pathogens11080939







