Helicobacter pylori CagA Protein Regulating the Biological Characteristics of Gastric Cancer through the miR-155-5p/SMAD2/SP1 axis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Construction of CagA Eukaryotic Expression Plasmid
2.2. Cell Culture and Transfection
2.3. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.4. Western Blot
2.5. RNA Immunoprecipitation (RIP)
2.6. CCK8 Assay
2.7. Wound Healing Assay
2.8. Transwell Assays
2.9. Statistical Analysis
3. Results
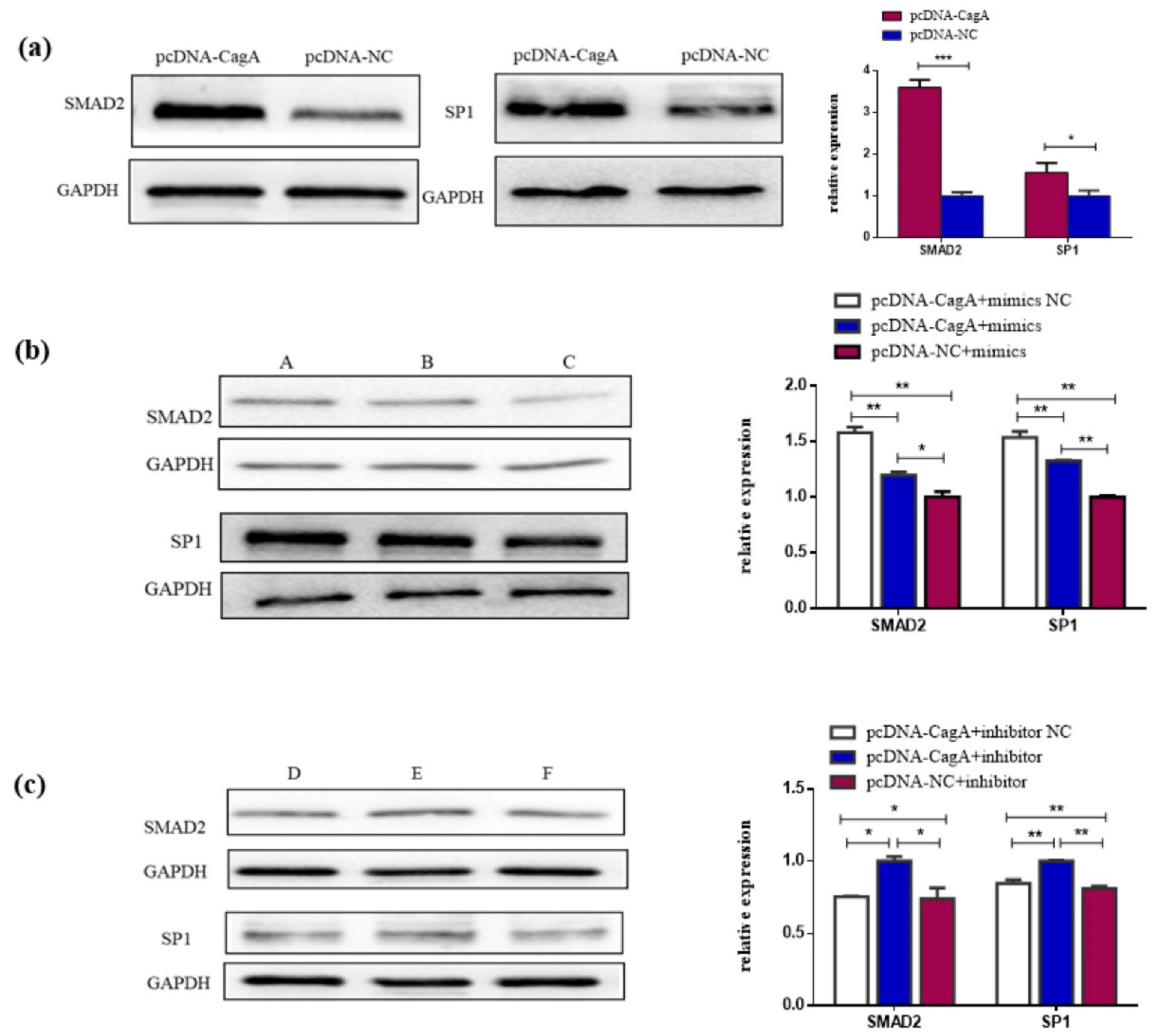
3.1. CagA Promoted the Proliferation, Migration, and Invasion of GC Cells
3.2. miR-155-Inhibited the Proliferation, Migration, and Invasion of GC Cells
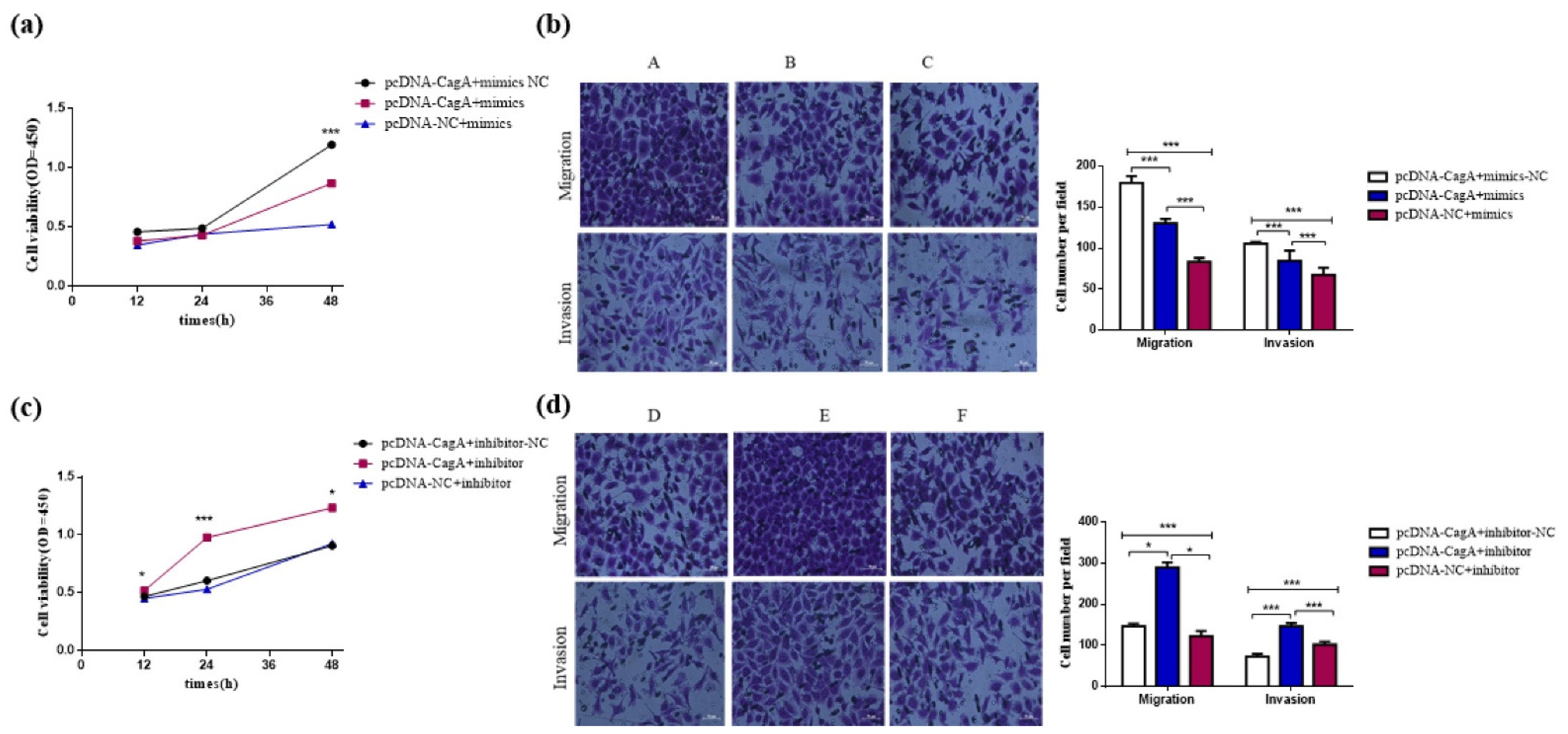
3.3. CagA Promoted the Proliferation, Migration, and Invasion of GC Cells by Regulating miR-155-5p
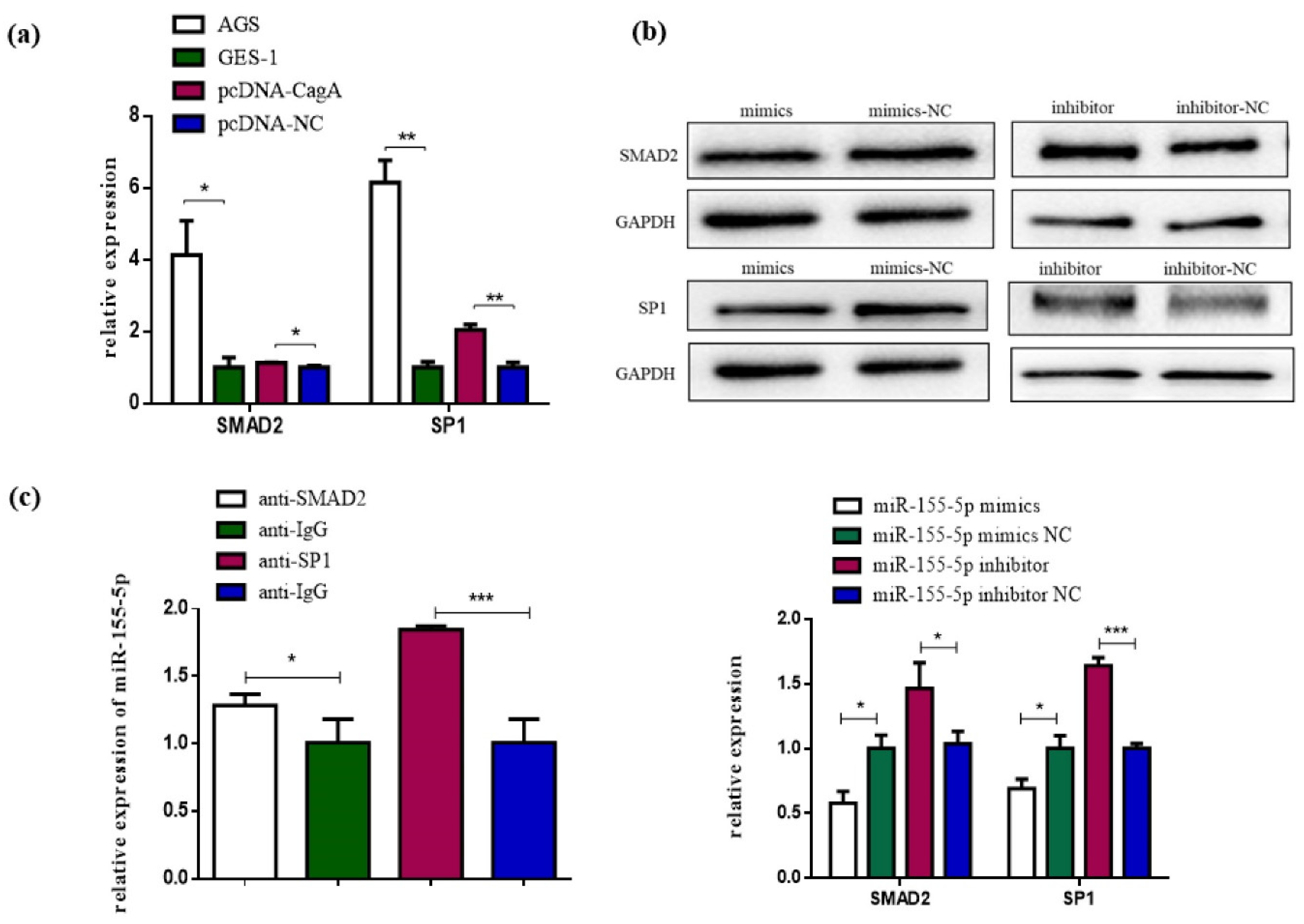
3.4. SP1 and SMAD2 Were Targets of miR-155-5p
3.5. CagA Promotes the Proliferation, Migration, and Invasion of GC Cells through the miR-155-5p-SP1/SMAD2 axis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ishaq, S.; Nunn, L. Helicobacter pylori and gastric cancer: A state of the art review. Gastroenterol. Hepatol. Bed Bench 2015, 8, 6–14. [Google Scholar]
- Plummer, M.; Franceschi, S.; Vignat, J.; Forman, D.; de Martel, C. Global burden of gastric cancer attributable to Helicobacter pylori. Int. J. Cancer 2015, 136, 487–490. [Google Scholar] [CrossRef]
- Backert, S.; Tegtmeyer, N. Type IV Secretion and Signal Transduction of Helicobacter pylori CagA through Interactions with Host Cell Receptors. Toxins 2017, 9, 115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bravo, D.; Hoare, A.; Soto, C.; Valenzuela, M.A.; Quest, A.F. Helicobacter pylori in human health and disease: Mechanisms for local gastric and systemic effects. World J. Gastroenterol. 2018, 24, 3071–3089. [Google Scholar] [CrossRef] [PubMed]
- Plummer, M.; de Martel, C.; Vignat, J.; Ferlay, J.; Bray, F.; Franceschi, S. Global burden of cancers attributable to infections in 2012: A synthetic analysis. Lancet Glob. Health 2016, 4, 609–616. [Google Scholar] [CrossRef] [Green Version]
- Zhang, R.G.; Duan, G.C.; Fan, Q.T.; Chen, S.Y. Role of Helicobacter pylori infection in pathogenesis of gastric carcinoma. World J. Gastrointest. Pathophysiol. 2016, 7, 97–107. [Google Scholar] [CrossRef]
- Lee, Y.C.; Chen, T.H.; Chiu, H.M.; Shun, C.T.; Chiang, H.; Liu, T.Y.; Wu, M.S.; Lin, J.T. The benefit of mass eradication of Helicobacter pylori infection: A community-based study of gastric cancer prevention. Gut 2013, 62, 676–682. [Google Scholar] [CrossRef] [Green Version]
- Baj, J.; Forma, A.; Sitarz, M.; Portincasa, P.; Garruti, G.; Krasowska, D.; Maciejewski, R. Helicobacter pylori Virulence Factors-Mechanisms of Bacterial Pathogenicity in the Gastric Microenvironment. Cells 2020, 10, 27. [Google Scholar] [CrossRef]
- Alipour, M. Molecular Mechanism of Helicobacter pylori-Induced Gastric Cancer. J. Gastrointest. Cancer 2021, 52, 23–30. [Google Scholar] [CrossRef]
- Xu, Z.; Li, Z.; Wang, W.; Xia, Y.; He, Z.; Li, B.; Wang, S.; Huang, X.; Sun, G.; Xu, J.; et al. MIR-1265 regulates cellular proliferation and apoptosis by targeting calcium binding protein 39 in gastric cancer and, thereby, impairing oncogenic autophagy. Cancer Lett. 2019, 449, 226–236. [Google Scholar] [CrossRef]
- Konishi, H.; Sato, H.; Takahashi, K.; Fujiya, M. Tumor-Progressive Mechanisms Mediating miRNA-Protein Interaction. Int. J. Mol. Sci. 2021, 22, 12303. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Xie, S.; Liu, M.; Chen, Z.; Liu, X.; Wang, L.; Li, D.; Zhou, Y. The clinical significance of downregulation of mir-124-3p, mir-146a-5p, mir-155-5p and mir-335-5p in gastric cancer tumorigenesis. Int. J. Oncol. 2014, 45, 197–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zare, A.; Alipoor, B.; Omrani, M.D.; Zali, M.R.; Malekpour Alamdari, N.; Ghaedi, H. Decreased miR-155-5p, miR-15a, and miR-186 Expression in Gastric Cancer Is Associated with Advanced Tumor Grade and Metastasis. Iran. Biomed. J. 2019, 23, 338–343. [Google Scholar] [CrossRef] [Green Version]
- Zhu, M.; Wang, M.; Yang, F.; Tian, Y.; Cai, J.; Yang, H.; Fu, H.; Mao, F.; Zhu, W.; Qian, H.; et al. miR-155-5p inhibition promotes the transition of bone marrow mesenchymal stem cells to gastric cancer tissue derived MSC-like cells via NF-κB p65 activation. Oncotarget 2016, 7, 16567–16580. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Zhang, T.; Zhou, X.; Du, Z.; Chen, F.; Luo, J.; Liu, Q. The tumor suppressor role of miR-155-5p in gastric cancer. Oncol. Lett. 2018, 16, 2709–2714. [Google Scholar] [CrossRef] [Green Version]
- McGeary, S.E.; Lin, K.S.; Shi, C.Y.; Pham, T.M.; Bisaria, N.; Kelley, G.M.; Bartel, D.P. The biochemical basis of microRNA targeting efficacy. Science 2019, 366, eaav1741. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, 127–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.Y.; Lin, Y.C.; Cui, S.; Huang, Y.; Tang, Y.; Xu, J.; Bao, J.; Li, Y.; Wen, J.; Zuo, H.; et al. miRTarBase update 2022: An informative resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 2022, 50, 222–230. [Google Scholar] [CrossRef]
- Ansari, S.; Yamaoka, Y. Helicobacter pylori Virulence Factor Cytotoxin-Associated Gene A (CagA)-Mediated Gastric Pathogenicity. Int. J. Mol. Sci. 2020, 21, 7430. [Google Scholar] [CrossRef]
- He, H.; Liu, J.; Li, L.; Qian, G.; Hao, D.; Li, M.; Zhang, Y.; Hong, X.; Xu, J.; Yan, D. Helicobacter pylori CagA Interacts with SHP-1 to Suppress the Immune Response by Targeting TRAF6 for K63-Linked Ubiquitination. J. Immunol. 2021, 206, 1161–1170. [Google Scholar] [CrossRef]
- Satoshi, I.; Takuya, O.; Naoko, M.K.; Daisuke, K.; Kamrunnesa, T.; Weida, W.; Atsushi, T.K.; Takaya, K.C.; Akiko, K.; Nobumi, S.; et al. Helicobacter pylori CagA elicits BRCAness to induce genome instability that may underlie bacterial gastric carcinogenesis. Cell Host Microbe 2021, 29, 941–958. [Google Scholar]
- Lin, L.; Wei, H.; Yi, J.; Xie, B.; Chen, J.; Zhou, C.; Wang, L.; Yang, Y. Chronic CagA-positive Helicobacter pylori infection with MNNG stimulation synergistically induces mesenchymal and cancer stem cell-like properties in gastric mucosal epithelial cells. J. Cell Biochem. 2019, 120, 17635–17649. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Li, J.; Mu, H.; Guo, M.; Deng, H. MiR-503 suppresses cell proliferation and invasion of gastric cancer by targeting HMGA2 and inactivating WNT signaling pathway. Cancer Cell Int. 2019, 19, 164. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Huang, X.; Chen, G.; Wang, Y.; Liu, Y.; Xu, W.; Tang, S.; Guleng, B.; Liu, J.; Ren, J. miR-632 promotes gastric cancer progression by accelerating angiogenesis in a TFF1-dependent manner. BMC Cancer 2019, 19, 14. [Google Scholar] [CrossRef]
- Nicole, T.; Aileen, H.; Klemens, R.; Steffen, B. Helicobacter pylori CagA Induces Cortactin Y-470 Phosphorylation-Dependent Gastric Epithelial Cell Scattering via Abl, Vav2 and Rac1 Activation. Cancers 2021, 13, 4241. [Google Scholar]
- Lei, Q.Q.; Huang, Y.; Li, B.; Han, L.; Lv, C. MiR-155-5p promotes metastasis and epithelial-mesenchymal transition of renal cell carcinoma by targeting apoptosis-inducing factor. Int. J. Biol. Markers 2021, 36, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Cui, T.; Guo, W.; Wang, D.; Mao, L. MiR-155-5p accelerates the metastasis of cervical cancer cell via targeting TP53INP1. Onco Targets Ther. 2019, 12, 3181–3196. [Google Scholar]
- Wang, J.; Che, J. CircTP63 promotes hepatocellular carcinoma progression by sponging miR-155-5p and upregulating ZBTB18. Cancer Cell Int. 2021, 21, 156. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zheng, S.Y.; Yang, C.Q.; Ma, B.M.; Jiang, D. MiR-155-5p inhibits the proliferation and migration of VSMCs and HUVECs in atherosclerosis by targeting AKT1. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 2223–2233. [Google Scholar] [PubMed]
- Yang, F.; Xu, Y.; Liu, C.; Ma, C.; Zou, S.; Xu, X.; Jia, J.; Liu, Z. NF-κB/miR-223-3p/ARID1A axis is involved in Helicobacter pylori CagA-induced gastric carcinogenesis and progression. Cell Death Dis. 2018, 9, 12. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.Z. MicroRNAs as oncogenes and tumor suppressors. N. Engl. J. Med. 2005, 353, 1768–1771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cherukunnath, A.; Davargaon, R.S.; Ashraf, R.; Kamdar, U.; Srivastava, A.K.; Tripathi, P.P.; Chatterjee, N.; Kumar, S. KLF8 is activated by TGF-β1 via Smad2 and contributes to ovarian cancer progression. J. Cell. Biochem. 2022, 123, 921–934. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Gao, W.; Yuan, B.; Zhang, S.; Wang, K.; Du, T. TRIM22 inhibits the proliferation of gastric cancer cells through the Smad2 protein. Cell Death Discov. 2021, 7, 234. [Google Scholar] [CrossRef] [PubMed]
- Niu, Q.; Dong, Z.; Liang, M.; Luo, Y.; Lin, H.; Lin, M.; Zhong, X.; Yao, W.; Weng, J.; Zhou, X. Circular RNA hsa_circ_0001829 promotes gastric cancer progression through miR-155-5p/SMAD2 axis. J. Exp. Clin. Cancer Res. 2020, 39, 280. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yang, H.; Jia, Y.; Xu, Z.; Zhang, L.; Sun, M.; Fu, J. circRNA_0005529 facilitates growth and metastasis of gastric cancer via regulating miR-527/Sp1 axis. BMC Mol. Cell Biol. 2021, 22, 6. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Yuan, W.; Huang, C.; Luo, Q.; Xiao, R.; Chen, Z.H. LncRNA PCAT19 induced by SP1 and acted as oncogene in gastric cancer competitively binding to miR429 and upregulating DHX9. J. Cancer 2022, 13, 102–111. [Google Scholar] [CrossRef]
- Xu, X.W.; Pan, C.W.; Yang, X.M.; Zhou, L.; Zheng, Z.Q.; Li, D.C. SP1 reduces autophagic flux through activating p62 in gastric cancer cells. Mol. Med. Rep. 2018, 17, 4633–4638. [Google Scholar] [CrossRef] [Green Version]
- Shi, S.; Zhang, Z.G. Role of Sp1 expression in gastric cancer: A meta-analysis and bioinformatics analysis. Oncol. Lett. 2019, 18, 4126–4135. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, L.; Jiang, F.; Shen, X. Helicobacter pylori CagA Protein Regulating the Biological Characteristics of Gastric Cancer through the miR-155-5p/SMAD2/SP1 axis. Pathogens 2022, 11, 846. https://doi.org/10.3390/pathogens11080846
Wu L, Jiang F, Shen X. Helicobacter pylori CagA Protein Regulating the Biological Characteristics of Gastric Cancer through the miR-155-5p/SMAD2/SP1 axis. Pathogens. 2022; 11(8):846. https://doi.org/10.3390/pathogens11080846
Chicago/Turabian StyleWu, Leilei, Fei Jiang, and Xiaobing Shen. 2022. "Helicobacter pylori CagA Protein Regulating the Biological Characteristics of Gastric Cancer through the miR-155-5p/SMAD2/SP1 axis" Pathogens 11, no. 8: 846. https://doi.org/10.3390/pathogens11080846
APA StyleWu, L., Jiang, F., & Shen, X. (2022). Helicobacter pylori CagA Protein Regulating the Biological Characteristics of Gastric Cancer through the miR-155-5p/SMAD2/SP1 axis. Pathogens, 11(8), 846. https://doi.org/10.3390/pathogens11080846




