Corynebacterium striatum—Got Worse by a Pandemic?
Abstract
:1. Introduction
2. Results
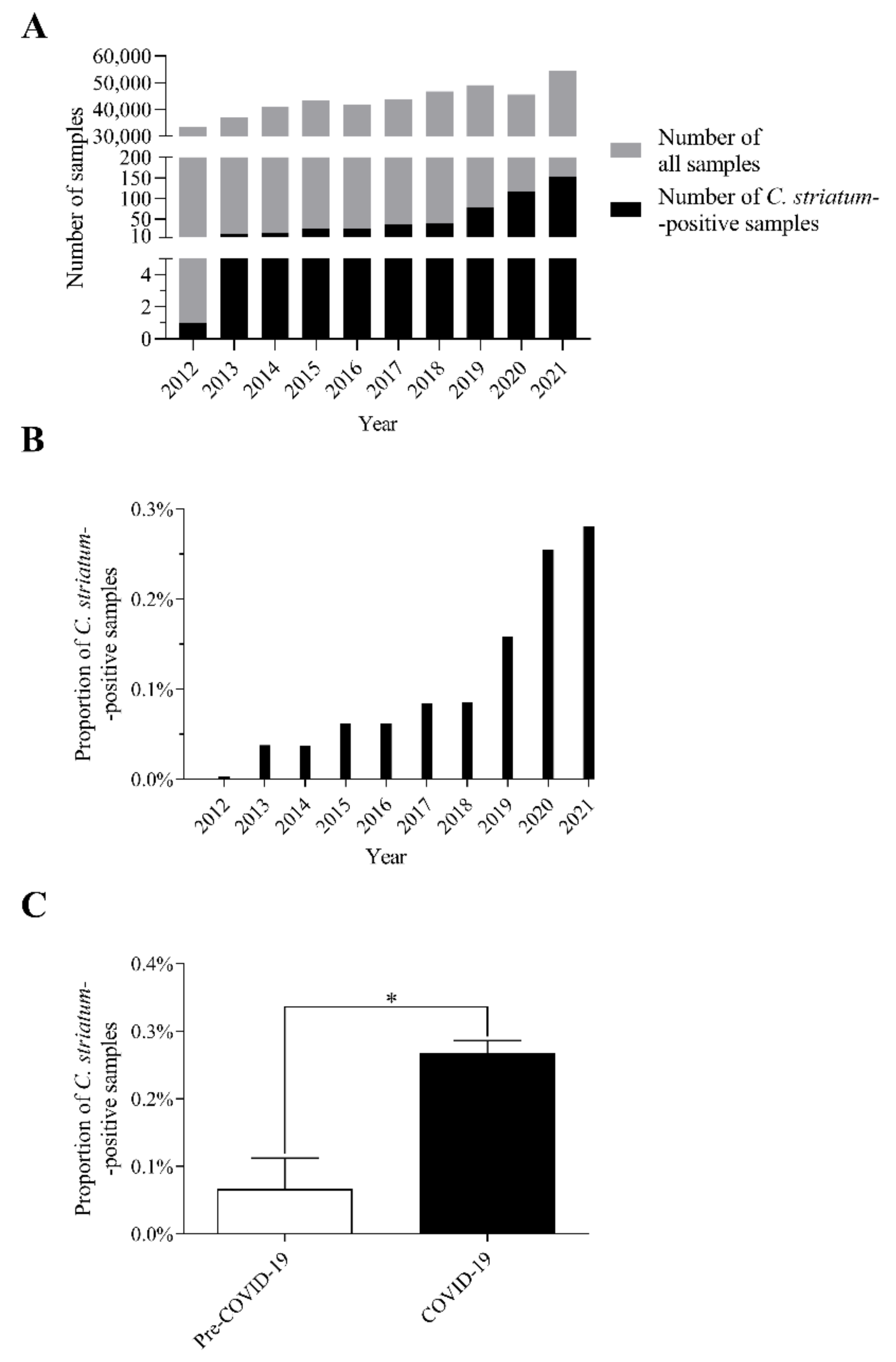
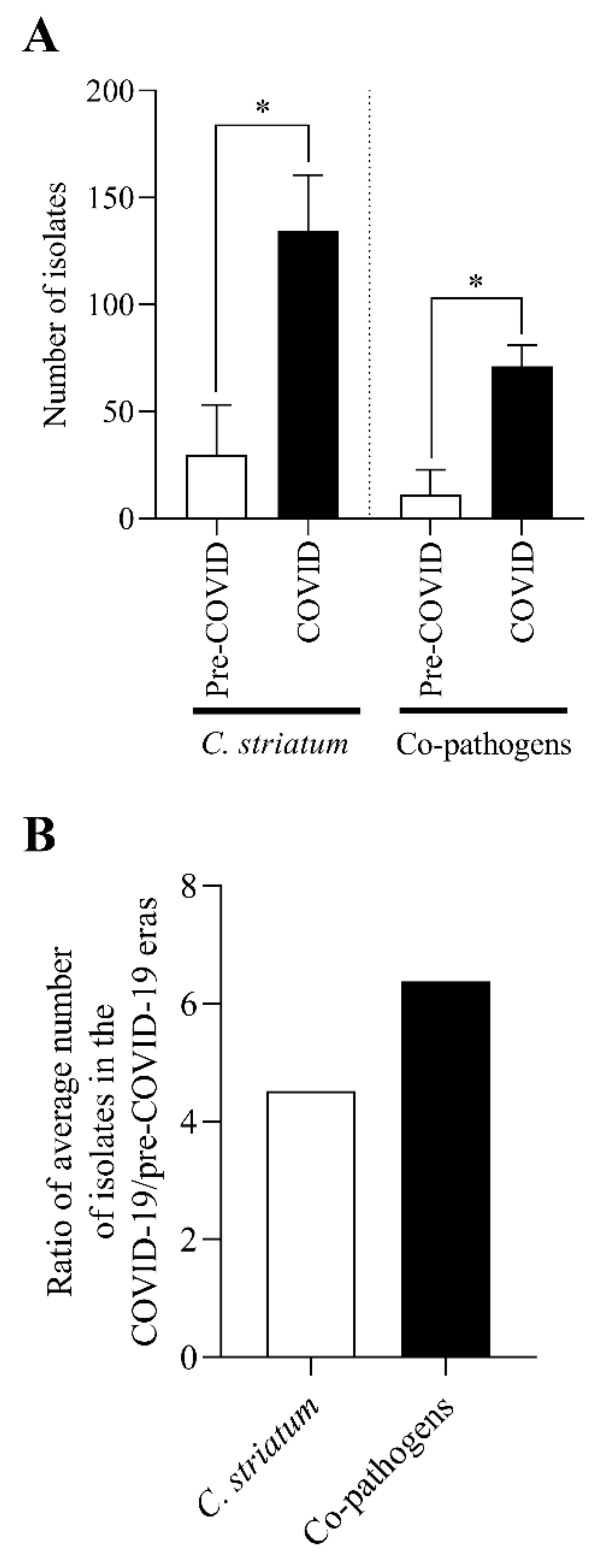
2.1. The Number of C. striatum Isolates Has Increased Remarkably over the Last 10 Years
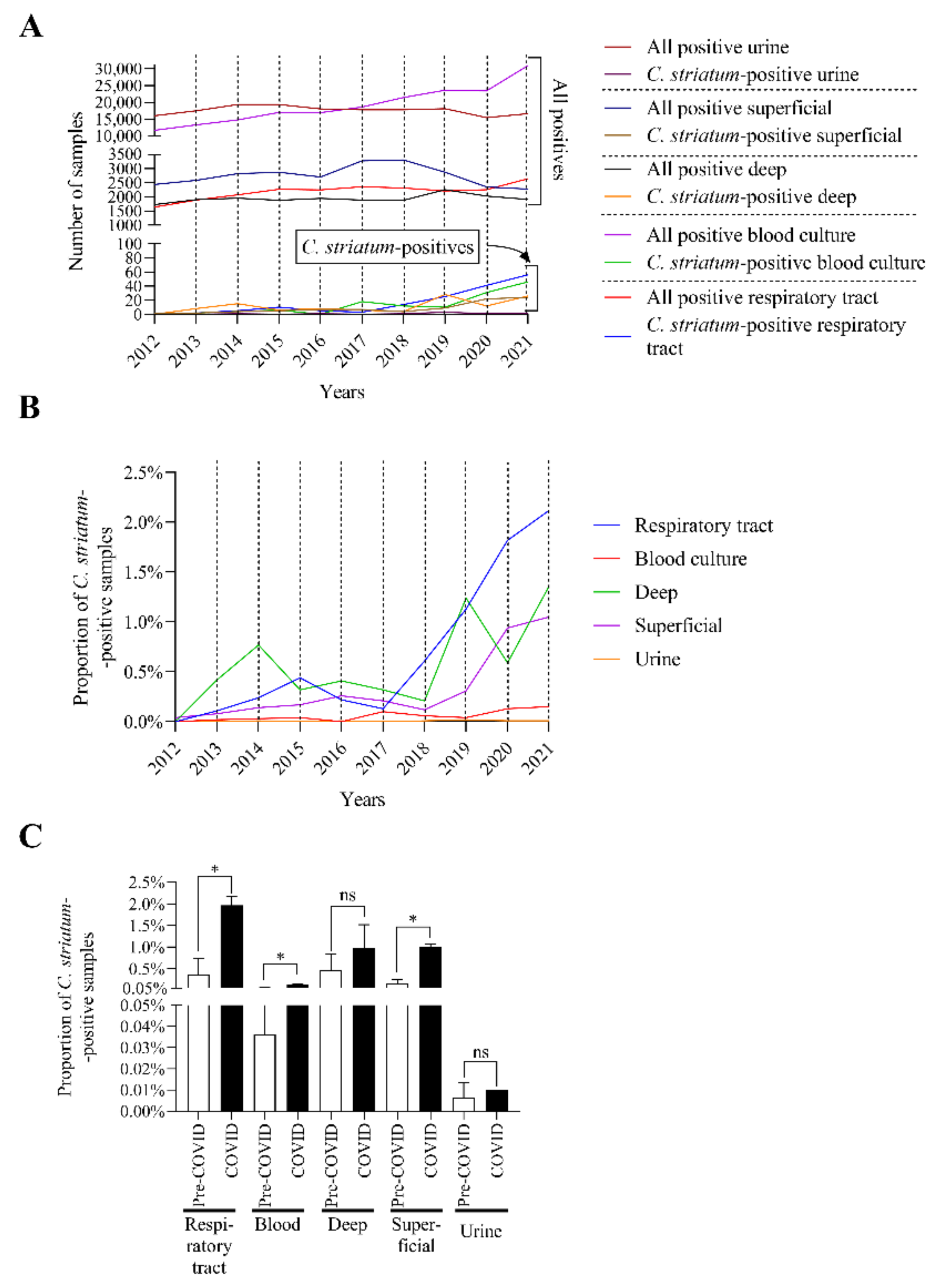
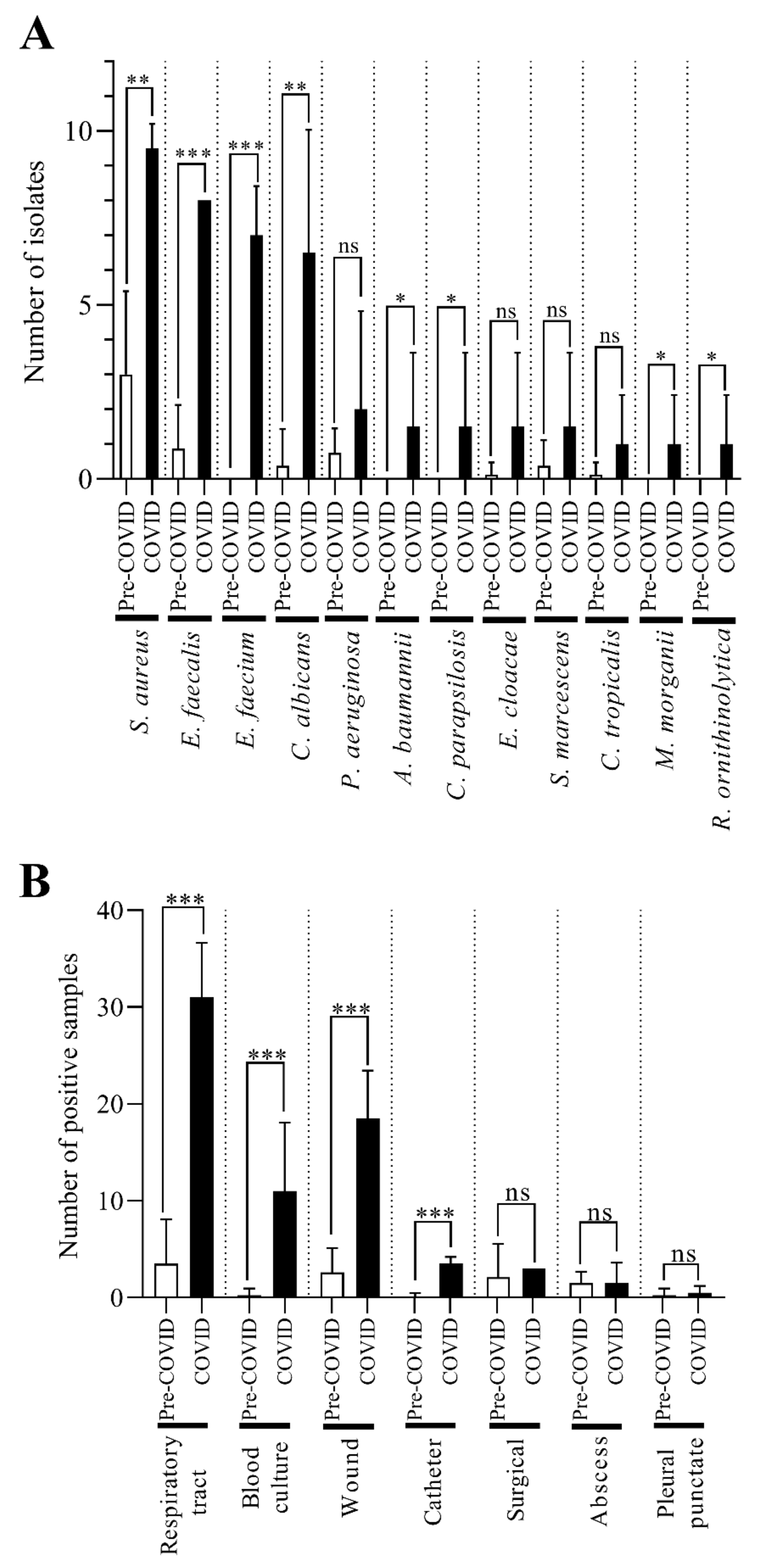
2.2. C. striatum Was Significantly more Frequent in Some Sample Types in the COVID-19 Era
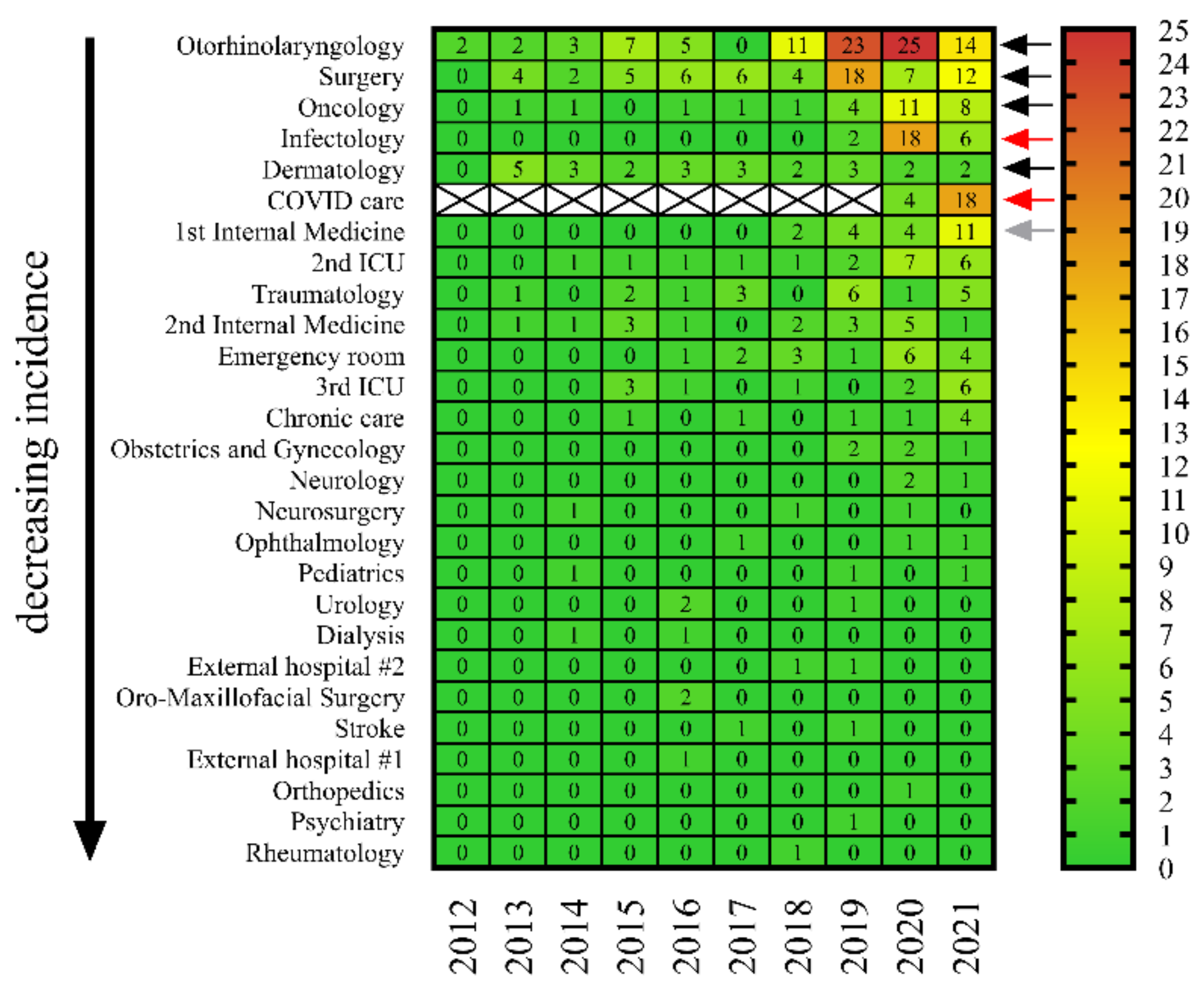
2.3. C. striatum Strains Were Consistently Present in Some Wards, including COVID-19 Care Sites
2.4. One-Third of C. striatum Strains Were Isolated from Cases Associated with Viral Infection in 2020 and 2021
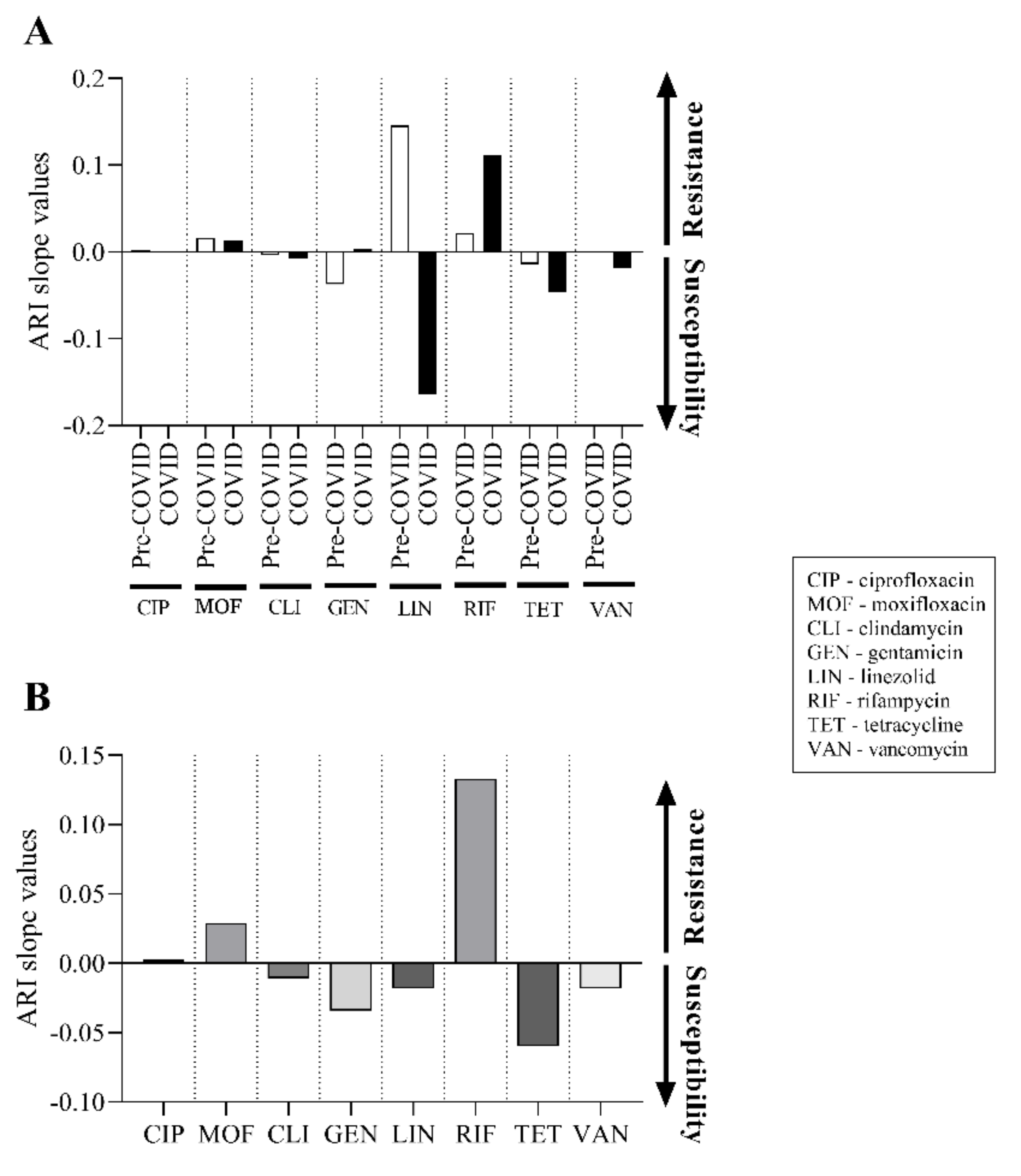
2.5. There Was No Significant Change in the Antibiotic Susceptibility of C. striatum Isolates between 2012 and 2021
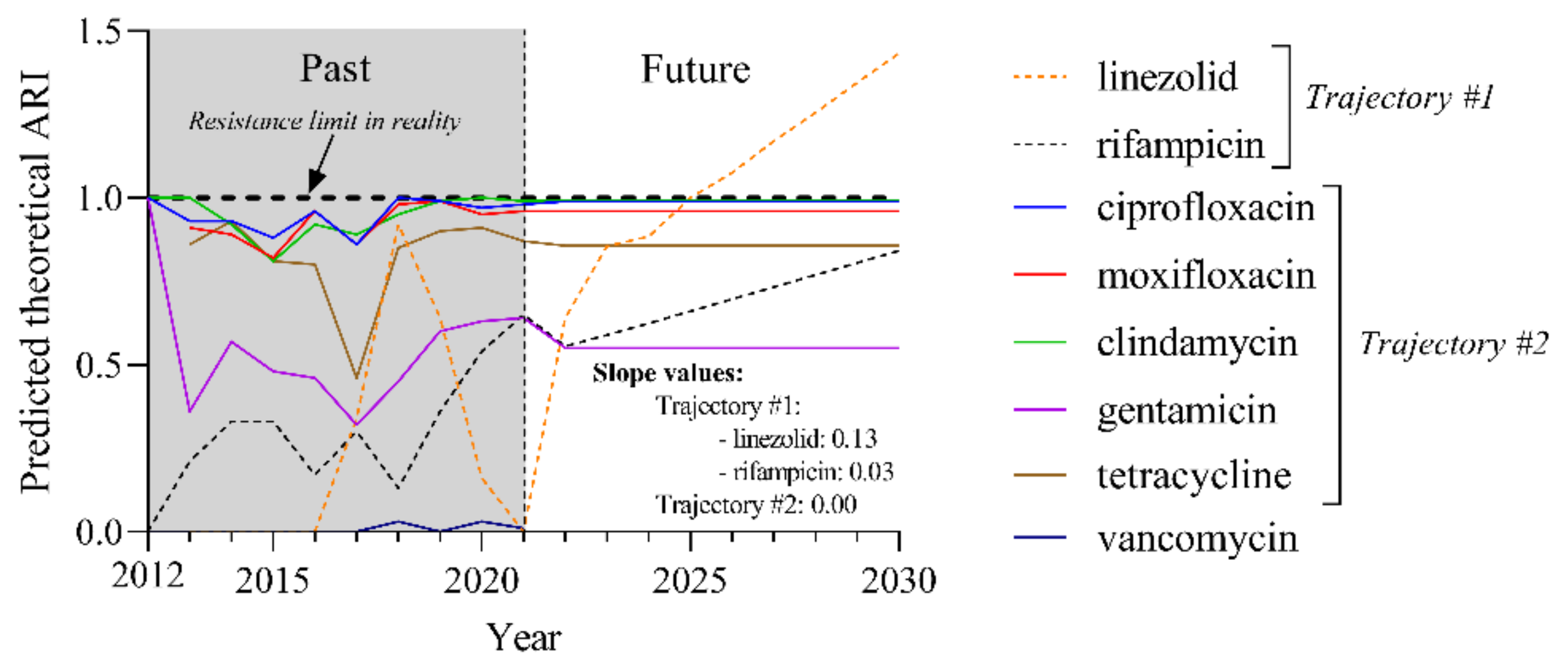
2.6. Predicted Theoretical ARI Values for Currently Used Agents Project Two Trajectories until 2030
2.7. The C. striatum Occurred with Diverse and Changing Trends of Co-Pathogens in the COVID-19 Era
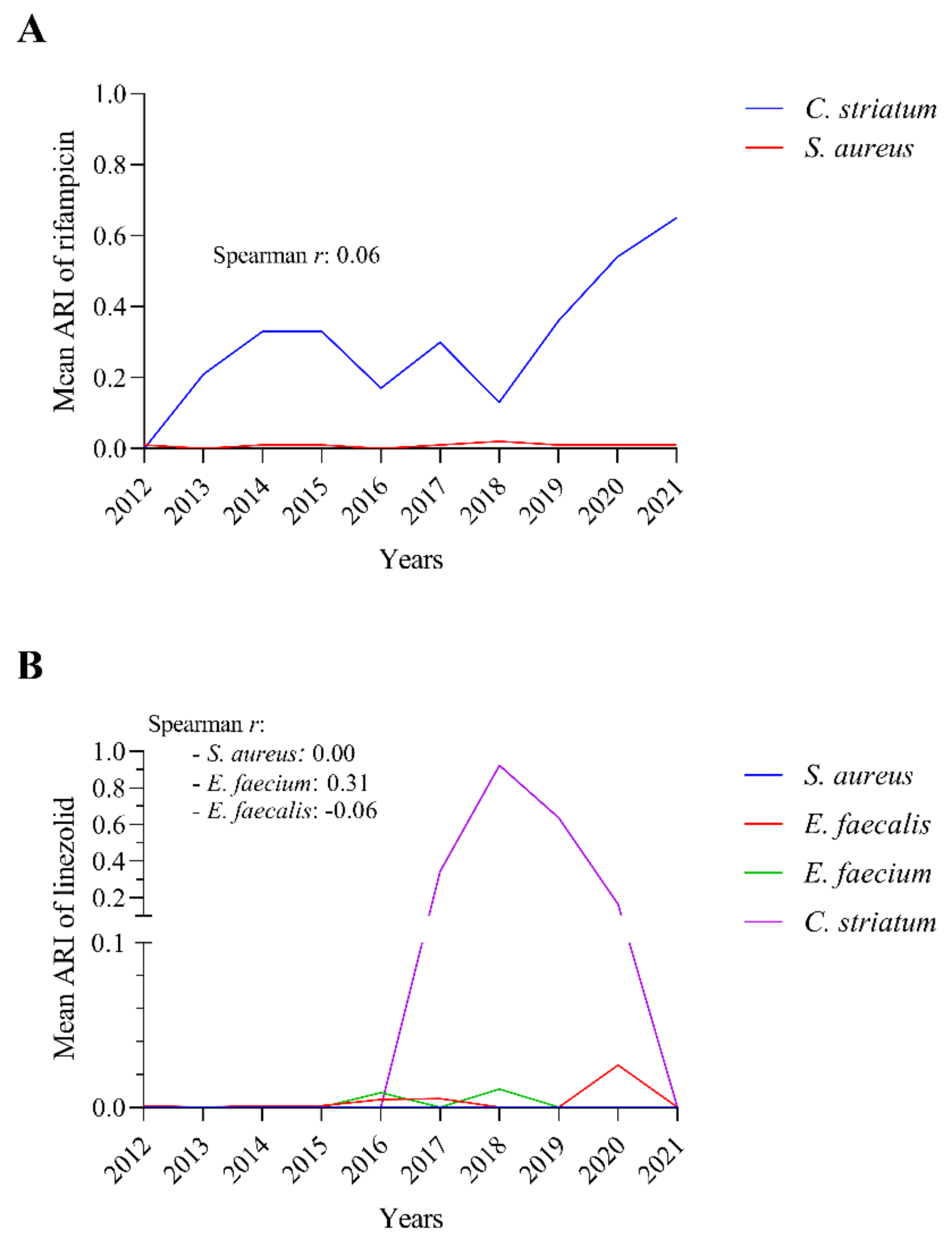
2.8. The Rifampicin and Linezolid Resistance of C. striatum Is Probably Not Due to the Most Commonly Isolated Co-Pathogenic Bacteria
3. Discussion
4. Materials and Methods
4.1. Study Setting
4.2. Microbiological Data Set
4.3. Calculating Antibiotic Resistance Index
4.4. Statistical Analysis
4.5. Time Series Forecasting Using Machine Learning
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kim, R.; Reboli, A.C. Other Coryneform Bacteria and Rhodococci. In Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases; Elsevier Inc.: Philadelphia, PA, USA, 2015; Volume 2, pp. 2373–2382. [Google Scholar]
- Asgin, N.; Otlu, B. Antimicrobial Resistance and Molecular Epidemiology of Corynebacterium striatum Isolated in a Tertiary Hospital in Turkey. Pathogens 2020, 9, 136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Funke, G.; Bernard, K.A. Coryneform Gram-Positive Rods. In Manual of Clinical Microbiology; American Society for Microbiology Press: Washington, DC, USA, 2015; Volume 1, pp. 474–503. [Google Scholar]
- Charalampous, T.; Alcolea-Medina, A.; Snell, L.B.; Williams, T.G.S.; Batra, R.; Alder, C.; Telatin, A.; Camporota, L.; Meadows, C.I.S.; Wyncoll, D.; et al. Evaluating the Potential for Respiratory Metagenomics to Improve Treatment of Secondary Infection and Detection of Nosocomial Transmission on Expanded COVID-19 Intensive Care Units. Genome Med. 2021, 13, 182. [Google Scholar] [CrossRef] [PubMed]
- McMullen, A.R.; Anderson, N.; Wallace, M.A.; Shupe, A.; Burnham, C.-A.D. When Good Bugs Go Bad: Epidemiology and Antimicrobial Resistance Profiles of Corynebacterium striatum, an Emerging Multidrug-Resistant, Opportunistic Pathogen. Antimicrob. Agents Chemother. 2017, 61, e01111-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shariff, M.; Aditi, A.; Beri, K. Corynebacterium striatum: An Emerging Respiratory Pathogen. J. Infect. Dev. Ctries. 2018, 12, 581–586. [Google Scholar] [CrossRef]
- Fernández Guerrero, M.L.; Molins, A.; Rey, M.; Romero, J.; Gadea, I. Multidrug-Resistant Corynebacterium striatum Endocarditis Successfully Treated with Daptomycin. Int. J. Antimicrob. Agents 2012, 40, 373–374. [Google Scholar] [CrossRef]
- Oliva, A.; Belvisi, V.; Iannetta, M.; Andreoni, C.; Mascellino, M.T.; Lichtner, M.; Vullo, V.; Mastroianni, C.M. Pacemaker Lead Endocarditis Due to Multidrug-Resistant Corynebacterium striatum Detected with Sonication of the Device. J. Clin. Microbiol. 2010, 48, 4669–4671. [Google Scholar] [CrossRef] [Green Version]
- Boltin, D.; Katzir, M.; Bugoslavsky, V.; Yalashvili, I.; Brosh-Nissimov, T.; Fried, M.; Elkayam, O. Corynebacterium striatum—A Classic Pathogen Eluding Diagnosis. Eur. J. Intern. Med. 2009, 20, e49–e52. [Google Scholar] [CrossRef]
- Campanile, F.; Carretto, E.; Barbarini, D.; Grigis, A.; Falcone, M.; Goglio, A.; Venditti, M.; Stefani, S. Clonal Multidrug-Resistant Corynebacterium striatum Strains, Italy. Emerg. Infect. Dis. 2009, 15, 75–78. [Google Scholar] [CrossRef]
- De Arriba, J.J.; Blanch, J.J.; Mateos, F.; Martínez-Alfaro, E.; Solera, J. Corynebacterium striatum First Reported Case of Prosthetic Valve Endocarditis. J. Infect. 2002, 44, 193. [Google Scholar] [CrossRef]
- Melero-Bascones, M.; Munoz, P.; Rodriguez-Creixems, M.; Bouza, E. Corynebacterium striatum: An Undescribed Agent of Pacemaker-Related Endocarditis. Clin. Infect. Dis. 1996, 22, 576–577. [Google Scholar] [CrossRef] [Green Version]
- McElvania TeKippe, E.; Thomas, B.S.; Ewald, G.A.; Lawrence, S.J.; Burnham, C.-A.D. Rapid Emergence of Daptomycin Resistance in Clinical Isolates of Corynebacterium striatum… A Cautionary Tale. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 2199–2205. [Google Scholar] [CrossRef] [Green Version]
- Wong, K.Y.; Chan, Y.C.; Wong, C.Y. Corynebacterium striatum as an Emerging Pathogen. J. Hosp. Infect. 2010, 76, 371–372. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Y.; Du, X.; Cui, J.; Wang, K.; Zhang, L.; Han, Y. Rapid Transmission of Multidrug-Resistant Corynebacterium striatum among Susceptible Patients in a Tertiary Hospital in China. J. Infect. Dev. Ctries. 2016, 10, 1299–1305. [Google Scholar] [CrossRef] [Green Version]
- Werth, B.J.; Hahn, W.O.; Butler-Wu, S.M.; Rakita, R.M. Emergence of High-Level Daptomycin Resistance in Corynebacterium striatum in Two Patients with Left Ventricular Assist Device Infections. Microb. Drug Resist. 2016, 22, 233–237. [Google Scholar] [CrossRef] [Green Version]
- Hahn, W.O.; Werth, B.J.; Butler-Wu, S.M.; Rakita, R.M. Multidrug-Resistant Corynebacterium striatum Associated with Increased Use of Parenteral Antimicrobial Drugs. Emerg. Infect. Dis. 2016, 22, 1908. [Google Scholar] [CrossRef] [Green Version]
- Silva-Santana, G.; Silva, C.M.F.; Olivella, J.G.B.; Silva, I.F.; Fernandes, L.M.O.; Sued-Karam, B.R.; Santos, C.S.; Souza, C.; Mattos-Guaraldi, A.L. Worldwide Survey of Corynebacterium striatum Increasingly Associated with Human Invasive Infections, Nosocomial Outbreak, and Antimicrobial Multidrug-Resistance, 1976–2020. Arch. Microbiol. 2021, 203, 1863–1880. [Google Scholar] [CrossRef]
- Yoon, S.; Kim, H.; Lee, Y.; Kim, S. Bacteremia Caused by Corynebacterium amycolatum with a Novel Mutation in GyrA Gene That Confers High-Level Quinolone Resistance. Ann. Lab. Med. 2011, 31, 47–48. [Google Scholar] [CrossRef] [Green Version]
- Reddy, B.; Chaudhury, A.; Kalawat, U.; Jayaprada, R.; Reddy, G.; Ramana, B. Isolation, Speciation, and Antibiogram of Clinically Relevant Non-Diphtherial Corynebacteria (Diphtheroids). Indian J. Med. Microbiol. 2012, 30, 52–57. [Google Scholar] [CrossRef]
- Leyton, B.; Ramos, J.N.; Baio, P.V.P.; Veras, J.F.C.; Souza, C.; Burkovski, A.; Mattos-Guaraldi, A.L.; Vieira, V.V.; Abanto Marin, M. Treat Me Well or Will Resist: Uptake of Mobile Genetic Elements Determine the Resistome of Corynebacterium striatum. Int. J. Mol. Sci. 2021, 22, 7499. [Google Scholar] [CrossRef]
- Ramos, J.N.; Souza, C.; Faria, Y.V.; da Silva, E.C.; Veras, J.F.C.; Baio, P.V.P.; Seabra, S.H.; de Oliveira Moreira, L.; Hirata Júnior, R.; Mattos-Guaraldi, A.L.; et al. Bloodstream and Catheter-Related Infections Due to Different Clones of Multidrug-Resistant and Biofilm Producer Corynebacterium striatum. BMC Infect. Dis. 2019, 19, 672. [Google Scholar] [CrossRef]
- De Souza, C.; Faria, Y.V.; de Oliveira Sant’Anna, L.; Viana, V.G.; Seabra, S.H.; de Souza, M.C.; Vieira, V.V.; Hirata Júnior, R.; de Oliveira Moreira, L.; de Mattos-Guaraldi, A.L. Biofilm Production by Multiresistant Corynebacterium striatum Associated with Nosocomial Outbreak. Mem. Inst. Oswaldo Cruz 2015, 110, 242–248. [Google Scholar] [CrossRef] [Green Version]
- Mhade, S.; Panse, S.; Tendulkar, G.; Awate, R.; Narasimhan, Y.; Kadam, S.; Yennamalli, R.M.; Kaushik, K.S. AMPing Up the Search: A Structural and Functional Repository of Antimicrobial Peptides for Biofilm Studies, and a Case Study of Its Application to Corynebacterium striatum, an Emerging Pathogen. Front. Cell. Infect. Microbiol. 2021, 11, 803774. [Google Scholar] [CrossRef]
- De Souza, C.; Mota, H.F.; Faria, Y.V.; de Oliveira Cabral, F.; de Oliveira, D.R.; de Oliveira Sant’Anna, L.; Nagao, P.E.; da Silva Santos, C.; Moreira, L.O.; Mattos-Guaraldi, A.L. Resistance to Antiseptics and Disinfectants of Planktonic and Biofilm-Associated Forms of Corynebacterium striatum. Microb. Drug Resist. 2020, 26, 1546–1558. [Google Scholar] [CrossRef]
- Pollard, C.A.; Morran, M.P.; Nestor-Kalinoski, A.L. The COVID-19 Pandemic: A Global Health Crisis. Physiol. Genom. 2020, 52, 549–557. [Google Scholar] [CrossRef]
- Ripa, M.; Galli, L.; Poli, A.; Oltolini, C.; Spagnuolo, V.; Mastrangelo, A.; Muccini, C.; Monti, G.; De Luca, G.; Landoni, G.; et al. Secondary Infections in Patients Hospitalized with COVID-19: Incidence and Predictive Factors. Clin. Microbiol. Infect. 2021, 27, 451–457. [Google Scholar] [CrossRef]
- Fattorini, L.; Creti, R.; Palma, C.; Pantosti, A. Unit of Antibiotic Resistance and Special Pathogens; Unit of Antibiotic Resistance and Special Pathogens of the Department of Infectious Diseases, Istituto Superiore di Sanità, Rome Bacterial Coinfections in COVID-19: An Underestimated Adversary. Ann. Ist. Super. Sanita 2020, 56, 359–364. [Google Scholar] [CrossRef]
- De Socio, G.V.; Rubbioni, P.; Botta, D.; Cenci, E.; Belati, A.; Paggi, R.; Pasticci, M.B.; Mencacci, A. Measurement and Prediction of Antimicrobial Resistance in Bloodstream Infections by ESKAPE Pathogens and Escherichia Coli. J. Glob. Antimicrob. Resist. 2019, 19, 154–160. [Google Scholar] [CrossRef]
- Coronavirus (COVID-19) Hospitalizations—Our World in Data. Available online: https://ourworldindata.org/covid-hospitalizations (accessed on 23 January 2022).
- Marino, A.; Campanella, E.; Stracquadanio, S.; Ceccarelli, M.; Zagami, A.; Nunnari, G.; Cacopardo, B. Corynebacterium striatum Bacteremia during SARS-CoV2 Infection: Case Report, Literature Review, and Clinical Considerations. Infect. Dis. Rep. 2022, 14, 383–390. [Google Scholar] [CrossRef]
- Neemuchwala, A.; Soares, D.; Ravirajan, V.; Marchand-Austin, A.; Kus, J.V.; Patel, S.N. In Vitro Antibiotic Susceptibility Pattern of Non-Diphtheriae Corynebacterium Isolates in Ontario, Canada, from 2011 to 2016. Antimicrob. Agents Chemother. 2018, 62, e01776-17. [Google Scholar] [CrossRef] [Green Version]
- Olender, A. Mechanisms of Antibiotic Resistance in Corynebacterium spp. Causing Infections in People. In Antibiotic Resistant Bacteria—A Continuous Challenge in the New Millennium; Pana, M., Ed.; InTech, 2012; ISBN 978-953-51-0472-8. Available online: https://www.intechopen.com/chapters/34699 (accessed on 23 May 2022).
- Parasher, A. COVID-19: Current Understanding of Its Pathophysiology, Clinical Presentation and Treatment. Postgrad. Med. J. 2021, 97, 312–320. [Google Scholar] [CrossRef]
- Ortiz-Pérez, A.; Martín-De-Hijas, N.Z.; Esteban, J.; Fernández-Natal, M.I.; García-Cía, J.I.; Fernández-Roblas, R. High Frequency of Macrolide Resistance Mechanisms in Clinical Isolates of Corynebacterium Species. Microb. Drug Resist. 2010, 16, 273–277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adékambi, T.; Drancourt, M.; Raoult, D. The RpoB Gene as a Tool for Clinical Microbiologists. Trends Microbiol. 2009, 17, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Kornblum, D.; Godefroy, N.; Monsel, G.; Robert, J.; Caumes, E.; Pourcher, V.; Klement-Frutos, E. Corynebacterium striatum Thrombophlebitis: A Nosocomial Multidrug-Resistant Disease? Access Microbiol. 2021, 3, 000307. [Google Scholar] [CrossRef] [PubMed]
- Alibi, S.; Ferjani, A.; Boukadida, J.; Cano, M.E.; Fernández-Martínez, M.; Martínez-Martínez, L.; Navas, J. Occurrence of Corynebacterium striatum as an Emerging Antibiotic-Resistant Nosocomial Pathogen in a Tunisian Hospital. Sci. Rep. 2017, 7, 9704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chapartegui-González, I.; Fernández-Martínez, M.; Rodríguez-Fernández, A.; Rocha, D.J.P.; Aguiar, E.R.G.R.; Pacheco, L.G.C.; Ramos-Vivas, J.; Calvo, J.; Martínez-Martínez, L.; Navas, J. Antimicrobial Susceptibility and Characterization of Resistance Mechanisms of Corynebacterium urealyticum Clinical Isolates. Antibiotics 2020, 9, 404. [Google Scholar] [CrossRef]
- EUCAST: Clinical Breakpoints and Dosing of Antibiotics. Available online: https://www.eucast.org/clinical_breakpoints/ (accessed on 24 January 2022).
- Martínez-Martínez, L.; Suárez, A.I.; Winstanley, J.; Ortega, M.C.; Bernard, K. Phenotypic Characteristics of 31 Strains of Corynebacterium striatum Isolated from Clinical Samples. J. Clin. Microbiol. 1995, 33, 2458–2461. [Google Scholar] [CrossRef] [Green Version]
- Milosavljevic, M.N.; Milosavljevic, J.Z.; Kocovic, A.G.; Stefanovic, S.M.; Jankovic, S.M.; Djesevic, M.; Milentijevic, M.N. Antimicrobial Treatment of Corynebacterium striatum Invasive Infections: A Systematic Review. Rev. Inst. Med. Trop. São Paulo 2021, 63, e49. [Google Scholar] [CrossRef]
- Hashemian, S.M.R.; Farhadi, T.; Ganjparvar, M. Linezolid: A Review of Its Properties, Function, and Use in Critical Care. Drug Des. Devel. Ther. 2018, 12, 1759–1767. [Google Scholar] [CrossRef]
- Noussair, L.; Salomon, E.; El Sayed, F.; Duran, C.; Bouchand, F.; Roux, A.-L.; Gaillard, J.-L.; Bauer, T.; Rottman, M.; Dinh, A. Monomicrobial Bone and Joint Infection Due to Corynebacterium striatum: Literature Review and Amoxicillin-Rifampin Combination as Treatment Perspective. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 1269–1278. [Google Scholar] [CrossRef]
- Shah, M.; Murillo, J.L. Successful Treatment of Corynebacterium striatum Endocarditis with Daptomycin plus Rifampin. Ann. Pharmacother. 2005, 39, 1741–1744. [Google Scholar] [CrossRef]
- Folliero, V.; Dell’Annunziata, F.; Roscetto, E.; Cammarota, M.; De Filippis, A.; Schiraldi, C.; Catania, M.R.; Casolaro, V.; Perrella, A.; Galdiero, M.; et al. Niclosamide as a Repurposing Drug against Corynebacterium striatum Multidrug-Resistant Infections. Antibiotics 2022, 11, 651. [Google Scholar] [CrossRef]
- Cimolai, N. The Complexity of Co-Infections in the Era of COVID-19. SN Compr. Clin. Med. 2021, 3, 1502–1514. [Google Scholar] [CrossRef]
- Bardi, T.; Pintado, V.; Gomez-Rojo, M.; Escudero-Sanchez, R.; Azzam Lopez, A.; Diez-Remesal, Y.; Martinez Castro, N.; Ruiz-Garbajosa, P.; Pestaña, D. Nosocomial Infections Associated to COVID-19 in the Intensive Care Unit: Clinical Characteristics and Outcome. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 495–502. [Google Scholar] [CrossRef]
- EUCAST: Disk Diffusion Methodology. Available online: https://www.eucast.org/ast_of_bacteria/disk_diffusion_methodology/ (accessed on 2 June 2022).
- Moez, A. PyCaret: An Open Source, Low-Code Machine Learning Library in Python. Available online: https://pycaret.org/ (accessed on 24 April 2022).

| Submitting Diagnoses | Occurrence | Frequency |
|---|---|---|
| Viral Pneumonia | 14 | 8.24% |
| Viral Infection | 10 | 5.88% |
| Bacterial infections | 8 | 4.71% |
| Dyspnoea | 8 | 4.71% |
| Fractura colli femoris medialis | 7 | 4.12% |
| Coronavirus Infection | 7 | 4.12% |
| Subarachnoid haemorrhage from the middle cerebral artery | 7 | 4.12% |
| Septicaemia | 6 | 3.53% |
| Malignant neoplasm of larynx | 6 | 3.53% |
| Local infections of the skin and subcutaneous tissues | 5 | 2.94% |
| Pain | 5 | 2.94% |
| Severe respiratory Failure | 5 | 2.94% |
| Chronic renal failure | 5 | 2.94% |
| Malignant neoplasm of the glottis | 4 | 2.35% |
| Gastrointestinal bleeding | 4 | 2.35% |
| Malignant neoplasm of the hypopharynx | 4 | 2.35% |
| Other Viral Pneumonia | 4 | 2.35% |
| Aortic (valve) stenosis | 3 | 1.76% |
| COVID-19 with Detected Virus | 3 | 1.76% |
| Hodgkin’s disease. lymphocyte predominance | 2 | 1.18% |
| Congestive heart failure | 2 | 1.18% |
| Hepatorenal syndrome | 2 | 1.18% |
| Optic nerve sheath inflammation [neuromyelitis optica devic] | 2 | 1.18% |
| Contusio cerebri | 2 | 1.18% |
| Atherosclerosis of the limbal arteries | 2 | 1.18% |
| Stage III malignancy of the hypopharynx | 2 | 1.18% |
| Ulceration of the lower limb. | 2 | 1.18% |
| Haematoma parietis abdominis | 1 | 0.59% |
| Stage II malignant neoplasm of larynx | 1 | 0.59% |
| Malignant neoplasm of the upper limb. connective tissue. and soft tissues of the shoulder | 1 | 0.59% |
| Postoperative subglottic stenosis | 1 | 0.59% |
| Malignant neoplasm of the root of the tongue | 1 | 0.59% |
| Tumor of the larynx of uncertain and unknown behavior | 1 | 0.59% |
| Acute inflammation of the vagina | 1 | 0.59% |
| Stage III malignant neoplasm of the glottis | 1 | 0.59% |
| Staphylococcal infection | 1 | 0.59% |
| Malignant neoplasm of intra-abdominal lymph nodes | 1 | 0.59% |
| Staphylococcus aureus as a cause of other diseases | 1 | 0.59% |
| Background Retinopathy and retinal lesions | 1 | 0.59% |
| Oedema | 1 | 0.59% |
| Severe pansinusitis | 1 | 0.59% |
| Fractura costarum | 1 | 0.59% |
| Parotid effusion | 1 | 0.59% |
| Laryngeal stenosis | 1 | 0.59% |
| Burn involving less than 10% of the body surface | 1 | 0.59% |
| Urinary tract infection at unspecified site | 1 | 0.59% |
| Ear discharge | 1 | 0.59% |
| Aryepiglottic folds anterior to hypopharynx. malignant swelling III | 1 | 0.59% |
| Stage III malignant neoplasm of pharynx | 1 | 0.59% |
| Dermatopolymyositis | 1 | 0.59% |
| Bacterial pneumonia | 1 | 0.59% |
| Pain localised to other parts of the abdomen | 1 | 0.59% |
| Hypertensive heart disease (congestive) without heart failure | 1 | 0.59% |
| Insulin-dependent diabetes with ocular complications | 1 | 0.59% |
| Non-Hodgkin lymphoma. large cell (diffuse) | 1 | 0.59% |
| Cardiac arrest with successful resuscitation | 1 | 0.59% |
| Malignant tumor of the throat | 1 | 0.59% |
| Non-defined dementia | 1 | 0.59% |
| Obesity | 1 | 0.59% |
| Malignant tumor of the lung | 1 | 0.59% |
| Postoperative abnormality of the eye and its appendages | 1 | 0.59% |
| Heart failure | 1 | 0.59% |
| Non-toxic thyroid nodule | 1 | 0.59% |
| Other and abdominal pain | 1 | 0.59% |
| Stage III malignant neoplasm of the posterior wall of the hypopharynx | 1 | 0.59% |
| Malignancy of the pyriform sinus | 1 | 0.59% |
| Total | 170 | 100.00% |
| COVID-19-Associated | 51 | 30% |
| Not COVID-19-associated | 119 | 70% |
| Total | 170 | 100% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Orosz, L.; Sóki, J.; Kókai, D.; Burián, K. Corynebacterium striatum—Got Worse by a Pandemic? Pathogens 2022, 11, 685. https://doi.org/10.3390/pathogens11060685
Orosz L, Sóki J, Kókai D, Burián K. Corynebacterium striatum—Got Worse by a Pandemic? Pathogens. 2022; 11(6):685. https://doi.org/10.3390/pathogens11060685
Chicago/Turabian StyleOrosz, László, József Sóki, Dávid Kókai, and Katalin Burián. 2022. "Corynebacterium striatum—Got Worse by a Pandemic?" Pathogens 11, no. 6: 685. https://doi.org/10.3390/pathogens11060685
APA StyleOrosz, L., Sóki, J., Kókai, D., & Burián, K. (2022). Corynebacterium striatum—Got Worse by a Pandemic? Pathogens, 11(6), 685. https://doi.org/10.3390/pathogens11060685








