Occurrence and Phylogenetic Analysis of Avian Coronaviruses in Domestic Pigeons (Columba livia domestica) in Poland between 2016 and 2020
Abstract
:1. Introduction
2. Results
2.1. Prevalence of Coronaviral Genetic Material in Investigated Pigeons
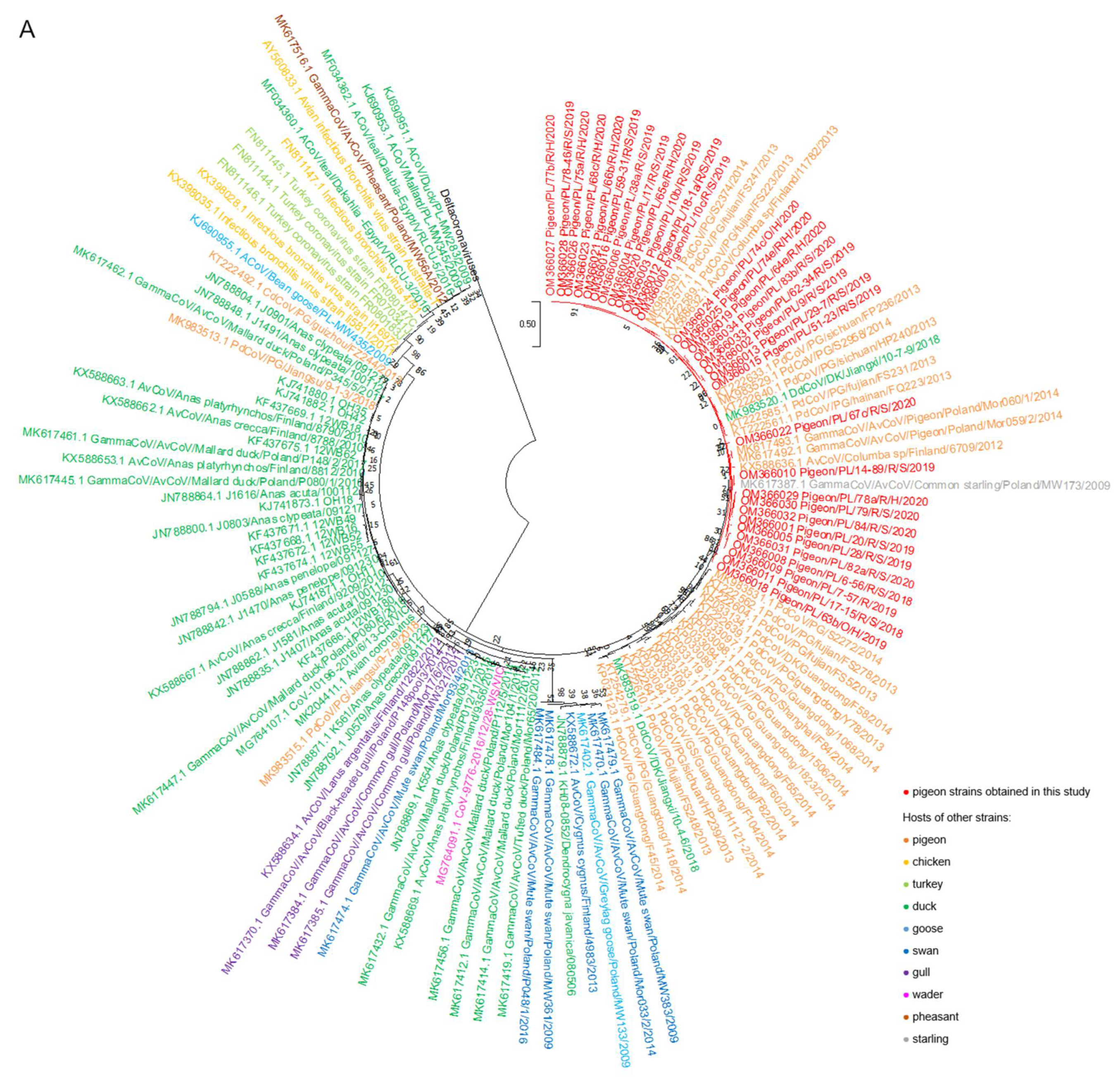
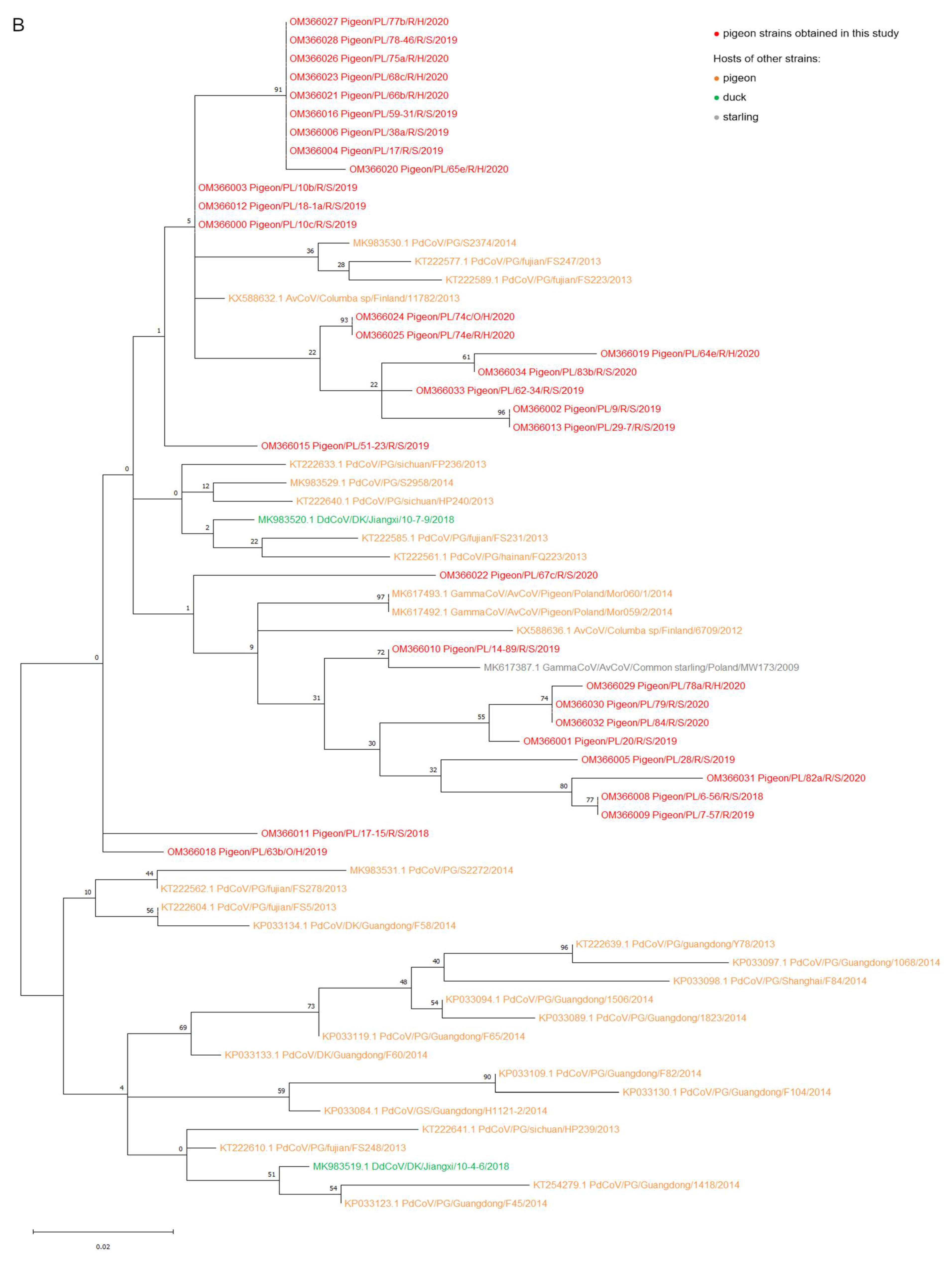
2.2. Phylogenetic Analysis
3. Discussion
4. Materials and Methods
4.1. Ethical Statement
4.2. Sample Collection
4.3. Molecular Detection of Coronaviruses
4.4. Sequence and Phylogenetic Analysis
4.5. Nucleotide Sequence Accession Numbers
4.6. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- ICTV Virus Taxonomy: 2020 Release. Available online: https://talk.ictvonline.org/taxonomy (accessed on 11 February 2022).
- Hepojoki, S.; Lindh, E.; Vapalahti, O.; Huovilainen, A. Prevalence and genetic diversity of coronaviruses in wild birds, Finland. Infect. Ecol. Epidemiol. 2017, 7, 1408360. [Google Scholar] [CrossRef] [PubMed]
- Wille, M.; Holmes, E.C. Wild birds as reservoirs for diverse and abundant gamma- and deltacoronaviruses. FEMS Microbiol. Rev. 2020, 44, 631–644. [Google Scholar] [CrossRef] [PubMed]
- Qian, D.H.; Zhu, G.J.; Wu, L.Z.; Hua, G.X. Isolation and characterization of a coronavirus from pigeons with pancreatitis. Am. J. Vet. Res. 2006, 67, 1575–1579. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.P.; Wong, E.Y.M.; Tsang, C.C.; Ahmed, S.S.; Au-Yeung, R.K.H.; Yuen, K.Y.; Wernery, U.; Woo, P.C.Y. Discovery and Sequence Analysis of Four Deltacoronaviruses from Birds in the Middle East Reveal Interspecies Jumping with Recombination as a Potential Mechanism for Avian-to-Avian and Avian-to-Mammalian Transmission. J. Virol. 2018, 92, e00265-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pantin-Jackwood, M.J.; Day, J.M.; Jackwood, M.W.; Spackman, E. Enteric viruses detected by molecular methods in commercial chicken and turkey flocks in the United States between 2005 and 2006. Avian Dis. 2008, 52, 235–244. [Google Scholar] [CrossRef] [Green Version]
- Domańska-Blicharz, K.; Bocian, Ł.; Lisowska, A.; Jacukowicz, A.; Pikuła, A.; Minta, Z. Cross-sectional survey of selected enteric viruses in Polish turkey flocks between 2008 and 2011. BMC Vet. Res. 2017, 13, 108. [Google Scholar] [CrossRef] [Green Version]
- Shapiro, M.D.; Domyan, E.T. Domestic pigeons. Curr. Biol. 2013, 23, R302–R303. [Google Scholar] [CrossRef] [Green Version]
- The Act of 11 March 2004 on the Protection of Animal Health and Combating Infectious Animal Diseases. Available online: http://isap.sejm.gov.pl/isap.nsf/DocDetails.xsp?id=WDU20040690625 (accessed on 15 February 2022). (In Polish)
- Terrestrial Code Online Access—OIE—World Organisation for Animal Health. Available online: https://www.oie.int/en/what-we-do/standards/codes-and-manuals/terrestrial-code-online-access (accessed on 15 February 2022).
- Coletti, M.; Franciosini, M.P.; Asdrubali, G.; Passamonti, F. Atrophy of the primary lymphoid organs of meat pigeons in Italy associated with circoviruslike particles in the bursa of Fabricius. Avian Dis. 2000, 44, 454–459. [Google Scholar] [CrossRef]
- Haunshi, S.; Basumatary, R.; Girish, P.S.; Doley, S.; Bardoloi, R.K.; Kumar, A. Identification of chicken, duck, pigeon and pig meat by species-specific markers of mitochondrial origin. Meat Sci. 2009, 83, 454–459. [Google Scholar] [CrossRef]
- Bu, Z.; Li, B.L.; Zhao, Z.H.; Wang, Q.; Chen, H.S.; Chen, W.B.; Wang, C. Resources of meat-type pigeon and the current breeding situation in China. China Anim. Husb. Vet. Med. 2010, 37, 116–119. (In Chinese) [Google Scholar]
- Ye, M.; Xu, M.; Chen, C.; He, Y.; Ding, M.; Ding, X.; Wei, W.; Yang, S.; Zhou, B. Expression analyses of candidate genes related to meat quality traits in squabs from two breeds of meat-type pigeon. J. Anim. Physiol. Anim. Nutr. 2018, 102, 727–735. [Google Scholar] [CrossRef] [PubMed]
- Gazda, M.A.; Andrade, P.; Afonso, S.; Dilyte, J.; Archer, J.P.; Lopes, R.J.; Faria, R.; Carneiro, M. Signatures of Selection on Standing Genetic Variation Underlie Athletic and Navigational Performance in Racing Pigeons. Mol. Biol. Evol. 2018, 35, 1176–1189. [Google Scholar] [CrossRef]
- Felippe, P.A.; da Silva, L.H.; Santos, M.M.; Spilki, F.R.; Arns, C.W. Genetic diversity of avian infectious bronchitis virus isolated from domestic chicken flocks and coronaviruses from feral pigeons in Brazil between 2003 and 2009. Avian Dis. 2010, 54, 1191–1196. [Google Scholar] [CrossRef] [PubMed]
- Pohl, R. The histopathogenesis of the nephrosis-nephritis syndrome. Avian Pathol. 1974, 3, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Cavanagh, D. Coronavirus avian infectious bronchitis virus. Vet. Res. 2007, 38, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Nagaraja, K.V.; Pomeroy, B.S. Coronaviral Enteritis of Turkeys (Bluecomb Disease). In Diseases of Poultry, 10th ed.; Calnek, B.W., Barnes, H.J., Beard, C.W., McDougald, L.R., Saif, Y.M., Eds.; Iowa State University Press: Ames, IA, USA, 1997; pp. 686–692. [Google Scholar]
- Cavanagh, D. Coronaviruses in poultry and other birds. Avian Pathol. 2005, 34, 439–448. [Google Scholar] [CrossRef]
- Chu, D.K.; Leung, C.Y.; Gilbert, M.; Joyner, P.H.; Ng, E.M.; Tse, T.M.; Guan, Y.; Peiris, J.S.; Poon, L.L. Avian coronavirus in wild aquatic birds. J. Virol. 2011, 85, 12815–12820. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.R.; Oem, J.K. Surveillance of avian coronaviruses in wild bird populations of Korea. J. Wildl. Dis. 2014, 50, 964–968. [Google Scholar] [CrossRef] [Green Version]
- De Sales Lima, F.E.; Gil, P.; Pedrono, M.; Minet, C.; Kwiatek, O.; Campos, F.S.; Spilki, F.R.; Roehe, P.M.; Franco, A.C.; Maminiaina, O.F.; et al. Diverse gammacoronaviruses detected in wild birds from Madagascar. Eur. J. Wildl. Res. 2015, 61, 635–639. [Google Scholar] [CrossRef] [Green Version]
- Wille, M.; Muradrasoli, S.; Nilsson, A.; Järhult, J.D. High Prevalence and Putative Lineage Maintenance of Avian Coronaviruses in Scandinavian Waterfowl. PLoS ONE 2016, 11, e0150198. [Google Scholar] [CrossRef] [Green Version]
- Chamings, A.; Nelson, T.M.; Vibin, J.; Wille, M.; Klaassen, M.; Alexandersen, S. Detection and characterisation of coronaviruses in migratory and non-migratory Australian wild birds. Sci. Rep. 2018, 8, 5980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbosa, C.M.; Durigon, E.L.; Thomazelli, L.M.; Ometto, T.; Marcatti, R.; Nardi, M.S.; de Aguiar, D.M.; Pinho, J.B.; Petry, M.V.; Neto, I.S.; et al. Divergent coronaviruses detected in wild birds in Brazil, including a central park in São Paulo. Braz. J. Microbiol. 2019, 50, 547–556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Domańska-Blicharz, K.; Miłek-Krupa, J.; Pikuła, A. Diversity of Coronaviruses in Wild Representatives of the Aves Class in Poland. Viruses 2021, 13, 1497. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Giner, A.; Blond, F.; Bru, T.; Vila, R. Bronquitis Infecciosa Aviar: Genotipos Identificados en España del 2012 al 2016. LIV Symposium Científico de Avicultura. 2017. Available online: https://www.wpsa-aeca.es/articulo.php?id_articulo=4535 (accessed on 26 May 2021). (In Spanish).
- Stenzel, T.A.; Pestka, D.; Tykałowski, B.; Śmiałek, M.; Koncicki, A. Epidemiological investigation of selected pigeon viral infections in Poland. Vet. Rec. 2012, 171, 562. [Google Scholar] [CrossRef]
- Rubbenstroth, D.; Peus, E.; Schramm, E.; Kottmann, D.; Bartels, H.; McCowan, C.; Schulze, C.; Akimkin, V.; Fischer, N.; Wylezich, C.; et al. Identification of a novel clade of group A rotaviruses in fatally diseased domestic pigeons in Europe. Transbound. Emerg. Dis. 2019, 66, 552–561. [Google Scholar] [CrossRef]
- Zhuang, Q.; Liu, S.; Zhang, X.; Jiang, W.; Wang, K.; Wang, S.; Peng, C.; Hou, G.; Li, J.; Yu, X.; et al. Surveillance and taxonomic analysis of the coronavirus dominant in pigeons in China. Transbound. Emerg. Dis. 2020, 67, 1981–1990. [Google Scholar] [CrossRef]
- Jonassen, C.M.; Kofstad, T.; Larsen, I.L.; Løvland, A.; Handeland, K.; Follestad, A.; Lillehaug, A. Molecular identification and characterization of novel coronaviruses infecting graylag geese (Anser anser), feral pigeons (Columbia livia) and mallards (Anas platyrhynchos). J. Gen. Virol. 2005, 86, 1597–1607. [Google Scholar] [CrossRef]
- Ling, Y.; Chen, H.; Chen, X.; Yang, X.; Yang, J.; Bavoil, P.M.; He, C. Epidemiology of Chlamydia psittaci Infection in Racing Pigeons and Pigeon Fanciers in Beijing, China. Zoonoses Public Health 2015, 62, 401–406. [Google Scholar] [CrossRef]
- Eckerle, L.D.; Lu, X.; Sperry, S.M.; Choi, L.; Denison, M.R. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J. Virol. 2007, 81, 12135–12144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woo, P.C.; Lau, S.K.; Huang, Y.; Yuen, K.Y. Coronavirus diversity, phylogeny and interspecies jumping. Exp. Biol. Med. 2009, 234, 1117–1127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanjuán, R.; Nebot, M.R.; Chirico, N.; Mansky, L.M.; Belshaw, R. Viral mutation rates. J. Virol. 2010, 84, 9733–9748. [Google Scholar] [CrossRef] [Green Version]
- Nikolaidis, M.; Markoulatos, P.; Van de Peer, Y.; Oliver, S.G.; Amoutzias, G.D. The Neighborhood of the Spike Gene Is a Hotspot for Modular Intertypic Homologous and Nonhomologous Recombination in Coronavirus Genomes. Mol. Biol. Evol. 2022, 39, msab292. [Google Scholar] [CrossRef] [PubMed]
- Nikolaidis, M.; Papakyriakou, A.; Chlichlia, K.; Markoulatos, P.; Oliver, S.G.; Amoutzias, G.D. Comparative Analysis of SARS-CoV-2 Variants of Concern, Including Omicron, Highlights Their Common and Distinctive Amino Acid Substitution Patterns, Especially at the Spike ORF. Viruses 2022, 14, 707. [Google Scholar] [CrossRef] [PubMed]
- Mihindukulasuriya, K.A.; Wu, G.; St Leger, J.; Nordhausen, R.W.; Wang, D. Identification of a novel coronavirus from a beluga whale by using a panviral microarray. J. Virol. 2008, 82, 5084–5088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lednicky, J.A.; Tagliamonte, M.S.; White, S.K.; Elbadry, M.A.; Alam, M.M.; Stephenson, C.J.; Bonny, T.S.; Loeb, J.C.; Telisma, T.; Chavannes, S.; et al. Independent infections of porcine deltacoronavirus among Haitian children. Nature 2021, 600, 133–137. [Google Scholar] [CrossRef]
- Werner, O.; Römer-Oberdörfer, A.; Köllner, B.; Manvell, R.J.; Alexander, D.J. Characterization of avian paramyxovirus type 1 strains isolated in Germany during 1992 to 1996. Avian Pathol. 1999, 28, 79–88. [Google Scholar] [CrossRef] [Green Version]
- Kommers, G.D.; King, D.J.; Seal, B.S.; Brown, C.C. Virulence of Pigeon-Origin Newcastle Disease Virus Isolates for Domestic Chickens. Avian Dis. 2001, 45, 906–921. [Google Scholar] [CrossRef]
- Mansour, S.M.; El Bakrey, R.M.; Ali, H.; Knudsen, D.E.; Eid, A.A. Natural infection with highly pathogenic avian influenza virus H5N1 in domestic pigeons (Columba livia) in Egypt. Avian Pathol. 2014, 43, 319–324. [Google Scholar] [CrossRef]
- Shao, H.; Li, J.; Yuan, H.; Ji, L.; Zhang, J.; Jin, W.; Qian, K.; Ye, J.; Qin, A. Isolation and Molecular Characteristics of a CIAV Isolate from Pigeons, China. Front. Vet. Sci. 2021, 8, 669154. [Google Scholar] [CrossRef] [PubMed]

| CoV | |||
|---|---|---|---|
| Group of Pigeons | n | n+ | % |
| Adult | 116 | 23 | 19.83 a |
| Young | 99 | 34 | 34.34 b |
| Asymptomatic | 87 | 17 | 19.54 |
| Symptomatic | 128 | 40 | 31.25 |
| Adult asymptomatic | 73 | 14 | 19.18 |
| Adult symptomatic | 43 | 9 | 20.93 |
| Young asymptomatic | 14 | 3 | 21.43 |
| Young symptomatic | 85 | 31 | 36.47 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Łukaszuk, E.; Dziewulska, D.; Stenzel, T. Occurrence and Phylogenetic Analysis of Avian Coronaviruses in Domestic Pigeons (Columba livia domestica) in Poland between 2016 and 2020. Pathogens 2022, 11, 646. https://doi.org/10.3390/pathogens11060646
Łukaszuk E, Dziewulska D, Stenzel T. Occurrence and Phylogenetic Analysis of Avian Coronaviruses in Domestic Pigeons (Columba livia domestica) in Poland between 2016 and 2020. Pathogens. 2022; 11(6):646. https://doi.org/10.3390/pathogens11060646
Chicago/Turabian StyleŁukaszuk, Ewa, Daria Dziewulska, and Tomasz Stenzel. 2022. "Occurrence and Phylogenetic Analysis of Avian Coronaviruses in Domestic Pigeons (Columba livia domestica) in Poland between 2016 and 2020" Pathogens 11, no. 6: 646. https://doi.org/10.3390/pathogens11060646
APA StyleŁukaszuk, E., Dziewulska, D., & Stenzel, T. (2022). Occurrence and Phylogenetic Analysis of Avian Coronaviruses in Domestic Pigeons (Columba livia domestica) in Poland between 2016 and 2020. Pathogens, 11(6), 646. https://doi.org/10.3390/pathogens11060646






