Dynamics of Mycobacterium tuberculosis Lineages in Oman, 2009 to 2018
Abstract
1. Introduction
2. Material and Methods
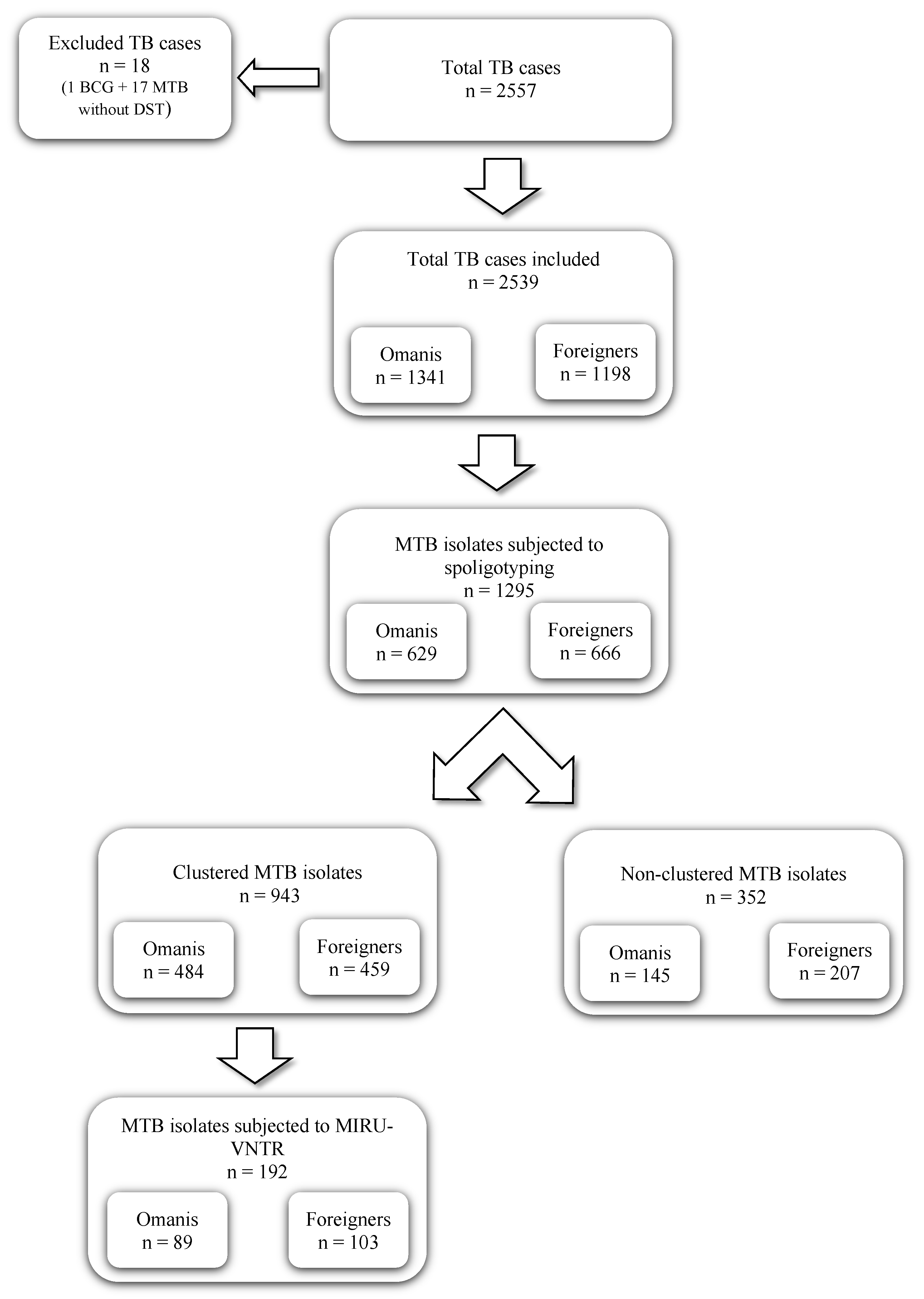
2.1. Mycobacterium tuberculosis Isolates
2.2. Spoligotyping and MIRU-VNTR Typing
2.3. Data Analysis
3. Results
3.1. Study Subjects
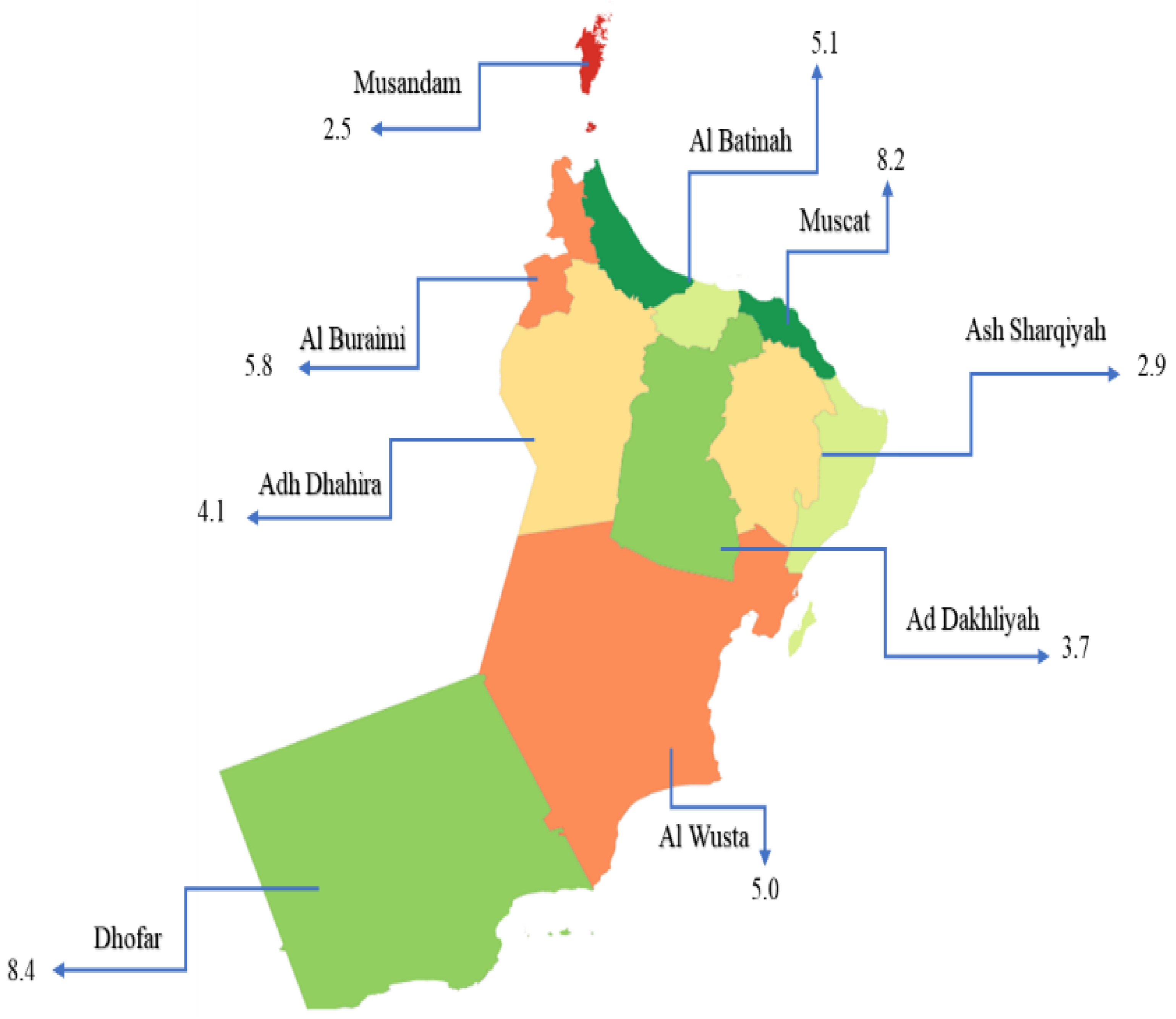
3.2. Spatiotemporal Distribution of TB, 2009–2018, in Oman
3.3. Distribution of M. tuberculosis Lineages
3.4. Distribution of M. tuberculosis Lineages among Omanis and Foreigners
3.5. Clustering of M. tuberculosis Lineages
3.6. MIRU-VNTR Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO. Global Tuberculosis Report 2020; WHO: Geneva, Switzerland, 2020. [Google Scholar]
- WHO. Global Tuberculosis Report 2021; WHO: Geneva, Switzerland, 2021. [Google Scholar]
- Statista. Incidence Rate of Tuberculosis in the Gulf Cooperation Council in 2018, by Country. Available online: https://www.statista.com/statistics/681396/gcc-incidence-rate-of-tuberculosis-inhabitants-by-country/ (accessed on 30 April 2022).
- Ahmed, M.M.; Velayati, A.A.; Mohammed, S.H. Epidemiology of Multidrug-Resistant, Extensively Drug Resistant, and Totally Drug Resistant Tuberculosis in Middle East Countries. Int. J. Mycobacteriology 2016, 5, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Al Awaidy, S.T. Tuberculosis Elimination in Oman: Winning the War on the Disease. ERJ Open Res. 2018, 4, 121–2018. [Google Scholar] [CrossRef] [PubMed]
- Al Abri, S.; Kasaeva, T.; Migliori, G.B.; Goletti, D.; Zenner, D.; Denholm, J.; Al Maani, A.; Cirillo, D.M.; Schön, T.; Lillebæk, T.; et al. Tools to Implement the World Health Organization End TB Strategy: Addressing Common Challenges in High and Low Endemic Countries. Int. J. Infect. Dis. 2020, 92, S60–S68. [Google Scholar] [CrossRef] [PubMed]
- Chaabna, K.; Cheema, S.; Mamtani, R. Migrants, Healthy Worker Effect, and Mortality Trends in the Gulf Cooperation Council Countries. PLoS ONE 2017, 12, e0179711. [Google Scholar] [CrossRef] [PubMed]
- Al-Maniri, A.; Singh, J.P.; Al-Rawas, O.; Al Busaidi, S.; Al Balushi, L.; Ahmed, I.; Al Mahruqi, S.; Haile, M.; Diwan, V.K.; Hoffner, S. A Snapshot of the Biodiversity and Clustering of Mycobacterium Tuberculosis in Oman Using Spoligotyping. Int. J. Tuberc. Lung Dis. 2010, 8, 994–1000. [Google Scholar]
- NCSI Population. Available online: https://data.gov.om/OMPOP2016/population (accessed on 30 April 2022).
- Warren, R.; de Kock, M.; Engelke, E.; Myburgh, R.; Gey van Pittius, N.; Victor, T.; van Helden, P. Safe Mycobacterium Tuberculosis DNA Extraction Method That Does Not Compromise Integrity. J. Clin. Microbiol. 2006, 44, 254–256. [Google Scholar] [CrossRef] [PubMed]
- Kamerbeek, J.; Schouls, L.; Kolk, A.; van Agterveld, M.; van Soolingen, D.; Kuijper, S.; Bunschoten, A.; Molhuizen, H.; Shaw, R.; Goyal, M.; et al. Simultaneous Detection and Strain Differentiation of Mycobacterium Tuberculosis for Diagnosis and Epidemiology. J. Clin. Microbiol. 1997, 35, 907–914. [Google Scholar] [CrossRef]
- Demay, C.; Liens, B.; Burguière, T.; Hill, V.; Couvin, D.; Millet, J.; Mokrousov, I.; Sola, C.; Zozio, T.; Rastogi, N. SITVITWEB—A Publicly Available International Multimarker Database for Studying Mycobacterium Tuberculosis Genetic Diversity and Molecular Epidemiology. Infect. Genet. Evol. 2012, 12, 755–766. [Google Scholar] [CrossRef]
- Supply, P.; Allix, C.; Lesjean, S.; Cardoso-Oelemann, M.; Rüsch-Gerdes, S.; Willery, E.; Savine, E.; De Haas, P.; Van Deutekom, H.; Roring, S. Proposal for Standardization of Optimized Mycobacterial Interspersed Repetitive Unit-Variable-Number Tandem Repeat Typing of Mycobacterium Tuberculosis. J. Clin. Microbiol. 2006, 44, 4498–4510. [Google Scholar] [CrossRef]
- Gwitira, I.; Karumazondo, N.; Shekede, M.D.; Sandy, C.; Siziba, N.; Chirenda, J. Spatial Patterns of Pulmonary Tuberculosis (TB) Cases in Zimbabwe from 2015 to 2018. PLoS ONE 2021, 16, e0249523. [Google Scholar] [CrossRef]
- van Deutekom, H.; Hoijng, S.P.; de Haas, P.E.W.; Langendam, M.W.; Horsman, A.; van Soolingen, D.; Coutinho, R.A. Clustered Tuberculosis Cases. Am. J. Respir. Crit. Care Med. 2004, 169, 806–810. [Google Scholar] [CrossRef] [PubMed]
- Weniger, T.; Krawczyk, J.; Supply, P.; Niemann, S.; Harmsen, D. MIRU-VNTRplus: A Web Tool for Polyphasic Genotyping of Mycobacterium Tuberculosis Complex Bacteria. Nucleic Acids Res. 2010, 38, W326–W331. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Al Mayahi, Z.K.; AlAufi, I.; Al Ghufaili, B.; Al Balushi, Z.; Al Mughazwi, Z.; Mohammed, E.; Essa, R.; Yousif, H.M.; Al Mayahi, A.K.; Al Hattali, A.; et al. Epidemiological Profile and Surveillance Activity of Tuberculosis in South Batinah, Oman, 2017 and 2018. Int. J. Mycobacteriology 2020, 9, 39–47. [Google Scholar]
- Sultana, Z.Z.; Hoque, F.U.; Beyene, J.; Akhlak-Ul-Islam, M.; Khan, M.H.R.; Ahmed, S.; Hawlader, D.H.; Hossain, A. HIV Infection and Multidrug Resistant Tuberculosis: A Systematic Review and Meta-Analysis. BMC Infect. Dis. 2021, 21, 51. [Google Scholar] [CrossRef]
- Han, Z.; Li, J.; Sun, G.; Gu, K.; Zhang, Y.; Yao, H.; Jiang, Y. Transmission of Multidrug-Resistant Tuberculosis in Shimen Community in Shanghai, China: A Molecular Epidemiology Study. BMC Infect. Dis. 2021, 21, 1118. [Google Scholar] [CrossRef]
- Trauer, J.M.; Dodd, P.J.; Gomes, M.G.M.; Gomez, G.B.; Houben, R.M.G.J.; McBryde, E.S.; Melsew, Y.A.; Menzies, N.A.; Arinaminpathy, N.; Shrestha, S.; et al. The Importance of Heterogeneity to the Epidemiology of Tuberculosis. Clin. Infect. Dis. 2019, 69, 159–166. [Google Scholar] [CrossRef]
- Gutierrez, M.C.; Ahmed, N.; Willery, E.; Narayanan, S.; Hasnain, S.E.; Chauhan, D.S.; Katoch, V.M.; Vincent, V.; Locht, C.; Supply, P. Predominance of Ancestral Lineages of Mycobacterium Tuberculosis in India. Emerg. Infect. Dis. 2006, 12, 1367–1374. [Google Scholar] [CrossRef]
- Parwati, I.; van Crevel, R.; van Soolingen, D. Possible Underlying Mechanisms for Successful Emergence of the Mycobacterium Tuberculosis Beijing Genotype Strains. Lancet Infect. Dis. 2010, 10, 103–111. [Google Scholar] [CrossRef]
- Devi, K.R.; Pradhan, J.; Bhutia, R.; Dadul, P.; Sarkar, A.; Gohain, N.; Narain, K. Molecular Diversity of Mycobacterium Tuberculosis Complex in Sikkim, India and Prediction of Dominant Spoligotypes Using Artificial Intelligence. Sci. Rep. 2021, 11, 7365. [Google Scholar] [CrossRef]
- Uddin, M.K.M.; Ather, M.F.; Rahman, A.; Nasrin, R.; Rahman, S.M.M.; Kabir, S.; Chedid, C.; Ahmed, S.; Banu, S. Genetic Diversity and Characterization of M. Tuberculosis Isolates Causing Extrapulmonary Tuberculosis in Bangladesh. Infect. Genet. Evol. 2021, 95, 105052. [Google Scholar] [CrossRef]
- Chihota, V.N.; Niehaus, A.; Streicher, E.M.; Wang, X.; Sampson, S.L.; Mason, P.; Källenius, G.; Mfinanga, S.G.; Pillay, M.; Klopper, M.; et al. Geospatial Distribution of Mycobacterium Tuberculosis Genotypes in Africa. PLoS ONE 2018, 13, e0200632. [Google Scholar]
- Al-Hajoj, S.; Varghese, B.; Shoukri, M.M.; Al-Omari, R.; Al-Herbwai, M.; Alrabiah, F.; Alrajhi, A.A.; Abuljadayel, N.; Al-Thawadi, S.; Zumla, A.; et al. Epidemiology of Antituberculosis Drug Resistance in Saudi Arabia: Findings of the First National Survey. Antimicrob. Agents Chemother. 2013, 57, 2161–2166. [Google Scholar] [CrossRef] [PubMed]
- Wippel, S. Oman and the Indian Ocean Rim–Economic Integration across Conventional Meta-Regions. In Regionalizing Oman; Springer: Berlin/Heidelberg, Germany, 2013; pp. 159–183. [Google Scholar]
- Gĩthĩnji, M. wa Dhows to Planes: Trade Relations between the Arabian Gulf and Africa and Their Impacts on Development. J. Afr. Dev. 2010, 12, 131–154. [Google Scholar] [CrossRef]
- Dayalan, D. Ancient Seaports on the Western Coast of India: The Hub of the Maritime Silk Route Network. Acta Via Serica 2018, 3, 49–72. [Google Scholar]
- Awadalla, P.; Walliker, D.; Babiker, H.; Mackinnon, M. The Question of Plasmodium Falciparum Population Structure. Trends Parasitol. 2001, 17, 351–353. [Google Scholar] [CrossRef]
- Fenner, L.; Gagneux, S.; Helbling, P.; Battegay, M.; Rieder, H.L.; Pfyffer, G.E.; Zwahlen, M.; Furrer, H.; Siegrist, H.H.; Fehr, J. Mycobacterium Tuberculosis Transmission in a Country with Low Tuberculosis Incidence: Role of Immigration and HIV Infection. J. Clin. Microbiol. 2012, 50, 388–395. [Google Scholar] [CrossRef]
- Gagneux, S.; Small, P.M. Global Phylogeography of Mycobacterium Tuberculosis and Implications for Tuberculosis Product Development. Lancet Infect. Dis. 2007, 7, 328–337. [Google Scholar] [CrossRef]
- Barniol, J.; Niemann, S.; Louis, V.R.; Brodhun, B.; Dreweck, C.; Richter, E.; Becher, H.; Haas, W.; Junghanss, T. Transmission Dynamics of Pulmonary Tuberculosis between Autochthonous and Immigrant Sub-Populations. BMC Infect. Dis. 2009, 9, 197. [Google Scholar] [CrossRef]
- Dahle, U.R.; Eldholm, V.; Winje, B.A.; Mannsåker, T.; Heldal, E. Impact of Immigration on the Molecular Epidemiology of Mycobacterium Tuberculosis in a Low-Incidence Country. Am. J. Respir. Crit. Care Med. 2007, 176, 930–935. [Google Scholar] [CrossRef]
- Lillebaek, T.; Andersen, A.B.; Bauer, J.; Dirksen, A.; Glismann, S.; de Haas, P.; Kok-Jensen, A. Risk of Mycobacterium Tuberculosis Transmission in a Low-Incidence Country Due to Immigration from High-Incidence Areas. J. Clin. Microbiol. 2001, 39, 855–861. [Google Scholar] [CrossRef]
- Varghese, B.; Supply, P.; Allix-Béguec, C.; Shoukri, M.; Al-Omari, R.; Herbawi, M.; Al-Hajoj, S. Admixed Phylogenetic Distribution of Drug Resistant Mycobacterium Tuberculosis in Saudi Arabia. PLoS ONE 2013, 8, e55598. [Google Scholar] [CrossRef] [PubMed]
- Marx, F.M.; Fiebig, L.; Hauer, B.; Brodhun, B.; Glaser-Paschke, G.; Magdorf, K.; Haas, W. Higher Rate of Tuberculosis in Second Generation Migrants Compared to Native Residents in a Metropolitan Setting in Western Europe. PLoS ONE 2015, 10, e0119693. [Google Scholar] [CrossRef] [PubMed]
- Aldridge, R.W.; Zenner, D.; White, P.J.; Williamson, E.J.; Muzyamba, M.C.; Dhavan, P.; Mosca, D.; Thomas, H.L.; Lalor, M.K.; Abubakar, I.; et al. Tuberculosis in Migrants Moving from High-Incidence to Low-Incidence Countries: A Population-Based Cohort Study of 519 955 Migrants Screened before Entry to England, Wales, and Northern Ireland. Lancet 2016, 388, 2510–2518. [Google Scholar] [CrossRef]

| Total | 2009 n = 136 | 2010 n = 35 | 2011 n = 50 | 2012 n = 56 | 2013 n = 255 | 2014 n = 156 | 2015 n = 150 | 2016 n = 148 | 2017 n = 195 | 2018 n = 114 | p-Value | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EAI | 318 (30.9%) | 44 (32.4%) | 7 (20.0%) | 9 (18.0%) | 22 (39.3%) | 61 (23.9%) | 39 (25.0%) | 41 (27.3%) | 41 (27.7%) | 31 (15.9%) | 23 (20.2%) | 0.019 |
| Beijing | 88 (8.5%) | 3 (2.2%) | 3 (8.6%) | 8 (16.0%) | 4 (7.1%) | 14 (5.5%) | 9 (5.8%) | 9 (6.0%) | 15 (10.1%) | 19 (9.7%) | 4 (3.5%) | 0.022 |
| CAS | 310 (30.1%) | 32 (23.5%) | 7 (20.0%) | 11 (22.0%) | 18 (32.1%) | 67 (26.3%) | 39 (25.0%) | 37 (24.7%) | 28 (18.9%) | 50 (25.6%) | 21 (18.4%) | 0.629 |
| T | 154 (14.9%) | 12 (8.8%) | 6 (17.1%) | 6 (12.0%) | 4 (7.1%) | 28 (11.0%) | 22 (14.1%) | 21 (14.0%) | 18 (12.2%) | 20 (10.3%) | 17 (14.9%) | 0.613 |
| LAM | 64 (6.2%) | 15 (11.0%) | 1 (2.9%) | 3 (6.0%) | 3 (5.4%) | 13 (5.1%) | 7 (4.5%) | 6 (4.0%) | 5 (3.4%) | 6 (3.1%) | 5 (4.4%) | 0.585 |
| H | 42 (4.1%) | 4 (2.9%) | 2 (5.7%) | 0 | 2 (3.6%) | 13 (5.1%) | 4 (2.6%) | 4 (2.7%) | 6 (4.1%) | 5 (2.6%) | 2 (1.8%) | 0.320 |
| Others 1 | 54 (4.2%) | 6 (4.4%) | 1 (2.9%) | 2 (4.0%) | 1 (1.8%) | 9 (3.5%) | 5 (3.2%) | 8 (5.3%) | 3 (2.0%) | 10 (5.1%) | 9 (7.9%) | 0.011 |
| Orphan | 265 (20.5%) | 20 (14.7%) | 8 (22.9%) | 11 (22.0%) | 2 (3.6%) | 50 (19.6%) | 31 (19.9%) | 24 (16.0%) | 32 (21.6%) | 54 (27.7%) | 33 (28.9%) | 0.002 |
| Foreigners n (%) | Omani n (%) | Total | p-Value * | |
|---|---|---|---|---|
| EAI | 145 (21.8%) | 173 (27.5%) | 318 (24.5%) | 0.017 |
| Beijing | 56 (8.4%) | 32 (5.1%) | 88 (6.8%) | 0.018 |
| CAS | 133 (20.0%) | 177 (28.1%) | 310 (23.9%) | 0.001 |
| T | 78 (11.7%) | 76 (12.1%) | 154 (11.9%) | 0.837 |
| H | 31 (4.7%) | 11 (1.7%) | 42 (3.2%) | 0.003 |
| LAM | 28 (4.2%) | 36 (5.7%) | 64 (4.9%) | 0.207 |
| Others | 40 (6.0%) | 14 (2.2%) | 54 (4.2%) | 0.001 |
| Orphan | 155 (23.3%) | 110 (17.5%) | 265 (20.5%) | 0.010 |
| Total | 666 | 629 | 1295 |
| Variable | Clustered | Non-Clustered | p-Value | |
|---|---|---|---|---|
| n (%) | n (%) | |||
| Year | 2009 | 73 (55.7) | 58 (44.3) | 0.385 |
| 2010 | 6 (17.1) | 29 (82.9) | ||
| 2011 | 20 (40.0) | 30 (60.0) | ||
| 2012 | 25 (44.6) | 31 (55.4) | ||
| 2013 | 151 (59.2) | 104 (40.8) | ||
| 2014 | 80 (53.7) | 69 (46.3) | ||
| 2015 | 95 (65.5) | 50 (34.5) | ||
| 2016 | 73 (56.1) | 57 (43.9) | ||
| 2017 | 87 (50.6) | 85 (49.4) | ||
| 2018 | 42 (40.0%) | 63 (60.0%) | ||
| Sex | Male | 556 (70.4) | 234 (29.6) | 0.279 |
| Female | 367 (73.3) | 134 (26.7) | ||
| Nationality | Omani | 469 (74.4) | 160 (31.2) | 0.021 |
| Foreigners | 458 (68.8) | 208 (25.6) | ||
| Age group | Child | 9 (52.9) | 8 (47.1) | 0.174 |
| Adult | 919 (71.8) | 366 (28.5) | ||
| Area of residence | Muscat | 375 (68.6) | 172 (31.4) | 0.300 |
| Al Batinah | 206 (74.9) | 69 (25.1) | ||
| Dhofar | 118 (77.1) | 35 (22.9) | ||
| Al Dakhilia | 59 (71.1) | 24 (28.9) | ||
| Al Sharqiya | 62 (70.5) | 26 (29.5) | ||
| Al Buraimi | 19 (61.3) | 12 (38.7) | ||
| Al Wusta | 7 (87.5) | 1 (12.5) | ||
| Al Dhahira | 35 (76.1) | 11 (23.9) | ||
| Musandam | 4 (80.0) | 1 (20.0) | ||
| DST profiles | Sensitive | 747 (71.6) | 297 (28.4) | 0.078 |
| MonoR | 125 (69.1) | 56 (30.9) | ||
| PolyR | 20 (76.9) | 6 (23.1) | ||
| MDR | 31 (88.6) | 4 (11.4) | ||
| Sub-lineages | CAS | 288 (89.7) | 33 (10.3) | <0.05 |
| EAI | 274 (83.8) | 53 (16.2) | ||
| Beijing | 82 (93.2) | 6 (6.8) | ||
| H | 26 (61.9) | 16 (38.1) | ||
| LAM | 54 (77.1) | 16 (22.9) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Mahrouqi, S.; Ahmed, R.; Al-Azri, S.; Al-Hamidhi, S.; Balkhair, A.A.; Al-Jardani, A.; Al-Fahdi, A.; Al-Balushi, L.; Al-Zadjali, S.; Adikaram, C.; et al. Dynamics of Mycobacterium tuberculosis Lineages in Oman, 2009 to 2018. Pathogens 2022, 11, 541. https://doi.org/10.3390/pathogens11050541
Al-Mahrouqi S, Ahmed R, Al-Azri S, Al-Hamidhi S, Balkhair AA, Al-Jardani A, Al-Fahdi A, Al-Balushi L, Al-Zadjali S, Adikaram C, et al. Dynamics of Mycobacterium tuberculosis Lineages in Oman, 2009 to 2018. Pathogens. 2022; 11(5):541. https://doi.org/10.3390/pathogens11050541
Chicago/Turabian StyleAl-Mahrouqi, Sara, Reham Ahmed, Saleh Al-Azri, Salama Al-Hamidhi, Abdullah A. Balkhair, Amina Al-Jardani, Amira Al-Fahdi, Laila Al-Balushi, Samia Al-Zadjali, Chamila Adikaram, and et al. 2022. "Dynamics of Mycobacterium tuberculosis Lineages in Oman, 2009 to 2018" Pathogens 11, no. 5: 541. https://doi.org/10.3390/pathogens11050541
APA StyleAl-Mahrouqi, S., Ahmed, R., Al-Azri, S., Al-Hamidhi, S., Balkhair, A. A., Al-Jardani, A., Al-Fahdi, A., Al-Balushi, L., Al-Zadjali, S., Adikaram, C., Al-Marhoubi, A., Gadalla, A., & Babiker, H. A. (2022). Dynamics of Mycobacterium tuberculosis Lineages in Oman, 2009 to 2018. Pathogens, 11(5), 541. https://doi.org/10.3390/pathogens11050541







