Microarray Profiling and Co-Expression Network Analysis of LncRNAs and mRNAs in Acute Respiratory Distress Syndrome Mouse Model
Abstract
:1. Introduction
2. Results
2.1. Establishment and Assessment of the ARDS Model
2.2. Differentially Expressed LncRNA and mRNAs in ALI
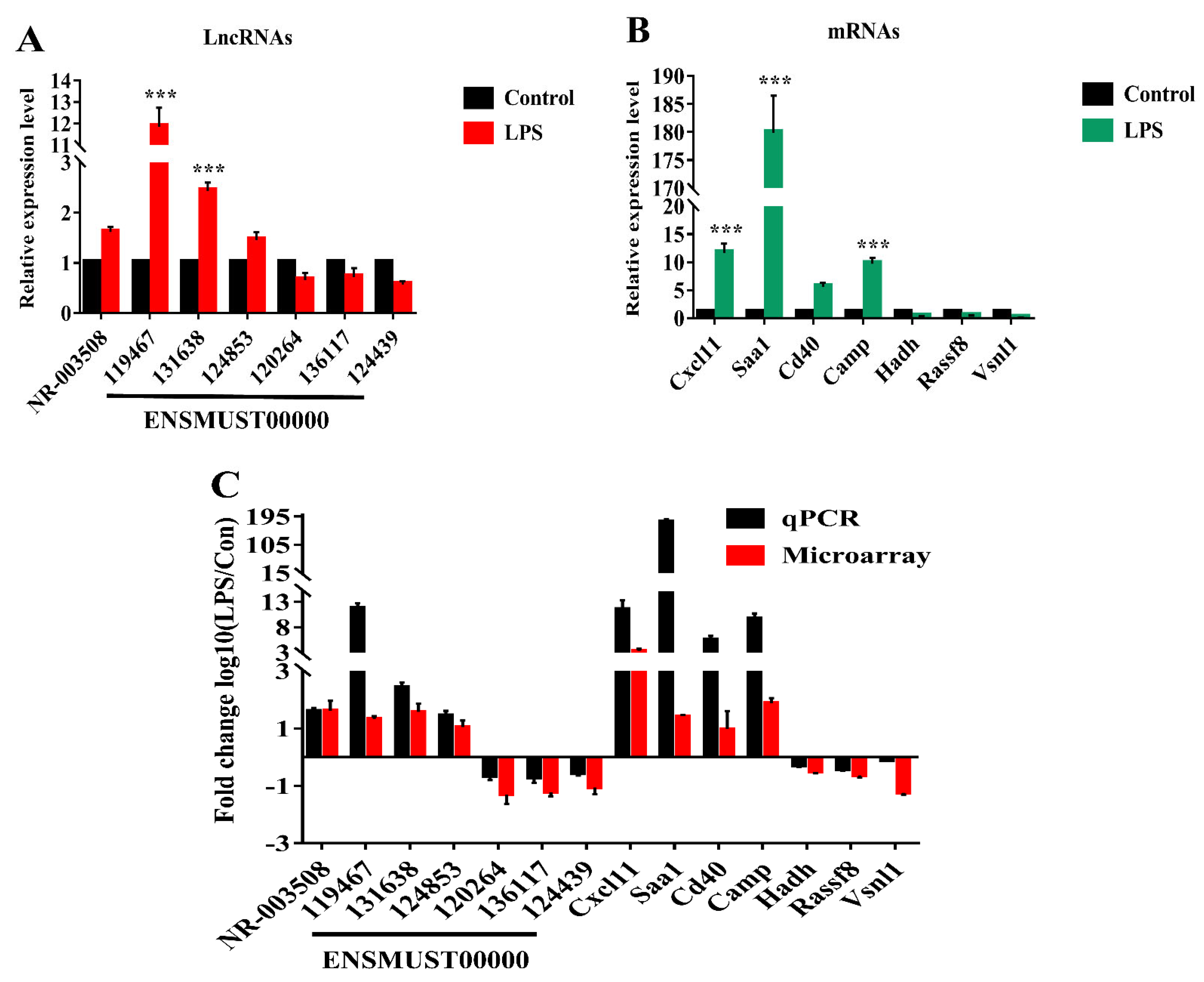
2.3. Verification of LncRNAs and mRNAs by qRT-PCR
2.4. GO and KEGG Enrichment Analysis of DE LncRNA Target Genes and DE mRNAs
2.5. Co-Expression Correlations between mRNAs and LncRNAs
2.6. si-RNA Targeting NR_003508 Decreased Cell Necroptosis in RAW264.7 Cell Induced by LPS
3. Discussion
4. Materials and Methods
4.1. Experimental Protocols for Acute Lung Injury Models
4.2. Bronchoalveolar Lavage Fluid (BALF) Collection and Analysis
4.3. Enzyme-Linked Immunosorbent Assay (ELISA) for Mouse Interleukin-6 (IL-6), Interleukin-1β (IL-1β), and Tumor Necrosis Factor-Alpha (TNF-α)
4.4. Histopathological Evaluation
4.5. An Analysis of Differentially Expressed LncRNAs and mRNAs in ALI
4.6. Purification of Total RNA from the Male Kunming Mouse Tissue and RAW264.7 Cells
4.7. Quantitative Real-Time PCR Validation
4.8. GO and KEGG Pathway Analysis
4.9. Analysis of the LncRNA-mRNA Co-Expression Network
4.10. RAW264.7 Cell Transfection
- Antisense (5′-3′): AAGUUGACUGAAUCAACGCTT
- Antisense (5′-3′): AAUUUCAGUGACCGUGUGCTT
- Antisense (5′-3′): AAUUUAUUUCUAGCAGUGUGC
4.11. Immunoblotting Analysis
4.12. Immunofluorescence
4.13. PI Staining
4.14. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Matute-Bello, G.; Frevert, C.W.; Martin, T.R. Animal models of acute lung injury. Am. J. Physiol. Cell. Mol. Physiol. 2008, 295, L379–L399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parekh, D.; Dancer, R.C.; Thickett, D.R. Acute lung injury. Clin. Med. 2011, 11, 615–618. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.Q.; Mo, Z.Z.; Liao, J.B.; Feng, X.X.; Liang, Y.Z.; Zhang, X.; Liu, Y.-H.; Chen, X.-Y.; Chen, Z.-W.; Su, Z.-R.; et al. Usnic acid protects LPS-induced acute lung injury in mice through attenuating inflammatory responses and oxidative stress. Int. Immunopharmacol. 2014, 22, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Glas, G.J.; Van Der Sluijs, K.F.; Schultz, M.J.; Hofstra, J.-J.H.; Van Der Poll, T.; Levi, M. Bronchoalveolar hemostasis in lung injury and acute respiratory distress syndrome. J. Thromb. Haemost. 2013, 11, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wu, H.; Nie, Y.C.; Chen, J.L.; Su, W.W.; Li, P.B. Naringin attenuates acute lung injury in LPS-treated mice by inhibiting NF-κB pathway. Int. Immunopharmacol. 2011, 11, 1606–1612. [Google Scholar] [CrossRef] [PubMed]
- Cockrell, A.S.; Yount, B.L.; Scobey, T.; Jensen, K.; Douglas, M.; Beall, A.; Tang, X.-C.; Marasco, W.A.; Heise, M.T.; Baric, R.S. A mouse model for MERS coronavirus-induced acute respiratory distress syndrome. Nat. Microbiol. 2016, 2, 16226. [Google Scholar] [CrossRef]
- MacLaren, G.; Fisher, D.; Brodie, D. Preparing for the Most Critically Ill Patients With COVID-19: The Potential Role of Extracorporeal Membrane Oxygenation. JAMA 2020, 323, 1245–1246. [Google Scholar] [CrossRef]
- Xu, Z.; Shi, L.; Wang, Y.; Zhang, J.; Huang, L.; Zhang, C.; Liu, S.; Zhao, P.; Liu, H.; Zhu, L.; et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020, 8, 420–422. [Google Scholar] [CrossRef]
- Li, H.; Shi, H.; Ma, N.; Zi, P.; Liu, Q.; Sun, R. BML-111 alleviates acute lung injury through regulating the expression of lncRNA MALAT1. Arch. Biochem. Biophys. 2018, 649, 15–21. [Google Scholar] [CrossRef]
- Ma, L.; Bajic, V.B.; Zhang, Z. On the classification of long non-coding RNAs. RNA Biol. 2013, 10, 925–933. [Google Scholar] [CrossRef]
- Sarropoulos, I.; Marin, R.; Cardoso-Moreira, M.; Kaessmann, H. Developmental dynamics of lncRNAs across mammalian organs and species. Nature 2019, 571, 510–514. [Google Scholar] [CrossRef] [PubMed]
- Maass, P.G.; Luft, F.C.; Bähring, S. Long non-coding RNA in health and disease. J. Mol. Med. 2014, 92, 337–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Chen, D.; Liu, H.; Yang, K. Increased expression of lncRNA CASC9 promotes tumor progression by suppressing autophagy-mediated cell apoptosis via the AKT/mTOR pathway in oral squamous cell carcinoma. Cell Death Dis. 2019, 10, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, P.S.; Mura, M.; Seth, R.; Liu, M. Acute lung injury and cell death: How many ways can cells die? Am. J. Physiol. Lung Cell. Mol. Physiol. 2008, 294, L632–L641. [Google Scholar] [CrossRef] [Green Version]
- Thompson, B.T.; Chambers, R.C.; Liu, K.D. Acute Respiratory Distress Syndrome. N. Engl. J. Med. 2017, 377, 562–572. [Google Scholar] [CrossRef] [PubMed]
- Kane, M.; Yadav, S.S.; Bitzegeio, J.; Kutluay, S.B.; Zang, T.; Wilson, S.; Schoggins, J.W.; Rice, C.M.; Yamashita, M.; Hatziioannou, T.; et al. MX2 is an interferon-induced inhibitor of HIV-1 infection. Nature 2013, 502, 563. [Google Scholar] [CrossRef] [Green Version]
- Kelly JF, C.; Davies, E.R.; Holgate, S.T.; Xu, X.; Whitsett, J.A.; Davies, D.E.; Haitchi, H.M. S89 Soluble adam33 augments the pulmonary immune response promoting allergic airway sensitivity. Thorax 2017, 72, A55. [Google Scholar]
- Rickard, J.A.; O’Donnell, J.A.; Evans, J.M.; Lalaoui, N.; Poh, A.R.; Rogers, T.; Vince, J.E.; Lawlor, K.E.; Ninnis, R.L.; Anderton, H.; et al. RIPK1 regulates RIPK3-MLKL-driven systemic inflammation and emergency hematopoiesis. Cell 2014, 157, 1175–1188. [Google Scholar] [CrossRef] [Green Version]
- Mulay, S.R.; Desai, J.; Kumar, S.V.; Eberhard, J.N.; Thomasova, D.; Romoli, S.; Grigorescu, M.; Kulkarni, O.P.; Popper, B.; Vielhauer, V.; et al. Cytotoxicity of crystals involves RIPK3-MLKL-mediated necroptosis. Nat. Commun. 2016, 7, 10274. [Google Scholar] [CrossRef]
- Jo, E.-K.; Kim, J.K.; Shin, D.-M.; Sasakawa, C. Molecular mechanisms regulating NLRP3 inflammasome activation. Cell. Mol. Immunol. 2016, 13, 148–159. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Wang, H.; Wang, Z.; He, S.; Chen, S.; Liao, D.; Wang, L.; Yan, J.; Liu, W.; Lei, X.; et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell 2012, 148, 213–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wheeler, A.P.; Bernard, G.R. Acute lung injury and the acute respiratory distress syndrome: A clinical review. Lancet 2007, 369, 1553–1564. [Google Scholar] [CrossRef]
- Luo, H.; Yang, H.; Lin, Y.; Zhang, Y.; Pan, C.; Feng, P.; Yu, Y.; Chen, X. LncRNA and mRNA profiling during activation of tilapia macrophages by HSP70 and Streptococcus agalactiae antigen. Oncotarget 2017, 8, 98455–98471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dickson, R.P.; Singer, B.; Newstead, M.W.; Falkowski, N.R.; Erb-Downward, J.R.; Standiford, T.J.; Huffnagle, G.B. Enrichment of the lung microbiome with gut bacteria in sepsis and the acute respiratory distress syndrome. Nat. Microbiol. 2016, 1, 16113. [Google Scholar] [CrossRef] [PubMed]
- Ware, L.B.; Matthay, M.A. The Acute Respiratory Distress Syndrome. N. Engl. J. Med. 2000, 342, 1334–1349. [Google Scholar] [CrossRef]
- Wang, J.; Shen, Y.-C.; Chen, Z.-N.; Yuan, Z.-C.; Wang, H.; Li, D.-J.; Liu, K.; Wen, F.-Q. Microarray profiling of lung long non-coding RNAs and mRNAs in lipopolysaccharide-induced acute lung injury mouse model. Biosci. Rep. 2019, 39, BSR20181634. [Google Scholar] [CrossRef] [Green Version]
- Martin, T.R.; Hagimoto, N.; Nakamura, M.; Matute-Bello, G. Apoptosis and epithelial injury in the lungs. Proc. Am. Thorac. Soc. 2005, 2, 214–220. [Google Scholar] [CrossRef]
- Teng, X.; Liao, J.; Zhao, L.; Dong, W.; Xue, H.; Bai, L.; Xu, S. Whole transcriptome analysis of the differential RNA profiles and associated competing endogenous RNA networks in LPS induced acute lung injury (ALI). PLoS ONE 2021, 16, e0251359. [Google Scholar] [CrossRef]
- Wong, J.; Garcia-Carbonell, R.; Zelic, M.; Ho, S.B.; Boland, B.S.; Yao, S.J.; Desai, S.A.; Das, S.; Planell, N.; Harris, P.A.; et al. RIPK1 Mediates TNF-Induced Intestinal Crypt Apoptosis During Chronic NF-kappaB Activation. Cell. Mol. Gastroenterol. Hepatol. 2019, 9, 295–312. [Google Scholar] [CrossRef] [Green Version]
- Willingham, S.B.; Allen, I.C.; Bergstralh, D.T.; Brickey, W.J.; Huang MT, H.; Taxman, D.J.; Duncan, J.A.; Ting, J.P.-Y. NLRP3 (NALP3, Cryopyrin) facilitates in vivo caspase-1 activation, necrosis, and HMGB1 release via inflammasome-dependent and -independent pathways. J. Immunol. 2009, 183, 2008–2015. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, X.; Ma, C.; Ma, Q.; Zhuang, P.; Deng, G. Microarray Profiling and Co-Expression Network Analysis of LncRNAs and mRNAs in Acute Respiratory Distress Syndrome Mouse Model. Pathogens 2022, 11, 532. https://doi.org/10.3390/pathogens11050532
Wu X, Ma C, Ma Q, Zhuang P, Deng G. Microarray Profiling and Co-Expression Network Analysis of LncRNAs and mRNAs in Acute Respiratory Distress Syndrome Mouse Model. Pathogens. 2022; 11(5):532. https://doi.org/10.3390/pathogens11050532
Chicago/Turabian StyleWu, Xiaoling, Chenjie Ma, Qinmei Ma, Peipei Zhuang, and Guangcun Deng. 2022. "Microarray Profiling and Co-Expression Network Analysis of LncRNAs and mRNAs in Acute Respiratory Distress Syndrome Mouse Model" Pathogens 11, no. 5: 532. https://doi.org/10.3390/pathogens11050532
APA StyleWu, X., Ma, C., Ma, Q., Zhuang, P., & Deng, G. (2022). Microarray Profiling and Co-Expression Network Analysis of LncRNAs and mRNAs in Acute Respiratory Distress Syndrome Mouse Model. Pathogens, 11(5), 532. https://doi.org/10.3390/pathogens11050532





