Type Strains of Entomopathogenic Nematode-Symbiotic Bacterium Species, Xenorhabdus szentirmaii (EMC) and X. budapestensis (EMA), Are Exceptional Sources of Non-Ribosomal Templated, Large-Target-Spectral, Thermotolerant-Antimicrobial Peptides (by Both), and Iodinin (by EMC)
Abstract
1. Introduction
2. Agricultural Aspects of Multidrug Resistance (MDR)
2.1. Antimicrobial Peptides as Tools to Beat MDR Pathogens
2.2. Changes in the Scope of the EPN/EPB Research Due to the Perspectives of EPB-Produced AMPs in Combatting MDR Pathogens
2.3. How Do Antibiotic-Producing EPN/EPB Symbioses Work?
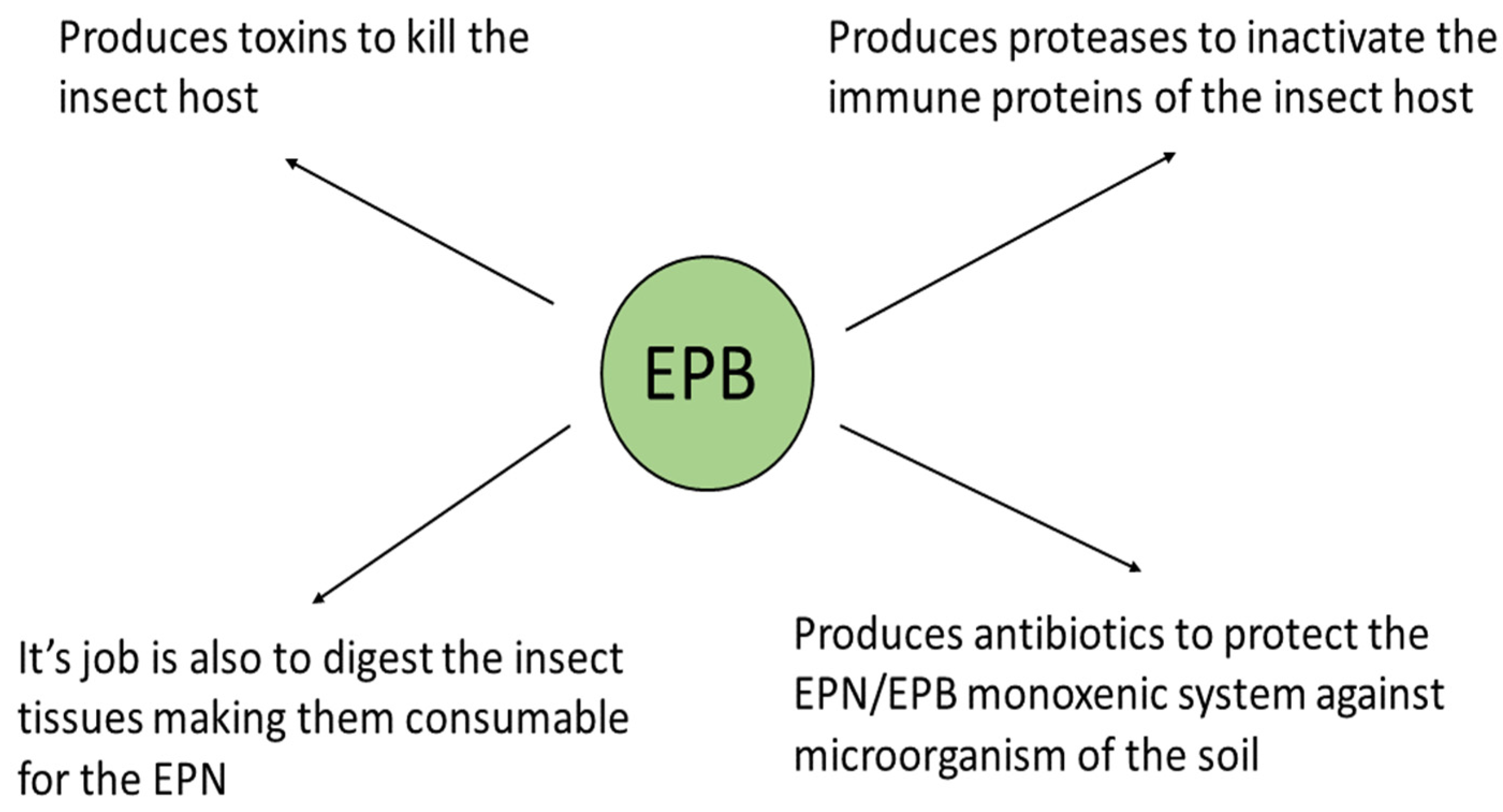
2.4. The Natural Role of the EPB
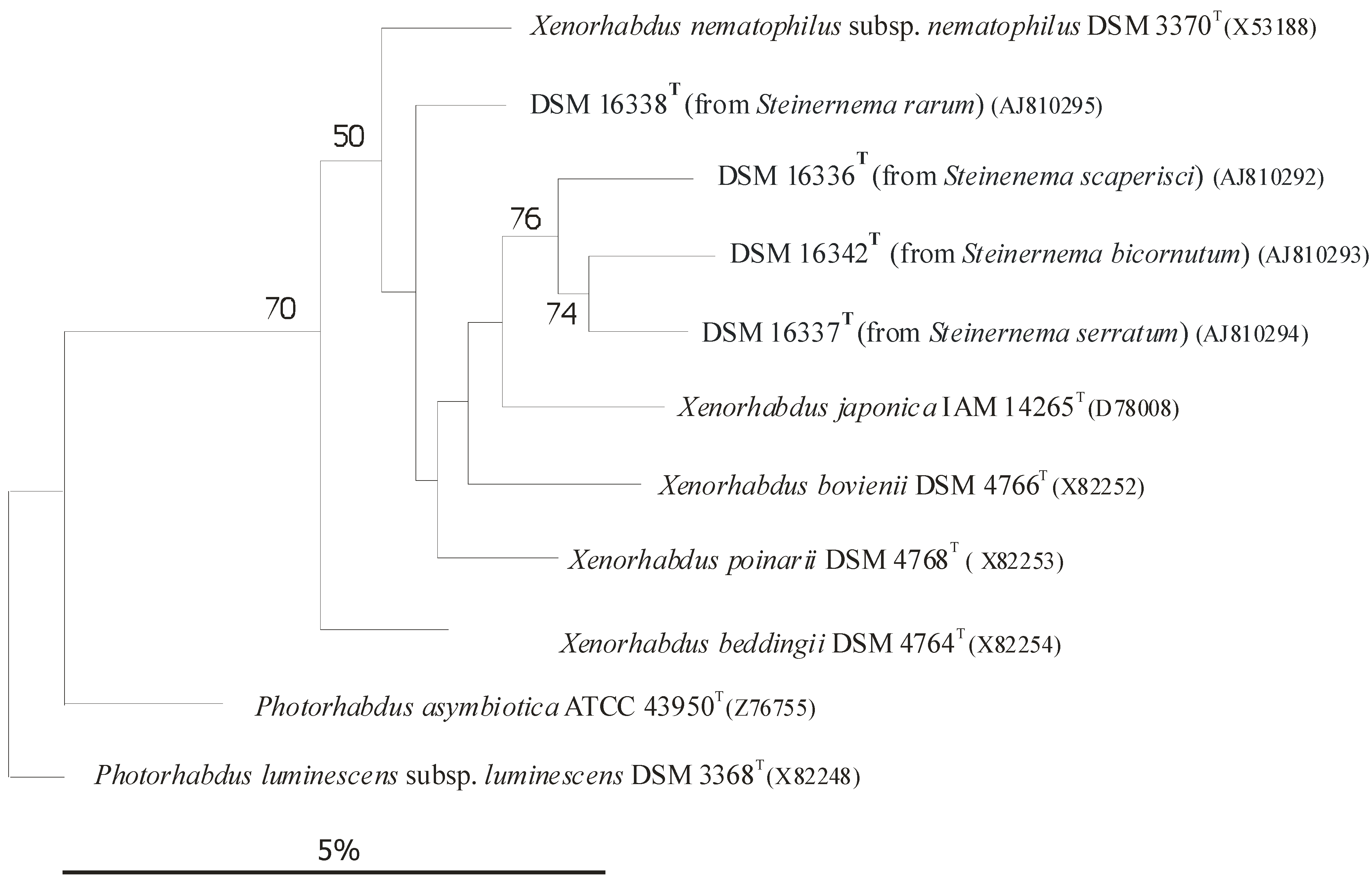
3. Coevolution and Co-Speciation of EPN/EPB Symbiotic Associations
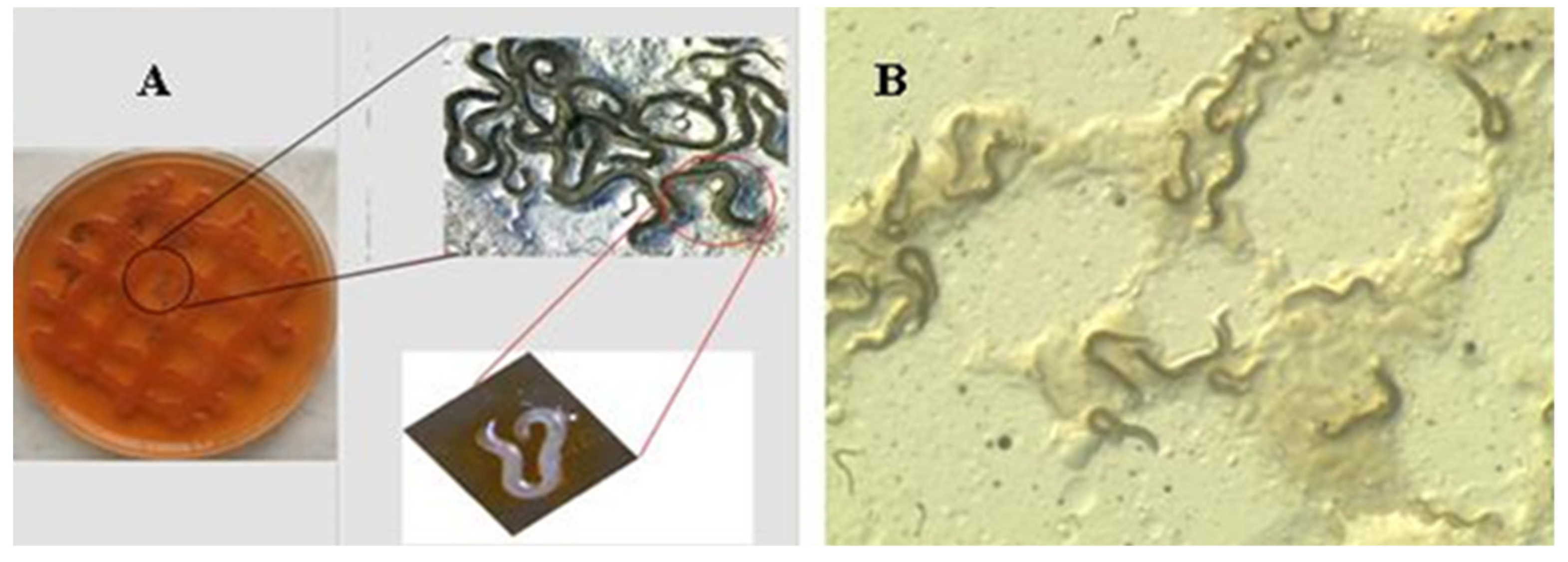
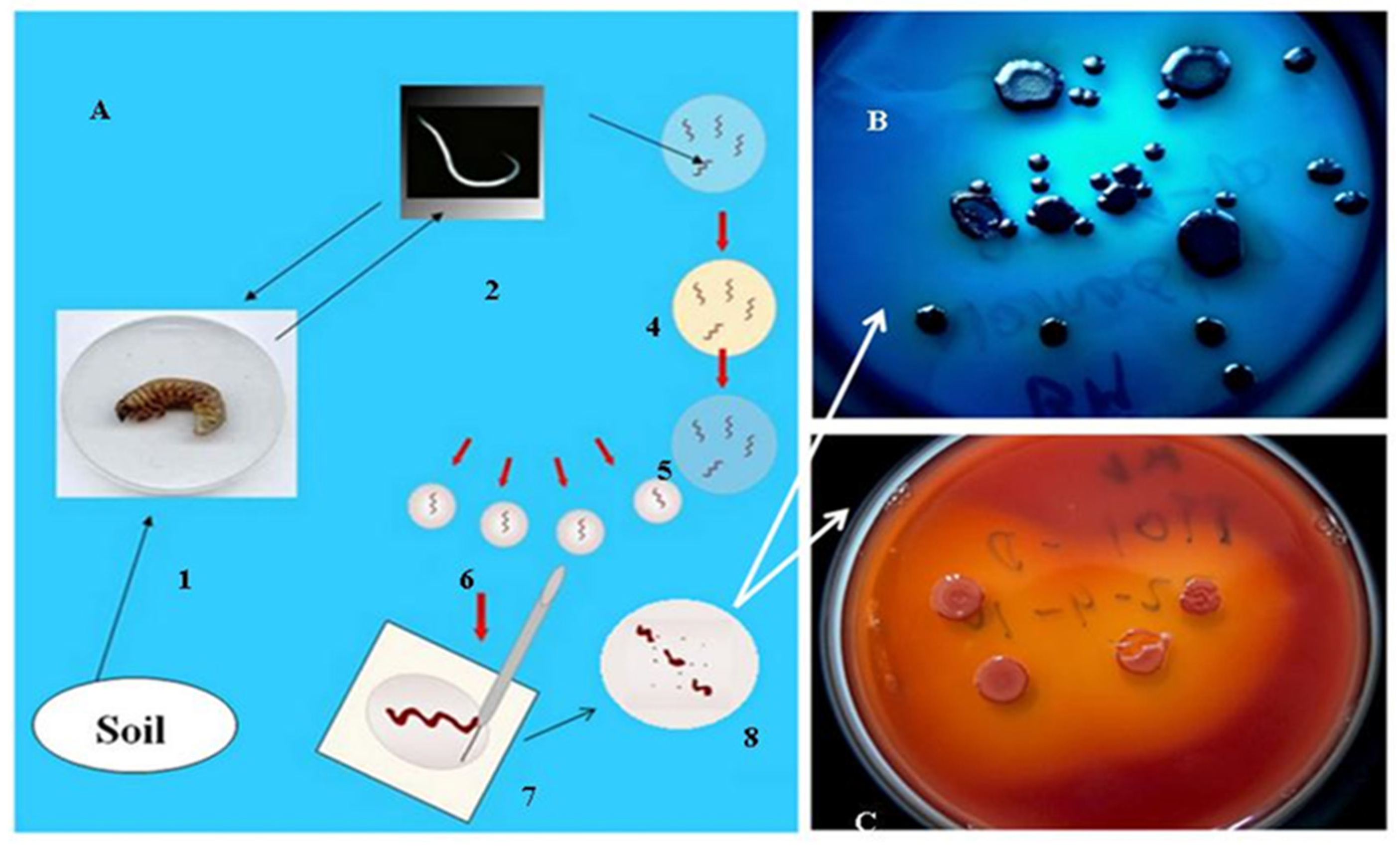
3.1. Gnotobiological Analysis as a Reliable Experimental Approach to Co-Speciation
3.2. Coevolution via Co-Speciation: Antimicrobial Active Peptides as Strategic Weapons Used in the Struggle to Conquest a Given Niche
3.2.1. Battle with the Insect Prey Using Toxins
3.2.2. Battle with EPN Competitors Using Rhabdopeptides
3.2.3. Battle between Competitor EPBs Using Xenorhabdicins
3.3. Antimicrobial Peptides from EMC and EMA, Fabclavines from both, and Phenazines from EMC
3.3.1. The Most Potent NRP-AMP Families of Xenorhabdus Origin
3.3.2. Iodinin and Phenazines
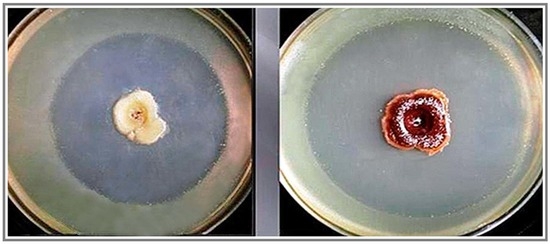
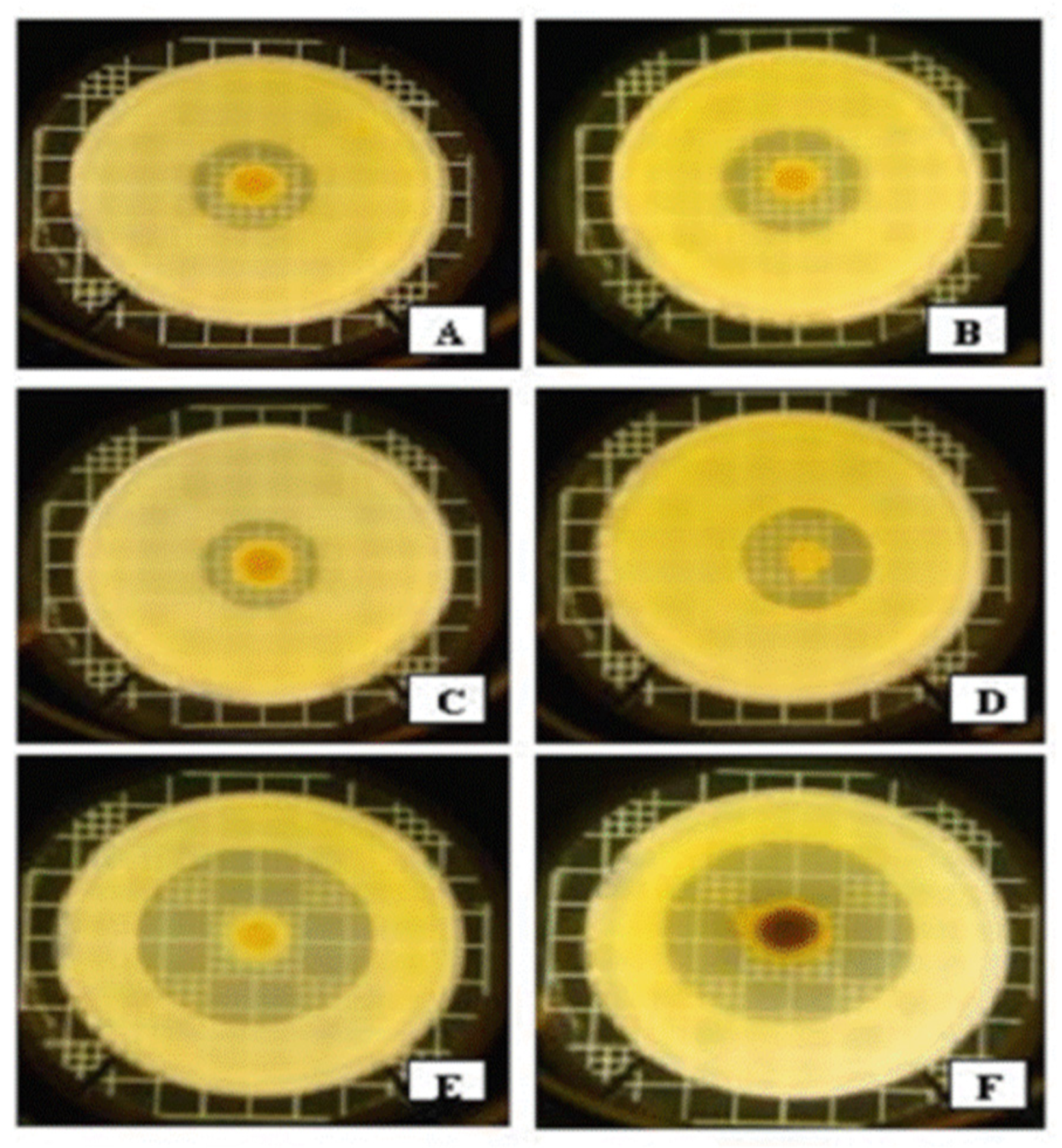
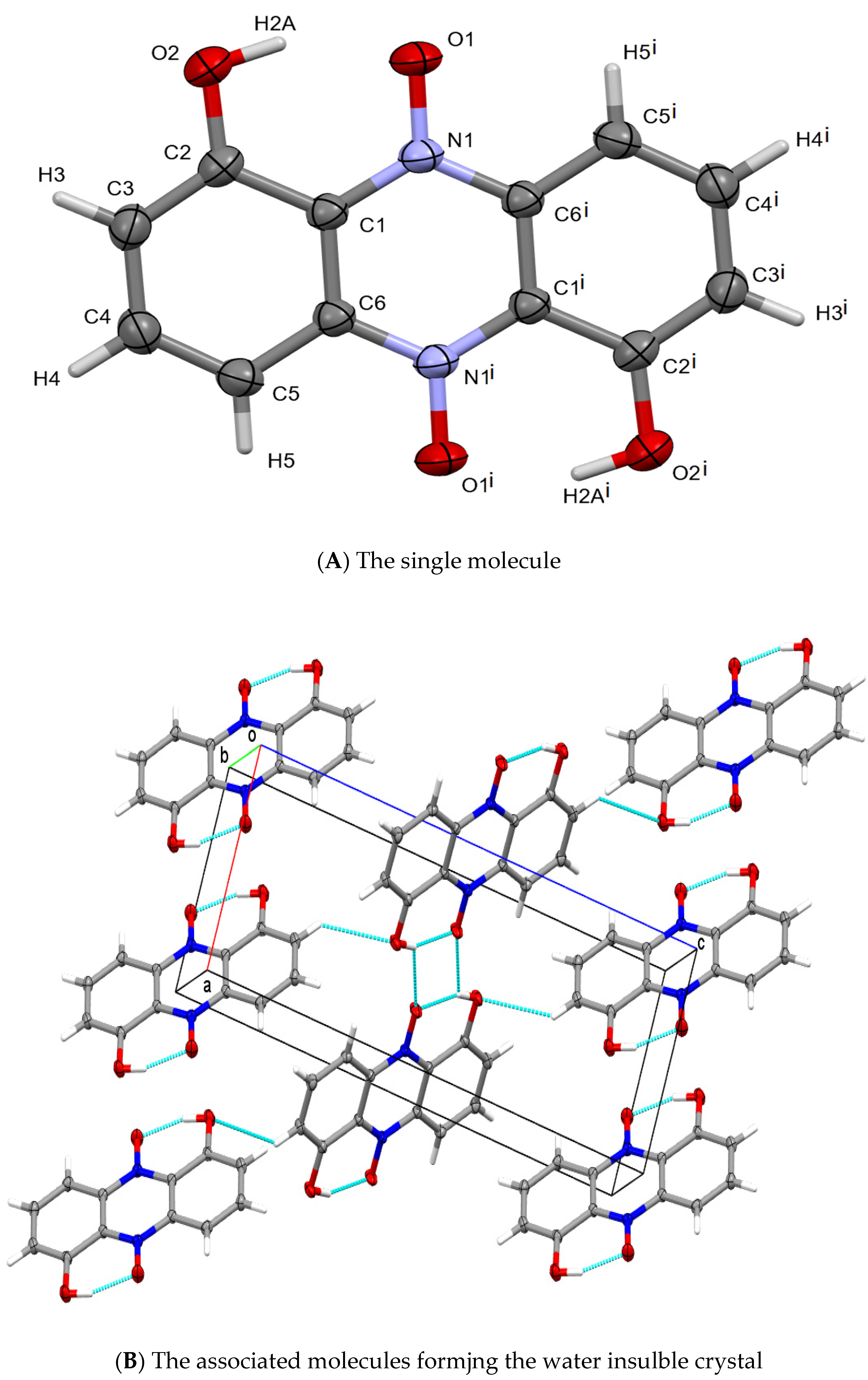
Basic Observation
Identification of the Material as Iodinin
Crystal Mutants in X. szentirmaii DSM16338T (EMC)
4. A Discussion: Discoveries and Evaluation
4.1. Strain and Genomic Information
4.1.1. Strain, and Genomic Information on Xenorhabdus szentirmaii
4.1.2. Strain, and Genomic Information on Xenorhabdus budapestensis
4.2. AMP Products of X. budapestensis and X. szentirmaii
4.2.1. A List of AMPs from Xenorhabdus Species Other Than X. budapestensis and X. szentirmaii
4.2.2. Antimicrobial Products of Xenorhabdus szentirmaii Other Than the Fabclavines and Phenazines
4.2.3. Antimicrobial Products of Strains of X. budapestensis Other Than DSM 16342T (EMA)
5. Conclusions
6. Closing Remark
Author Contributions
Funding
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix B
Appendix C
Appendix D
References
- Vivas, R.; Barbosa, A.A.T.; Dolabela, S.S.; Jain, S. Multidrug-resistant bacteria and alternative methods to control them: An Overview. Microb. Drug Resist. 2019, 25, 890–908. [Google Scholar] [CrossRef] [PubMed]
- Uddin, T.M.; Chakraborty, A.J.; Khusro, A.; Zidan, B.R.M.; Mitra, S.; Emran, T.B.; Dhama, K.; Ripon, M.K.H.; Gajdács, M.; Sahibzada, M.U.K.; et al. Antibiotic resistance in microbes: History, mechanisms, therapeutic strategies and future prospects. J. Infect. Public Health 2021, 14, 1750–1766. [Google Scholar] [CrossRef] [PubMed]
- Etesami, H.; Jeong, B.R. Silicon (Si): Review and future prospects on the action mechanisms in alleviating biotic and abiotic stresses in plants. Ecotoxicol. Environ. Saf. 2018, 147, 881–896. [Google Scholar] [CrossRef] [PubMed]
- Wiesner-Hanks, T.; Nelson, R. Multiple disease resistance in plants. Annu. Rev. Phytopathol. 2016, 54, 229–252. [Google Scholar] [CrossRef]
- Stockwell, V.O.; Duffy, B. Use of antibiotics in plant agriculture. Rev. Sci. Tech. 2012, 31, 199–210. [Google Scholar] [CrossRef]
- Forst, S.; Clarke, D. Bacteria-nematode symbiosis. In Entomopathogenic Nematology; Gaugler, R., Ed.; CABI Publishing: New York, NY, USA, 2002; pp. 57–77. [Google Scholar]
- Ötvös, L., Jr.; Wade, J.D. Current challenges in peptide-based drug discovery. Front. Chem. 2014, 2, 62. [Google Scholar] [CrossRef]
- Ogier, J.C.; Pagès, S.; Frayssinet, M.; Gaudriault, S. Entomopathogenic nematode-associated microbiota: From monoxenic paradigm to pathobiome. Microbiome 2020, 8, 25. [Google Scholar] [CrossRef]
- Lengyel, K.; Lang, E.; Fodor, A.; Szállás, E.; Schumann, P.; Stackebrandt, E. Description of four novel species of Xenorhabdus, family Enterobacteriaceae: Xenorhabdus budapestensis sp. nov., Xenorhabdus ehlersii sp. nov., Xenorhabdus innexi sp. nov., and Xenorhabdus szentirmaii sp. nov. Syst. Appl. Microbiol. 2005, 28, 115–122, Erratum in: Syst. Appl. Microbiol. 2007, 30, 83. [Google Scholar] [CrossRef]
- de Doucet, M.M.A. A new species of Neoaplectana Steiner, 1929 (Nematoda: Steinernematidae) from Cordoba, Argentina. Rev. Nematol. 1986, 9, 317–323. [Google Scholar]
- Tallósi, B.; Peters, A.; Ehlers, R.-U. Steinernema bicornutum sp. n. (Rhabditida: Steinernematidae) from Vojvodina, Yugoslavia. Russ. J. Nematol. 1995, 3, 71–80. [Google Scholar]
- Stackebrandt, E.; Mondotte, J.A.; Fazio, L.L.; Jetten, M. Authors Need to be Prudent When Assigning Names to Microbial Isolates. Curr. Microbiol. 2021, 78, 4005–4008. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, K.B.; Smart, G.C. Steinernema scapterisci n. sp. (Rhabditida: Steinernematidae). J. Nematol. 1990, 22, 187–199. [Google Scholar] [PubMed]
- Kim, I.H.; Aryal, S.K.; Aghai, D.T.; Casanova-Torres, Á.M.; Hillman, K.; Kozuch, M.P.; Mans, E.J.; Mauer, T.J.; Ogier, J.C.; Ensign, J.C.; et al. The insect pathogenic bacterium Xenorhabdus innexi has attenuated virulence in multiple insect model hosts yet encodes a potent mosquitocidal toxin. BMC Genom. 2017, 18, 927. [Google Scholar] [CrossRef] [PubMed]
- Bonifassi, E.; Fischer-Le Saux, M.; Boemare, N.; Lanois, A.; Laumond, C.; Smart, G. Gnotobiological study of infective juveniles and symbionts of Steinernema scapterisci: A model to clarify the concept of the natural occurrence of monoxenic associations in entomopathogenic nematodes. J. Invertebr. Pathol. 1999, 74, 164–172. [Google Scholar] [CrossRef]
- Kim, H.; Keum, S.; Hasan, A.; Kim, H.; Jung, Y.; Lee, D.; Kim, Y. Identification of an entomopathogenic bacterium, Xenorhabdus ehlersii KSY, from Steinernema longicaudum GNUS101 and its immunosuppressive activity against insect host by inhibiting eicosanoid biosynthesis. J. Invertebr. Pathol. 2018, 159, 6–17. [Google Scholar] [CrossRef]
- Shi, H.; Zeng, H.; Yang, X.; Zhao, J.; Chen, M.; Qiu, D. An insecticidal protein from Xenorhabdus ehlersii triggers prophenoloxidase activation and hemocyte decrease in Galleria mellonella. Curr. Microbiol. 2012, 64, 604–610. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, S.W.; Sachs, C.C.; Kegler, C.; Nollmann, F.I.; Karas, M.; Bode, H.B. Neutral loss fragmentation pattern based screening for arginine-rich natural products in Xenorhabdus and Photorhabdus. Anal. Chem. 2012, 84, 6948–6955. [Google Scholar] [CrossRef]
- Böszörményi, E.; Érsek, T.; Fodor, A.; Fodor, A.M.; Földes, L.S.; Hevesi, M.; Hogan, J.S.; Katona, Z.; Klein, M.G.; Kormány, A.; et al. Isolation and activity of Xenorhabdus antimicrobial compounds against the plant pathogens Erwinia amylovora and Phytophthora nicotianae. J. Appl. Microbiol. 2009, 107, 746–759. [Google Scholar] [CrossRef]
- Fuchs, S.W.; Grundmann, F.; Kurz, M.; Kaiser, M.; Bode, H.B. Fabclavines: Bioactive peptide-polyketide-polyamino hybrids from Xenorhabdus. Chembiochem 2014, 15, 512–516. [Google Scholar] [CrossRef]
- Wenski, S.L.; Kolbert, D.; Grammbitter, G.L.C.; Bode, H.B. Fabclavine biosynthesis in X. szentirmaii: Shortened derivatives and characterization of the thioester reductase FclG and the condensation domain-like protein FclL. J. Ind. Microbiol. Biotechnol. 2019, 46, 565–572. [Google Scholar] [CrossRef]
- Wenski, S.L.; Cimen, H.; Berghaus, N.; Fuchs, S.W.; Hazir, S.; Bode, H.B. Fabclavine diversity in Xenorhabdus bacteria. Beilstein J. Org. Chem. 2020, 16, 956–965. [Google Scholar] [CrossRef] [PubMed]
- Gualtieri, M.; Villain-Guillot, P.; Givaudan, A.; Pages, S. Nemaucin, an Antibiotic Produced by Entomopathogenic Xenorhabdus cabanillasii. France Patent WO2012085177A1, 28 June 2012. [Google Scholar]
- Fodor, A.; Forst, S.; Haynes, L.; Hevesi, M.; Hogan, J.A.; Klein, M.G.; Máthe-Fodor, A.; Stackebrandt, E.; Szentirmai, A.; Sztaricskai, F.; et al. New Perspectives of Xenorhabdus Antibiotics Research; Insect Pathogens and Insect Parasitic Nematodes IOBC/IOBC/WPRS Bulletin: Ales, France, 2008; Volume 31, pp. 157–164. [Google Scholar]
- Shi, Y.M.; Brachmann, A.O.; Westphalen, M.A.; Neubacher, N.; Tobias, N.J.; Bode, H.B. Dual phenazine gene clusters enable diversification during biosynthesis. Nat. Chem. Biol. 2019, 15, 331–339. [Google Scholar] [CrossRef] [PubMed]
- Upert, G.; Luther, A.; Obrecht, D.; Ermert, P. Emerging peptide antibiotics with therapeutic potential. Med. Drug Discov. 2021, 9, 100078. [Google Scholar] [CrossRef] [PubMed]
- Pál, C.; Papp, B.; Lázár, V. Collateral sensitivity of antibiotic-resistant microbes. Trends Microbiol. 2015, 23, 401–407. [Google Scholar] [CrossRef]
- Lázár, V.; Martins, A.; Spohn, R.; Daruka, L.; Grézal, G.; Fekete, G.; Számel, M.; Jangir, P.K.; Kintses, B.; Csörgő, B.; et al. Antibiotic-resistant bacteria show widespread collateral sensitivity to antimicrobial peptides. Nat. Microbiol. 2018, 3, 718–731. [Google Scholar] [CrossRef]
- Kintses, B.; Méhi, O.; Ari, E.; Számel, M.; Györkei, Á.; Jangir, P.K.; Nagy, I.; Pál, F.; Fekete, G.; Tengölics, R.; et al. Phylogenetic barriers to horizontal transfer of antimicrobial peptide resistance genes in the human gut microbiota. Nat. Microbiol. 2019, 4, 447–458. [Google Scholar] [CrossRef]
- Kintses, B.; Jangir, P.K.; Fekete, G.; Számel, M.; Méhi, O.; Spohn, R.; Daruka, L.; Martins, A.; Hosseinnia, A.; Gagarinova, A.; et al. Chemical-genetic profiling reveals limited cross-resistance between antimicrobial peptides with different modes of action. Nat. Commun. 2019, 10, 5731. [Google Scholar] [CrossRef]
- Fodor, A.; Abate, B.A.; Deák, P.; Fodor, L.; Gyenge, E.; Klein, M.G.; Koncz, Z.; Muvevi, J.; Ötvös, L.; Székely, G.; et al. Multidrug resistance (MDR) and collateral sensitivity in bacteria, with special attention to genetic and evolutionary aspects and to the perspectives of antimicrobial peptides-A Review. Pathogens 2020, 9, 522. [Google Scholar] [CrossRef]
- Baindara, P.; Ghosh, A.K.; Mandal, S.M. Coevolution of resistance against antimicrobial peptides. Microb. Drug Resist. 2020, 26, 880–899. [Google Scholar] [CrossRef]
- Khun, K.K.; Wilson, B.A.L.; Stevens, M.M.; Huwer, R.K.; Ash, G.J. Integration of entomopathogenic fungi into IPM programs: Studies involving weevils (Coleoptera: Curculionoidea) affecting horticultural crops. Insects 2020, 1, 659. [Google Scholar] [CrossRef]
- Mann, A.J.; Davis, T.S. Entomopathogenic fungi to control bark beetles: A review of ecological recommendations. Pest Manag. Sci. 2021, 77, 3841–3846. [Google Scholar] [CrossRef] [PubMed]
- Thomas, G.M.; Poinar, G.O., Jr. A new bacterium, Achromobacter nematophilus sp. nov. (Achromobacteriaceae: Eubacteriales) associated with a nematode. Int. J. Syst. Evol. Microbiol. 1965, 15, 249–254. [Google Scholar]
- Poinar, G.O., Jr.; Thomas, G.M. (Achromobacteraceae: Eubacteriales) in the development of the nematode, DD-136 (Neoaplectana sp. Steinernematidae). Parasitology 1966, 56, 385–390. [Google Scholar] [CrossRef] [PubMed]
- Kaya, H.K. Development of the DD-136 strain of Neoaplectana carpocapsae at constant temperatures. J. Nematol. 1977, 9, 346–349. [Google Scholar]
- Poinar, G.O., Jr.; Thomas, G.M.; Hess, R. Characteristics of the specific bacterium associated with Heterorhabditis bacteriophora (Heterorhabditidae: Rhabditida). Nematologica 1977, 23, 97–102. [Google Scholar]
- Thomas, G.M.; Poinar, G.O., Jr. Xenorhabdus gen. nov., a genus of entomopathogenic nematophilic bacteria of the family Enterobacteriaceae. Int. J. Syst. Bacteriol. 1979, 29, 352–360. [Google Scholar] [CrossRef]
- Bedding, R.A.; Molyneux, A.S.; Akhurst, R.J. Heterorhabditis spp., Neoaplectana spp., and Steinernema kraussei: Interspecific and intraspecific differences in infectivity for insects. Exp. Parasitol. 1983, 55, 249–257. [Google Scholar] [CrossRef]
- Ehlers, R.U. Mass production of entomopathogenic nematodes for plant protection. Appl. Microbiol. Biotechnol. 2001, 56, 623–633. [Google Scholar] [CrossRef]
- Samish, M.; Glazer, I. Entomopathogenic nematodes for the biocontrol of ticks. Trends Parasitol. 2001, 17, 368–371. [Google Scholar] [CrossRef]
- Shapiro-Ilan, D.I.; Gaugler, R. Production technology for entomopathogenic nematodes and their bacterial symbionts. J. Ind. Microbiol. Biotechnol. 2002, 28, 137–146. [Google Scholar] [CrossRef]
- Piñero, J.C.; Shapiro-Ilan, D.; Cooley, D.R.; Tuttle, A.F.; Eaton, A.; Drohan, P.; Leahy, K.; Zhang, A.; Hancock, T.; Wallingford, A.K.; et al. Toward the integration of an Attract-and-Kill approach with entomopathogenic nematodes to control multiple life stages of plum curculio (Coleoptera: Curculionidae). Insects 2020, 11, 375. [Google Scholar] [CrossRef] [PubMed]
- Torrini, G.; Paoli, F.; Mazza, G.; Simoncini, S.; Benvenuti, C.; Strangi, A.; Tarasco, E.; Barzanti, G.P.; Bosio, G.; Cutino, I.; et al. Evaluation of indigenous entomopathogenic nematodes as potential biocontrol agents against Popillia japonica (Coleoptera: Scarabaeidae) in Northern Italy. Insects 2020, 11, 804. [Google Scholar] [CrossRef] [PubMed]
- Fanelli, E.; Troccoli, A.; Tarasco, E.; De Luca, F. Molecular characterization and functional analysis of the Hb-hsp90-1 gene in relation to temperature changes in Heterorhabditis bacteriophora. Front. Physiol. 2021, 12, 615653. [Google Scholar] [CrossRef]
- Tarasco, E.; De Luca, F. Biological control and insect pathology. Insects 2021, 12, 291. [Google Scholar] [CrossRef] [PubMed]
- Bode, H.B. Entomopathogenic bacteria as a source of secondary metabolites. Curr. Opin. Chem. Biol. 2009, 13, 224–230. [Google Scholar] [CrossRef]
- Boucher, H.W.; Talbot, G.H.; Bradley, J.S.; Edwards, J.E.; Gilbert, D.; Rice, L.B.; Scheld, M.; Spellberg, B.; Bartlett, J. Bad bugs, no drugs: No ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009, 48, 1–12. [Google Scholar] [CrossRef]
- Nealson, K.H.; Eberhard, A.; Hastings, J.W. Catabolite repression of bacterial bioluminescence: Functional implications. Proc. Natl. Acad. Sci. USA. 1972, 69, 1073–1076. [Google Scholar] [CrossRef]
- Paul, W.J.; Frautschv, S.; Fenical, W.; Nealson, K.H. Antibiotics in microbial ecology: Isolation and structure assignment of several new antibacterial compounds for the insect symbiotic bacteria, Xenorhabdus spp. J. Chem. Ecol. 1981, 7, 589–597. [Google Scholar] [CrossRef]
- Akhurst, R.J. Antibiotic activity of Xenorhabdus spp., bacteria symbiotically associated with insect pathogenic nematodes of the families Heterorhabditidae and Steinernematidae. J. Gen. Microbiol. 1982, 128, 3061. [Google Scholar] [CrossRef]
- Akhurst, R.J. Neoaplectana species: Specificity of association with bacteria of the genus Xenorhabdus. Exp. Parasitol. 1983, 55, 258–263. [Google Scholar] [CrossRef]
- Akhurst, R.J.; Boemare, N.E. A numerical taxonomic study of the genus Xenorhabdus (Enterobacteriaceae) and proposed elevation of the subspecies of X. nematophilus to species. J. Gen. Microbiol. 1988, 134, 1835–1845. [Google Scholar] [CrossRef] [PubMed]
- Smigielski, A.J.; Akhurst, R.J.; Boemare, N.E. Phase variation in Xenorhabdus nematophilus and Photorhabdus luminescens: Differences in respiratory activity and membrane energization. Appl. Environ. Microbiol. 1994, 60, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Forst, S.; Nealson, K. Molecular biology of the symbiotic-pathogenic bacteria Xenorhabdus spp. and Photorhabdus spp. Microbiol. Rev. 1996, 60, 21–43. [Google Scholar] [CrossRef] [PubMed]
- Forst, S.; Dowds, B.; Boemare, N.; Stackebrandt, E. Xenorhabdus and Photorhabdus spp.: Bugs that kill bugs. Annu. Rev. Microbiol. 1997, 51, 47–72. [Google Scholar] [CrossRef] [PubMed]
- Völgyi, A.; Fodor, A.; Szentirmai, A.; Forst, S. Phase variation in Xenorhabdus nematophilus. Appl. Environ. Microbiol. 1998, 64, 1188–1193. [Google Scholar] [CrossRef]
- Szállás, E.; Koch, C.; Fodor, A.; Burghart, J.; Buss, O.; Szentirmai, A.; Nealson, K.H.; Stackebrandt, E. Phylogenetic evidence for the taxonomic heterogeneity of Photorhabdus luminescens. Int. J. Syst. Bacteriol. 1997, 47, 402–407. [Google Scholar] [CrossRef]
- Szállás, E.; Pukall, R.; Pamjav, H.; Kovács, G.; Buzás, Z.; Fodor, A.; Stackebrandt, E. Passengers who missed the train: Comparative sequence analysis, PhastSystem PAGE-PCR-RFLP and automated RiboPrint Phenotypes of Photorhabdus strains In Development in Entomopathogenic Nematode/Bacterial Research; Griffin, C.T., Burnell, A.M., Downes, M.J., Mulder, R., Eds.; European Commission Publications: Luxemburg, 2001; pp. 36–53. [Google Scholar]
- de Soete, G. A least square algorithm for fitting additive trees to proximity data. Psychometrika 1983, 48, 621–626. [Google Scholar] [CrossRef]
- Munro, H.N. (Ed.) Jukes and Cantor, Evolution of protein molecules. In Mammalian Protein Metabolism; Academic Press: New York, NY, USA, 1969; pp. 21–132. [Google Scholar]
- Felsenstein, J. PHYLIP Phylogeny Inference Package, Version 3.5.1; Department of Genetics, University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Triga, D.; Pamjav, H.; Vellai, T.; Fodor, A.; Buzás, Z. Gel electrophoretic restriction fragment length polymorphism analysis of DNA derived from individual nematodes, using the PhastSystem. Electrophoresis 1999, 20, 1274–1279. [Google Scholar] [CrossRef]
- Pamjav, H.; Triga, D.; Buzás, Z.; Vellai, T.; Lucskai, A.; Adams, B.; Reid, A.P.; Burnell, A.; Griffin, C.; Glazer, I.; et al. Novel application of PhastSystem polyacrylamide gel electrophoresis using restriction fragment length polymorphism--internal transcribed spacer patterns of individuals for molecular identification of entomopathogenic nematodes. Electrophoresis 1999, 20, 1266–1273. [Google Scholar] [CrossRef][Green Version]
- Tailliez, P.; Pagès, S.; Ginibre, N.; Boemare, N. New insight into diversity in the genus Xenorhabdus, including the description of ten novel species. Int. J. Syst. Evol. Microbiol. 2006, 56, 2805–2818. [Google Scholar] [CrossRef]
- Böszörményi, E. Entomopathogen Bacterium Antibiotic Activity and Symbiotic Capacity of Gnotobiological Analyses. Ph.D Thesis, Eötvös University, Budapest, Hungary, 2010. Available online: http://teo.elte.hu/minosites/tezis2010/burgettine_boszormenyi_e.pdf (accessed on 1 August 2019).
- Tailliez, P.; Laroui, C.; Ginibre, N.; Paule, A.; Pagès, S.; Boemare, N. Phylogeny of Photorhabdus and Xenorhabdus based on universally conserved protein-coding sequences and implications for the taxonomy of these two genera. Proposal of new taxa: X. vietnamensis sp. nov., P. luminescens subsp. caribbeanensis subsp. nov., P. luminescens subsp. hainanensis subsp. nov., P. temperata subsp. khanii subsp. nov., P. temperata subsp. tasmaniensis subsp. nov., and the reclassification of P. luminescens subsp. thracensis as P. temperata subsp. thracensis comb. nov. Int. J. Syst. Evol. Microbiol. 2010, 60, 1921–1937. [Google Scholar] [CrossRef] [PubMed]
- Machado, R.A.R.; Wüthrich, D.; Kuhnert, P.; Arce, C.C.M.; Thönen, L.; Ruiz, C.; Zhang, X.; Robert, C.A.M.; Karimi, J.; Kamali, S.; et al. Whole-genome-based revisit of Photorhabdus phylogeny: Proposal for the elevation of most Photorhabdus subspecies to the species level and description of one novel species Photorhabdus bodei sp. nov., and one novel subspecies Photorhabdus laumondii subsp. clarkei subsp. nov. Int. J. Syst. Evol. Microbiol. 2018, 68, 2664–2681. [Google Scholar] [CrossRef] [PubMed]
- Goodrich-Blair, H.; Clarke, D.J. Mutualism and pathogenesis in Xenorhabdus and Photorhabdus: Two roads to the same destination. Mol. Microbiol. 2007, 64, 260–268. [Google Scholar] [CrossRef] [PubMed]
- Morran, L.T.; Penley, M.J.; Byrd, V.S.; Meyer, A.J.; O’Sullivan, T.S.; Bashey, F.; Goodrich-Blair, H.; Lively, C.M. Nematode-bacteria mutualism: Selection within the mutualism supersedes selection outside of the mutualism. Evolution 2016, 70, 687–695. [Google Scholar] [CrossRef][Green Version]
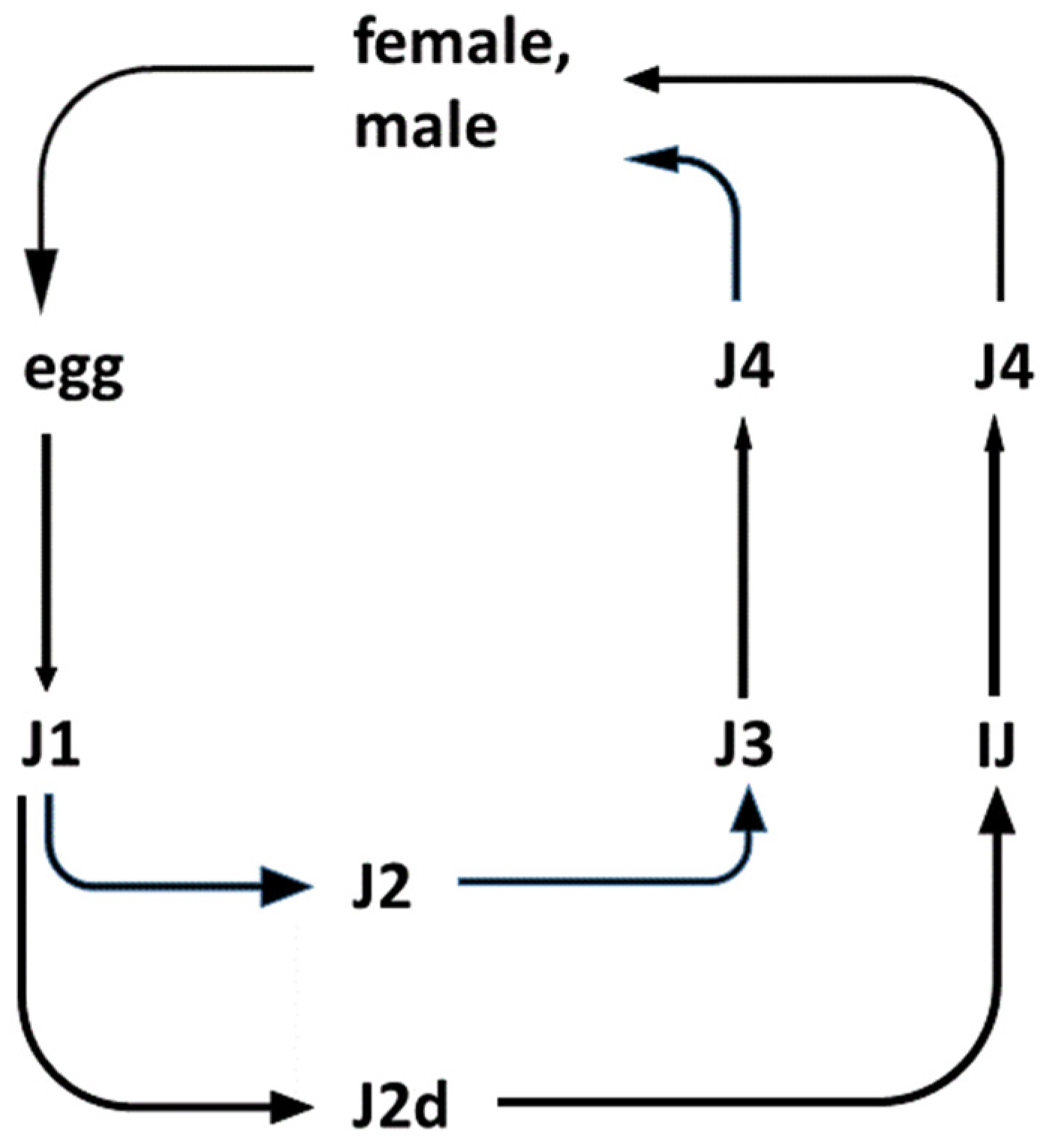
- Cassada, R.C.; Russell, R.L. The dauerlarva, a post-embryonic developmental variant of the nematode Caenorhabditis elegans. Dev. Biol. 1975, 46, 326–342. [Google Scholar] [CrossRef]
- ffrench-Constant, R.; Bowen, D. Photorhabdus toxins: Novel biological insecticides. Curr. Opin. Microbiol. 1999, 2, 284–288. [Google Scholar] [CrossRef]
- ffrench-Constant, R.; Dowling, A.; Waterfield, N.R. Insecticidal toxins from Photorhabdus bacteria and their potential use in agriculture. Toxicon 2007, 49, 436–451. [Google Scholar] [CrossRef]
- Kajla, M.K. Symbiotic bacteria as potential agents for mosquito control. Trends Parasitol. 2020, 36, 4–7. [Google Scholar] [CrossRef]
- Gerrard, J.; Waterfield, N.; Vohra, R.; ffrench-Constant, R. Human infection with Photorhabdus asymbiotica: An emerging bacterial pathogen. Microb. Infect. 2004, 6, 229–237. [Google Scholar] [CrossRef]
- Hapeshi, A.; Waterfield, N.R. Photorhabdus asymbiotica as an insect and human pathogen. In Current Topics in Microbiology and Immunology; Springer: Cham, Switzerlan, 2017; Volume 402, pp. 159–177. [Google Scholar] [CrossRef]
- Vlisidou, I.; Hapeshi, A.; Healey, J.R.; Smart, K.; Yang, G.; Waterfield, N.R. The Photorhabdus asymbiotica virulence cassettes deliver protein effectors directly into target eukaryotic cells. Elife 2019, 8, e46259. [Google Scholar] [CrossRef]
- Sicard, M.; Ferdy, J.B.; Pagès, S.; Le Brun, N.; Godelle, B.; Boemare, N.; Moulia, C. When mutualists are pathogens: An experimental study of the symbioses between Steinernema (entomopathogenic nematodes) and Xenorhabdus (bacteria). J. Evol. Biol. 2004, 17, 985–993. [Google Scholar] [CrossRef] [PubMed]
- Peat, S.M.; ffrench-Constant, R.H.; Waterfield, N.R.; Marokházi, J.; Fodor, A.; Adams, B.J. A robust phylogenetic framework for the bacterial genus Photorhabdus and its use in studying the evolution and maintenance of bioluminescence: A case for 16S, gyrB, and glnA. Mol. Phylogenet. Evol. 2010, 57, 728–740. [Google Scholar] [CrossRef] [PubMed]
- Ward, S.; Hogan, E.; Nelson, G.A. The initiation of spermiogenesis in the nematode Caenorhabditis elegans. Dev. Biol. 1983, 98, 70–79. [Google Scholar] [CrossRef]
- Fodor, A.; Vecseri, G.; Farkas, T. Caenorhabditis elegans as a Model for the Study of Entomopathogenic Nematodes. In Entomopathogenic Nematodes in Biological Control, 1st ed.; CRC Press: New York, NY, USA, 1990; 24p, ISBN 9781351071741. [Google Scholar]
- Han, R.; Ehlers, R.U. Pathogenicity, development, and reproduction of Heterorhabditis bacteriophora and Steinernema carpocapsae under axenic in vivo conditions. J. Invertebr. Pathol. 2000, 75, 55–58. [Google Scholar] [CrossRef] [PubMed]
- Hunt, P.R. The C. elegans model in toxicity testing. J. Appl. Toxicol. 2017, 37, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Brenner, S. The genetics of Caenorhabditis elegans. Genetics. 1974, 77, 71–94. [Google Scholar] [CrossRef]
- Fodor, A.; Varga, I.; Hevesi, M.; Máthé-Fodor, A.; Racsko, J.; Hogan, J.A. Novel anti-microbial peptides of Xenorhabdus origin against multidrug resistant plant pathogens. In A Search for Antibacterial Agents; Bobbarala, V., Ed.; IntechOpen: London, UK, 2012; pp. 3–32. Available online: https://www.intechopen.com/books/2129 (accessed on 28 February 2022). [CrossRef]
- Yang, J.; Zeng, H.M.; Lin, H.F.; Yang, X.F.; Liu, Z.; Guo, L.H.; Yuan, J.J.; Qiu, D.W. An insecticidal protein from Xenorhabdus budapestensis that results in prophenoloxidase activation in the wax moth, Galleria mellonella. J. Invertebr. Pathol. 2012, 110, 60–67. [Google Scholar] [CrossRef]
- Hemalatha, D.; Prabhu, S.; Rani, W.B.; Anandham, R. Isolation and characterization of toxins from Xenorhabdus nematophilus against Ferrisia virgata (Ckll.) on tuberose, Polianthes tuberosa. Toxicon 2018, 146, 42–49. [Google Scholar] [CrossRef]
- Mahmood, S.; Kumar, M.; Kumari, P.; Mahapatro, G.K.; Banerjee, N.; Sarin, N.B. Novel insecticidal chitinase from the insect pathogen Xenorhabdus nematophila. Int. J. Biol. Macromol. 2020, 159, 394–401. [Google Scholar] [CrossRef]
- da Silva, W.J.; Pilz-Júnior, H.L.; Heermann, R.; da Silva, O.S. The great potential of entomopathogenic bacteria Xenorhabdus and Photorhabdus for mosquito control: A review. Parasit Vectors 2020, 13, 376. [Google Scholar] [CrossRef]
- Alotaibi, S.S.; Darwish, H.; Alharthi, S.; Alghamdi, A.; Noureldeen, A.; Fallatah, A.M.; Fodor, A.; Al-Barty, A.; Albogami, B.; Baazeem, A. Control potentials of three entomopathogenic bacterial isolates for the carob moth, Ectomyelois ceratoniae (Lepidoptera: Pyralidae) in pomegranates. Agriculture 2021, 11, 1256. [Google Scholar] [CrossRef]
- Bi, Y.; Gao, C.; Yu, Z. Rhabdopeptides from Xenorhabdus budapestensis SN84 and their nematicidal activities against Meloidogyne incognita. J. Agric. Food Chem. 2018, 66, 3833–3839. [Google Scholar] [CrossRef] [PubMed]
- Thaler, J.O.; Baghdiguian, S.; Boemare, N. Purification and characterization of xenorhabdicin, a phage tail-like bacteriocin, from the lysogenic strain F1 of Xenorhabdus nematophilus. Appl. Environ. Microbiol. 1995, 61, 2049–2052. [Google Scholar] [CrossRef] [PubMed]
- Fodor, A.; Fodor, A.M.; Forst, S.; Hogan, J.S.; Klein, M.G.; Lengyel, K.; Sáringer, G.; Stackebrandt, E.; Taylor, R.A.J.; Lehoczky, E. Comparative analysis of antibacterial activities of Xenorhabdus species on related and nonrelated bacteria in vivo. J. Microbiol. Antimicrob. 2010, 2, 36–46. [Google Scholar]
- Tóth, E.M.; Márialigeti, K.; Fodor, A.; Lucskai, A.; Farkas, R. Evaluation of efficacy of entomopathogenic nematodes against larvae of Lucilia sericata (Meigen, 1826) (Diptera: Calliphoridae). Acta Vet. Hung. 2005, 53, 65–71. [Google Scholar] [CrossRef]
- Furgani, G.; Böszörményi, E.; Fodor, A.; Máthé-Fodor, A.; Forst, S.; Hogan, J.S.; Katona, Z.; Klein, M.G.; Stackebrandt, E.; Szentirmai, A.; et al. Xenorhabdus antibiotics: A comparative analysis and potential utility for controlling mastitis caused by bacteria. J. Appl. Microbiol. 2008, 104, 745–758. [Google Scholar] [CrossRef]
- Thappeta, K.R.V.; Ciezki, K.; Morales-Soto, N.; Wesener, S.; Goodrich-Blair, H.; Stock, S.P.; Forst, S. R-type bacteriocins of Xenorhabdus bovienii determine the outcome of interspecies competition in a natural host environment. Microbiology 2020, 166, 1074–1087. [Google Scholar] [CrossRef]
- Watzel, J.; Hacker, C.; Duchardt-Ferner, E.; Bode, H.B.; Wöhnert, J. A new docking domain type in the peptide-antimicrobial-Xenorhabdus peptide producing nonribosomal peptide synthetase from Xenorhabdus bovienii. ACS Chem. Biol. 2020, 15, 982–989. [Google Scholar] [CrossRef]
- Gualtieri, M.; Aumelas, A.; Thaler, J.O. Identification of a new antimicrobial lysine-rich cyclolipopeptide family from Xenorhabdus nematophila. J. Antibiot. 2009, 62, 295–302. [Google Scholar] [CrossRef]
- Tobias, N.J.; Heinrich, A.K.; Eresmann, H.; Wright, P.R.; Neubacher, N.; Backofen, R.; Bode, H.B. Photorhabdus-nematode symbiosis is dependent on hfq-mediated regulation of secondary metabolites. Environ. Microbiol. 2017, 19, 119–129. [Google Scholar] [CrossRef]
- Bode, E.; Heinrich, A.K.; Hirschmann, M.; Abebew, D.; Shi, Y.N.; Vo, T.D.; Wesche, F.; Shi, Y.M.; Grün, P.; Simonyi, S.; et al. Promoter activation in Δhfq mutants as an efficient tool for specialized metabolite production enabling direct bioactivity testing. Angew. Chem. Int. Ed. Engl. 2019, 58, 18957–18963. [Google Scholar] [CrossRef] [PubMed]
- McErlean, M.; Overbay, J.; Van Lanen, S. Refining and expanding nonribosomal peptide synthetase function and mechanism. J. Ind. Microbiol. Biotechnol. 2019, 46, 493–513. [Google Scholar] [CrossRef] [PubMed]
- Watzel, J.; Sarawi, S.; Duchardt-Ferner, E.; Bode, H.B.; Wöhnert, J. NMR resonance assignments for a docking domain pair with an attached thiolation domain from the PAX peptide-producing NRPS from Xenorhabdus cabanillasii. Biomol. NMR Assign. 2021, 15, 229–234. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, S.W.; Proschak, A.; Jaskolla, T.W.; Karas, M.; Bode, H.B. Structure elucidation and biosynthesis of lysine-rich cyclic peptides in Xenorhabdus nematophila. Org. Biomol. Chem. 2011, 9, 3130–3132. [Google Scholar] [CrossRef] [PubMed]
- Fodor, E.; Szállás, E.; Kiss, Z.; Fodor, A.; Horvath, L.I.; Chitwood, D.J.; Farkas, T. Composition and biophysical properties of lipids in Xenorhabdus nematophilus and Photorhabdus luminescens, symbiotic bacteria associated with entomopathogenic nematodes. Appl. Environ. Microbiol. 1997, 3, 2826–2831. [Google Scholar] [CrossRef]
- Mollah, M.M.I.; Roy, M.C.; Choi, D.Y.; Hasan, M.A.; Al Baki, M.A.; Yeom, H.S.; Kim, Y. Variations of indole metabolites and NRPS-PKS loci in two different virulent strains of Xenorhabdus hominickii. Front. Microbiol. 2020, 11, 583594. [Google Scholar] [CrossRef]
- Cowles, K.N.; Cowles, C.E.; Richards, G.R.; Martens, E.C.; Goodrich-Blair, H. The global regulator Lrp contributes to mutualism, pathogenesis and phenotypic variation in the bacterium Xenorhabdus nematophila. Cell Microbiol. 2007, 9, 1311–1323. [Google Scholar] [CrossRef]
- Cao, M.; Patel, T.; Rickman, T.; Goodrich-Blair, H.; Hussa, E.A. High levels of the Xenorhabdus nematophila transcription factor Lrp promote mutualism with the Steinernema carpocapsae nematode host. Appl. Environ. Microbiol. 2017, 83, e00276-17. [Google Scholar] [CrossRef]
- Engel, Y.; Windhorst, C.; Lu, X.; Goodrich-Blair, H.; Bode, H.B. The global regulators Lrp, LeuO, and HexA control secondary metabolism in entomopathogenic bacteria. Front. Microbiol. 2017, 8, 209. [Google Scholar] [CrossRef]
- Ziegler, C.A.; Freddolino, P.L. The leucine-responsive regulatory proteins/ feast-famine regulatory proteins: An ancient and complex class of transcriptional regulators in bacteria and archaea. Crit. Rev. Biochem. Mol. Biol. 2021, 56, 373–400. [Google Scholar] [CrossRef]
- Bhat, A.H.; Chaubey, A.K.; Půža, V. The first report of Xenorhabdus indica from Steinernema pakistanense: Co-phylogenetic study suggests co-speciation between X. indica and its steinernematid nematodes. J. Helminthol. 2019, 93, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Hurlbert, R.E.; Xu, J.; Small, C.L. Colonial and cellular polymorphism in Xenorhabdus luminescens. Appl Environ Microbiol. 1989, 55, 1136–1143. [Google Scholar] [CrossRef] [PubMed]
- Givaudan, A.; Baghdiguian, S.; Lanois, A.; Boemare, N. Swarming and swimming changes concomitant with phase variation in Xenorhabdus nematophilus. Appl. Environ. Microbiol. 1995, 1, 1408–1413. [Google Scholar] [CrossRef] [PubMed]
- Givaudan, A.; Lanois, A. FlhDC, the flagellar master operon of Xenorhabdus nematophilus: Requirement for motility, lipolysis, extracellular hemolysis, and full virulence in insects. J. Bacteriol. 2000, 182, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.J.; Boylan, B.; George, N.; Forst, S. Inactivation of ompR promotes precocious swarming and FlhDC expression in Xenorhabdus nematophila. J. Bacteriol. 2003, 185, 5290–5294. [Google Scholar] [CrossRef]
- Park, D.; Forst, S. Co-regulation of motility, exoenzyme and antibiotic production by the EnvZ-OmpR-FlhDC-FliA pathway in Xenorhabdus nematophila. Mol. Microbiol. 2006, 61, 1397–1412. [Google Scholar] [CrossRef]
- Chandra, H.; Khandelwal, P.; Khattri, A.; Banerjee, N. Type 1 fimbriae of insecticidal bacterium Xenorhabdus nematophila is necessary for growth and colonization of its symbiotic host nematode Steinernema carpocapsiae. Environ. Microbiol. 2008, 10, 1285–1295. [Google Scholar] [CrossRef]
- Hanson, A.W.; Hum, L.K. The crystal structure of. iodinin. Acta Cryst. 1969, 25, 768. [Google Scholar] [CrossRef]
- Myhren, L.E.; Nygaard, G.; Gausdal, G.; Sletta, H.; Teigen, K.; Degnes, K.F.; Zahlsen, K.; Brunsvik, A.; Bruserud, Ø.; Døskeland, S.O.; et al. Iodinin (1,6-dihydroxyphenazine 5,10-dioxide) from Streptosporangium sp. induces apoptosis selectively in myeloid leukemia cell lines and patient cells. Mar. Drugs 2013, 11, 332–349. [Google Scholar] [CrossRef]
- Sletta, H.; Degnes, K.F.; Herfindal, L.; Klinkenberg, G.; Fjærvik, E.; Zahlsen, K.; Brunsvik, A.; Nygaard, G.; Aachmann, F.L.; Ellingsen, T.E.; et al. Anti-microbial and cytotoxic 1,6-dihydroxyphenazine-5,10-dioxide (iodinin) produced by Streptosporangium sp. DSM 45942 isolated from the fjord sediment. Appl. Microbiol. Biotechnol. 2014, 98, 603–610. [Google Scholar] [CrossRef]
- Viktorsson, E.Ö.; Melling Grøthe, B.; Aesoy, R.; Sabir, M.; Snellingen, S.; Prandina, A.; Høgmoen Åstrand, O.A.; Bonge-Hansen, T.; Døskeland, S.O.; Herfindal, L.; et al. Total synthesis and antileukemic evaluations of the phenazine 5,10-dioxide natural products iodinin, myxin and their derivatives. Bioorg. Med. Chem. 2017, 25, 2285–2293. [Google Scholar] [CrossRef] [PubMed]
- Viktorsson, E.Ö.; Aesoy, R.; Støa, S.; Lekve, V.; Døskeland, S.O.; Herfindal, L.; Rongved, P. New prodrugs and analogs of the phenazine 5,10-dioxide natural products iodinin and myxin promote selective cytotoxicity towards human acute myeloid leukemia cells. RSC Med. Chem. 2021, 12, 767–778. [Google Scholar] [CrossRef] [PubMed]
- Prandina, A.; Herfindal, L.; Radix, S.; Rongved, P.; Døskeland, S.O.; Le Borgne, M.; Perret, F. Enhancement of iodinin solubility by encapsulation into cyclodextrin nanoparticles. J. Enzyme Inhib. Med. Chem. 2018, 33, 370–375. [Google Scholar] [CrossRef] [PubMed]
- SMART 5.630; Bruker Advanced X-ray Solutions. Bruker AXS Inc.: Madison, WI, USA, 2005.
- SAINT V8.40B; Bruker Advanced X-ray Solutions. Bruker AXS Inc.: Madison, WI, USA, 2020.
- Krause, L.; Herbst-Irmer, R.; Sheldrick, G.M.; Stalke, D. Comparison of silver and molybdenum microfocus X-ray sources for single-crystal structure determination. J. Appl. Crystallogr. 2015, 48 Pt 1, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Hübschle, C.B.; Sheldrick, G.M.; Dittrich, B. ShelXle: A Qt graphical user interface for SHELXL. J. Appl. Crystallogr. 2011, 44 Pt 6, 1281–1284. [Google Scholar] [CrossRef]
- SHELXTL Suite of Programs; Version 6.14, 2000–2003; Bruker Advanced X-ray Solutions; Bruker AXS Inc.: Madison, WI, USA, 2008.
- Sheldrick, G.M. A Short history of SHELX. Acta Crystallogr. 2008, 64, 112–122. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2018, 71, 3–8. [Google Scholar] [CrossRef]
- Lübben, J.; Wandtke, C.M.; Hübschle, C.B.; Ruf, M.; Sheldrick, G.M.; Dittrich, B. Aspherical scattering factors for SHELXL - model, implementation and application. Acta. Crystallogr. A Found Adv. 2019, 75 Pt 1, 50–62. [Google Scholar] [CrossRef]
- Pang, B.; Liu, T.; Zhang, W.; Ye, F.; Shang, C. Cloning and characterization of phzR gene from Pseudomonas aeruginosa. Curr. Microbiol. 2021, 78, 1482–1487. [Google Scholar] [CrossRef]
- Xiao, Y.; Meng, F.; Qiu, D.; Yang, X. Two novel antimicrobial peptides purified from the symbiotic bacteria Xenorhabdus budapestensis NMC-10. Peptides 2012, 35, 253–260. [Google Scholar] [CrossRef]
- Xi, X.; Lu, X.; Zhang, X.; Bi, Y.; Li, X.; Yu, Z. Two novel cyclic depsipeptides Xenematides F and G from the entomopathogenic bacterium Xenorhabdus budapestensis. J. Antibiot. 2019, 2, 736–743. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Qiu, D.; Wang, S. Complete genome sequence data of Xenorhabdus budapestensis strain C72, a candidate biological control agent from China. Plant Dis. 2021, 105, 3276–3278. [Google Scholar] [CrossRef] [PubMed]
- de Doucet, M.M.; Bertolotti, M.A.; Giayetto, A.L.; Miranda, M.B. Host range, specificity, and virulence of Steinernema feltiae, Steinernema rarum, and Heterorhabditis bacteriophora (Steinernematidae and Heterorhabditidae) from Argentina. J. Invertebr. Pathol. 1999, 73, 237–242. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, K.B.; Shapiro-Ilan, D.I.; Fuxa, J.R.; Wood, B.W.; Bertolotti, M.A.; Adams, B.J. Taxonomic and biological characterization of Stenernema rarum found in the Southeastern United States. J. Nematol. 2006, 38, 28–40. [Google Scholar] [PubMed]
- Castaneda-Alvarez, C.; Prodan, S.; Zamorano, A.; San-Blas, E.; Aballay, E. Xenorhabdus lircayensis sp. nov., the symbiotic bacterium associated with the entomopathogenic nematode Steinernema unicornum. Int. J. Syst. Evol. Microbiol. 2021, 71, 005151. [Google Scholar] [CrossRef] [PubMed]
- Gualtieri, M.; Ogier, J.C.; Pagès, S.; Givaudan, A.; Gaudriault, S. Draft genome sequence and annotation of the entomopathogenic bacterium Xenorhabdus szentirmaii Strain DSM16338. Genome Announc. 2014, 2, e00190-14. [Google Scholar] [CrossRef]
- Vozik, D.; Bélafi-Bakó, K.; Hevesi, M.; Böszöményi, E.; Fodor, A. Effectiveness of a peptide-rich fraction from Xenorhabdus budapestensis culture against fire blight disease on apple blossoms. Not. Bot. Horti Agrobo 2015, 43, 547–553. Available online: https://www.notulaebotanicae.ro/ndex.php/index (accessed on 22 February 2022). [CrossRef]
- Böszörményi, E.; Barcs, I.; Domján, G.; Bélafiné Bakó, K.; Fodor, A.; Makrai, L.; Vozik, D. Antimicrobial effect on some zoonotic bacteria, of the cell-free fermentation fluid and purified peptide fraction of the entomopathogenic bacterium, Xenorhabdus budapestensis. Orv. Hetil. 2015, 156, 1782–1786. (In Hungarian) [Google Scholar] [CrossRef]
- Brenner, D.J.; McWhorter, A.C.; Knutson, J.K.; Steigerwalt, A.G. Escherichia vulneris: A new species of Enterobacteriaceae associated with human wounds. J. Clin. Microbiol. 1982, 15, 1133–1140. [Google Scholar] [CrossRef]
- Vallenet, D.; Belda, E.; Calteau, A.; Cruveiller, S.; Engelen, S.; Lajus, A.; Le Fèvre, F.; Longin, C.; Mornico, D.; Roche, D.; et al. MicroScope—an integrated microbial resource for the curation and comparative analysis of genomic and metabolic data. Nucleic Acids Res. 2013, 41, D636–D647. [Google Scholar] [CrossRef]
- Dreyer, J.; Rautenbach, M.; Booysen, E.; van Staden, A.D.; Deane, S.M.; Dicks, L.M.T. Xenorhabdus khoisanae SB10 produces Lys-rich PAX lipopeptides and a Xenocoumacin in its antimicrobial complex. BMC Microbiol. 2019, 19, 132. [Google Scholar] [CrossRef] [PubMed]
- Brachmann, A.O.; Forst, S.; Furgani, G.M.; Fodor, A.; Bode, H.B. Xenofuranones A and B: Phenylpyruvate dimers from Xenorhabdus szentirmaii. J. Nat. Prod. 2006, 69, 1830–1832. [Google Scholar] [CrossRef] [PubMed]
- Brachmann, A.O. Isolation and Identification of Natural Products, and Biosynthetic Pathways from Photorhabdus and Xenorhabdus. Ph.D Thesis, Saarland University, Saarbrücken, Germany, 2009. [Google Scholar]
- Ohlendorf, B.; Simon, S.; Wiese, J.; Imhoff, J.F. Szentiamide, an N-formylated cyclic depsipeptide from Xenorhabdus szentirmaii DSM 16338T. Nat. Prod. Commun. 2011, 6, 1247–1250. [Google Scholar] [CrossRef] [PubMed]
- Nollmann, F.I.; Dowling, A.; Kaiser, M.; Deckmann, K.; Grösch, S.; Ffrench-Constant, R.; Bode, H.B. Synthesis of szentiamide, a depsipeptide from entomopathogenic Xenorhabdus szentirmaii with activity against Plasmodium falciparum. Beilstein J. Org. Chem. 2012, 8, 528–533. [Google Scholar] [CrossRef] [PubMed]
- Booysen, E.; Rautenbach, M.; Stander, M.A.; Dicks, L.M.T. Profiling the production of antimicrobial secondary metabolites by Xenorhabdus khoisanae J194 under different culturing conditions. Front. Chem. 2021, 9, 626653. [Google Scholar] [CrossRef]
- Houard, J.; Aumelas, A.; Noël, T.; Pages, S.; Givaudan, A.; Fitton-Ouhabi, V.; Villain-Guillot, P.; Gualtieri, M. Cabanillasin, a new antifungal metabolite, produced by entomopathogenic Xenorhabdus cabanillasii JM26. J. Antibiot. 2013, 66, 617–620. [Google Scholar] [CrossRef]
- Kronenwerth, M.; Bozhüyük, K.A.; Kahnt, A.S.; Steinhilber, D.; Gaudriault, S.; Kaiser, M.; Bode, H.B. Characterisation of taxlllaids A-G; natural products from Xenorhabdus indica. Chemistry 2014, 20, 17478–17487. [Google Scholar] [CrossRef]
- Reimer, D.; Nollmann, F.I.; Schultz, K.; Kaiser, M.; Bode, H.B. Xenortide Biosynthesis by entomopathogenic Xenorhabdus nematophila. J. Nat. Prod. 2014, 77, 1976–1980. [Google Scholar] [CrossRef]
- Esmati, N.; Maddirala, A.R.; Hussein, N.; Amawi, H.; Tiwari, A.K.; Andreana, P.R. Efficient syntheses and anti-cancer activity of xenortides A-D including ent/epi-stereoisomers. Org. Biomol. Chem. 2018, 16, 5332–5342. [Google Scholar] [CrossRef]
- Guo, S.; Zhang, S.; Fang, X.; Liu, Q.; Gao, J.; Bilal, M.; Wang, Y.; Zhang, X. Regulation of antimicrobial activity and xenocoumacins biosynthesis by pH in Xenorhabdus nematophila. Microb. Cell Fact. 2017, 16, 203. [Google Scholar] [CrossRef]
- Racine, E.; Gualtieri, M. From worms to drug candidate: The story of odilorhabdins, a new class of antimicrobial agents. Front. Microbiol. 2019, 10, 2893. [Google Scholar] [CrossRef] [PubMed]
- Pantel, L.; Florin, T.; Dobosz-Bartoszek, M.; Racine, E.; Sarciaux, M.; Serri, M.; Houard, J.; Campagne, J.M.; de Figueiredo, R.M.; Midrier, C.; et al. Odilorhabdins, antibacterial agents that cause miscoding by binding at a new ribosomal site. Mol. Cell 2018, 70, 83–94.e7. [Google Scholar] [CrossRef] [PubMed]
- Loza, E.; Sarciaux, M.; Ikaunieks, M.; Katkevics, M.; Kukosha, T.; Trufilkina, N.; Ryabova, V.; Shubin, K.; Pantel, L.; Serri, M.; et al. Structure-activity relationship studies on the inhibition of the bacterial translation of novel Odilorhabdins analogues. Bioorg. Med. Chem. 2020, 28, 115469. [Google Scholar] [CrossRef] [PubMed]
- Dreyer, J.; Malan, A.P.; Dicks, L.M.T. Bacteria of the genus Xenorhabdus, a novel source of bioactive compounds. Front. Microbiol. 2018, 9, 3177. [Google Scholar] [CrossRef]
- Zhao, L.; Kaiser, M.; Bode, H.B. Rhabdopeptide/Xenortide-like peptides from Xenorhabdus innexi with terminal amines showing potent antiprotozoal activity. Org. Lett. 2018, 20, 5116–5120. [Google Scholar] [CrossRef]
- Kim, S.K.; Flores-Lara, Y.; Stock, P.S. Morphology and ultrastructure of the bacterial receptacle in Steinernema nematodes (Nematoda: Steinernematidae). J. Invertebr. Pathol. 2012, 110, 366–374. [Google Scholar] [CrossRef]
- Furgani, G.M. Gnotobiological Analysis of Entomopathogenic Nematode/Bacterium Symbiotic Complexes. Ph.D. Thesis, . Eötvös University, Budapest, Hungary, 2008. [Google Scholar]
- Stock, S.P. Partners in crime: Symbiont-assisted resource acquisition in Steinernema entomopathogenic nematodes. Curr. Opin. Insect Sci. 2019, 32, 22–27. [Google Scholar] [CrossRef]
- Uribe-Lorío, L.; Mora, M.; Stock, S.P. Steinernema costaricense n. sp. and S. puntauvense n. sp. (Rhabditida: Steinernematidae), two new entomopathogenic nematodes from Costa Rica. Syst. Parasitol. 2007, 68, 167–182. [Google Scholar] [CrossRef]
- Ivanova, E.S.; Shepeleva, N.S.; Spiridonov, S.E. Morphological and molecular characterisation of Steinernema costaricense Uribe-Lorío, Mora and & Stock, 2007 (Panagrolaimorpha: Steinernematidae) isolate from Bush Augusta State Park, Missouri, USA. Syst. Parasitol. 2013, 85, 219–234. [Google Scholar] [CrossRef]
- Glaz, E.T.; Scheiber, E.; Gyimesi, J.; Horvath, I.; Steczek, K.; Szentirmai, A.; Bohus, G. A new trichothecin-like antifungal antibiotic. Nature 1959, 184 (Suppl. S12), 908. [Google Scholar] [CrossRef]
- Horváth, I.; Szentirmai, A. Inhibitory effect of fungistatic antibiotics on the production of amylase by Penicillium chrysogenum. Nature 1959, 184, 57–58. [Google Scholar] [CrossRef] [PubMed]
- Horvath, I.; Bajusz, S.; Szentirmai, A. Inhibitory action of amino-acid esters on the production of extracellular amylase by Penicillium chrysogenum. Nature 1959, 183, 477. [Google Scholar] [CrossRef] [PubMed]
- Szentirmai, A.; Horváth, I. Regulation of branched-chain amino acid biosynthesis. Acta Microbiol. Acad. Sci. Hung. 1976, 23, 137–149. [Google Scholar] [PubMed]
- Karaffa, L.; Sándor, E.; Kozma, J.; Kubicek, C.P.; Szentirmai, A. The role of the alternative respiratory pathway in the stimulation of cephalosporin C formation by soybean oil in Acremonium chrysogenum. Appl. Microbiol. Biotechnol. 1999, 51, 633–638. [Google Scholar] [CrossRef]
- Karaffa, L.; Sándor, E.; Fekete, E.; Szentirmai, A. The biochemistry of citric acid accumulation by Aspergillus niger. Acta Microbiol. Immunol. Hung. 2001, 48, 429–440. [Google Scholar] [CrossRef]
- Fekete, E.; Karaffa, L.; Sándor, E.; Bányai, I.; Seiboth, B.; Gyémánt, G.; Sepsi, A.; Szentirmai, A.; Kubicek, C.P. The alternative D-galactose degrading pathway of Aspergillus nidulans proceeds via L-sorbose. Arch. Microbiol. 2004, 181, 35–44. [Google Scholar] [CrossRef]
- Ilyés, H.; Fekete, E.; Karaffa, L.; Fekete, E.; Sándor, E.; Szentirmai, A.; Kubicek, C.P. CreA-mediated carbon catabolite repression of beta-galactosidase formation in Aspergillus nidulans is growth rate dependent. FEMS Microbiol. Lett. 2004, 235, 147–151. [Google Scholar] [CrossRef]
- Karaffa, L.; Fekete, E.; Gamauf, C.; Szentirmai, A.; Kubicek, C.P.; Seiboth, B. D-Galactose induces cellulase gene expression in Hypocrea jecorina at low growth rates. Microbiology 2006, 152, 1507–1514. [Google Scholar] [CrossRef]
- Fekete, E.; Seiboth, B.; Kubicek, C.P.; Szentirmai, A.; Karaffa, L. Lack of aldose 1-epimerase in Hypocrea jecorina (anamorph Trichoderma reesei): A key to cellulase gene expression on lactose. Proc. Natl. Acad. Sci. USA 2008, 105, 7141–7146. [Google Scholar] [CrossRef]
- Jónás, Á.; Fekete, E.; Flipphi, M.; Sándor, E.; Jäger, S.; Molnár, Á.P.; Szentirmai, A.; Karaffa, L. Extra- and intracellular lactose catabolism in Penicillium chrysogenum: Phylogenetic and expression analysis of the putative permease and hydrolase genes. J. Antibiot. 2014, 67, 489–497. [Google Scholar] [CrossRef]
- Embley, T.M.; Stackebrandt, E. The molecular phylogeny and systematics of the actinomycetes. Annu. Rev. Microbiol. 1994, 48, 257–289. [Google Scholar] [CrossRef] [PubMed]
- Venkateswaran, K.; Moser, D.P.; Dollhopf, M.E.; Lies, D.P.; Saffarini, D.A.; MacGregor, B.J.; Ringelberg, D.B.; White, D.C.; Nishijima, M.; Sano, H.; et al. Polyphasic taxonomy of the genus Shewanella and description of Shewanella oneidensis sp. nov. Int. J. Syst. Bacteriol. 1999, 49, 705–724. [Google Scholar] [CrossRef] [PubMed]
- Pütz, J.; Meinert, F.; Wyss, U.; Ehlers, R.U.; Stackebrandt, E. Development and application of oligonucleotide probes for molecular identification of Xenorhabdus species. Appl. Environ. Microbiol. 1990, 56, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Rainey, F.A.; Ehlers, R.U.; Stackebrandt, E. Inability of the polyphasic approach to systematics to determine the relatedness of the genera Xenorhabdus and Photorhabdus. Int. J. Syst. Bacteriol. 1995, 45, 379–381. [Google Scholar] [CrossRef][Green Version]
- Bode, H.B.; Reimer, D.; Fuchs, S.W.; Kirchner, F.; Dauth, C.; Kegler, C.; Lorenzen, W.; Brachmann, A.O.; Grün, P. Determination of the absolute configuration of peptide natural products by using stable isotope labeling and mass spectrometry. Chemistry 2012, 18, 2342–2348. [Google Scholar] [CrossRef]


| Bacterium EPB Species | Strain | Nematode | Genome Information | |||
|---|---|---|---|---|---|---|
| Ref | EPN Partner | Ref | Genome Announcement | Ref | ||
| Xenorhabdus budapestensis | DSM16342(T) | [9] | S. bicornutum Central Europe, 1995 | [11] | Bode H (https://www.genoscope.cns.fr/agc/microscope/home/index.php (accessed on 25 February 2022)). | - |
| D43, designated as HIP57 | [87] | S. bicornutum China, 2012 | [87] | - | - | |
| NMC-10 | [133] | S. bicornutum China, 2012 | [133] | - | - | |
| SN84 | [92] | S. bicornutum China, 2018 | [92] | - | - | |
| SN84 | [134] | [134] | - | - | ||
| C72 | [135] | S. bicornutum China, 2021 | [135] | https://www.ncbi.nlm.nih.gov/nuccore/2021543890/ (accessed on 25 February 2022). | [99] | |
| Xenorhabdus szentirmaii | DSM16338(T) | [7] | S. rarum, Cordoba, Argentina, South America | [8] | implemented in the public XenorhabduScope database (https://wwwgenoscope.cns.fr/agc/microscope/home/index.php (accessed on 25 February 2022)). | [116] |
| Xenorhabdus | Biosynthetic Operons, Antimicrobial Products | Reference | |
|---|---|---|---|
| Species | Strain | ||
| szentirmaii | [9] Lengyel et al., 2005 DSM16338T (EMC) | Draft Genome Annotation, | [139] |
| Iodinin | [24,86,132]; | ||
| Phenazine | [132] | ||
| Xenocoumacines | [145,146] | ||
| Szentiamide, | [147,148] | ||
| Fabclavine | [9,18,19,20,21,22] | ||
| budapestensis | [9] Lengyel et al., 2005 DSM16342T (EMA) | Fabclavine, Bicornutin | [7,18,19] |
| [135] Li et al., 2021 C72 | Genome Annotation | [135] | |
| [133] (Xiao et al., 2012) NMC10 | GP-19, EP-20 | [133] | |
| [134] Xi et al., 2021 SN84 | xenematides F xenematides G(depsipeptides | [134] | |
| [92] Bi et al., 2018 | Rhabdopeptides | [92] | |
| [87] Yang et al., 2012 | Insecticidal protein | [87] | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fodor, A.; Gualtieri, M.; Zeller, M.; Tarasco, E.; Klein, M.G.; Fodor, A.M.; Haynes, L.; Lengyel, K.; Forst, S.A.; Furgani, G.M.; et al. Type Strains of Entomopathogenic Nematode-Symbiotic Bacterium Species, Xenorhabdus szentirmaii (EMC) and X. budapestensis (EMA), Are Exceptional Sources of Non-Ribosomal Templated, Large-Target-Spectral, Thermotolerant-Antimicrobial Peptides (by Both), and Iodinin (by EMC). Pathogens 2022, 11, 342. https://doi.org/10.3390/pathogens11030342
Fodor A, Gualtieri M, Zeller M, Tarasco E, Klein MG, Fodor AM, Haynes L, Lengyel K, Forst SA, Furgani GM, et al. Type Strains of Entomopathogenic Nematode-Symbiotic Bacterium Species, Xenorhabdus szentirmaii (EMC) and X. budapestensis (EMA), Are Exceptional Sources of Non-Ribosomal Templated, Large-Target-Spectral, Thermotolerant-Antimicrobial Peptides (by Both), and Iodinin (by EMC). Pathogens. 2022; 11(3):342. https://doi.org/10.3390/pathogens11030342
Chicago/Turabian StyleFodor, András, Maxime Gualtieri, Matthias Zeller, Eustachio Tarasco, Michael G. Klein, Andrea M. Fodor, Leroy Haynes, Katalin Lengyel, Steven A. Forst, Ghazala M. Furgani, and et al. 2022. "Type Strains of Entomopathogenic Nematode-Symbiotic Bacterium Species, Xenorhabdus szentirmaii (EMC) and X. budapestensis (EMA), Are Exceptional Sources of Non-Ribosomal Templated, Large-Target-Spectral, Thermotolerant-Antimicrobial Peptides (by Both), and Iodinin (by EMC)" Pathogens 11, no. 3: 342. https://doi.org/10.3390/pathogens11030342
APA StyleFodor, A., Gualtieri, M., Zeller, M., Tarasco, E., Klein, M. G., Fodor, A. M., Haynes, L., Lengyel, K., Forst, S. A., Furgani, G. M., Karaffa, L., & Vellai, T. (2022). Type Strains of Entomopathogenic Nematode-Symbiotic Bacterium Species, Xenorhabdus szentirmaii (EMC) and X. budapestensis (EMA), Are Exceptional Sources of Non-Ribosomal Templated, Large-Target-Spectral, Thermotolerant-Antimicrobial Peptides (by Both), and Iodinin (by EMC). Pathogens, 11(3), 342. https://doi.org/10.3390/pathogens11030342