Prevalence, Infectious Characteristics and Genetic Diversity of Staphylococcus aureus and Methicillin-Resistant Staphylococcus aureus (MRSA) in Two Raw-Meat Processing Establishments in Northern Greece
Abstract
:1. Introduction
2. Results
2.1. Isolation of S. aureus
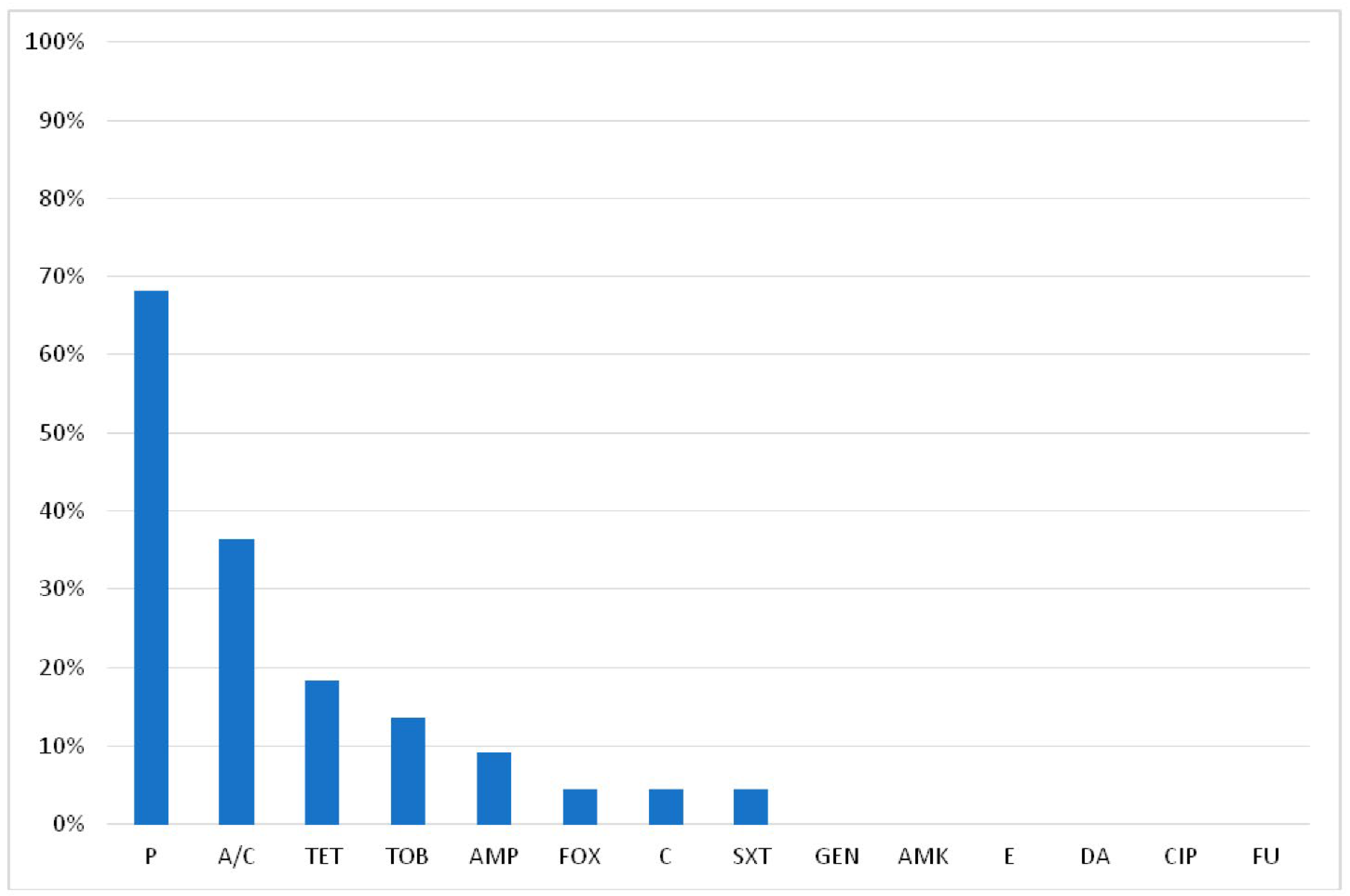
2.2. Antimicrobial Resistance of S. aureus
2.3. Carriage of the mecA, mecC, tst, lukF-PV and Enterotoxin Genes
2.4. Biofilm Formation Ability
2.5. Spa Typing
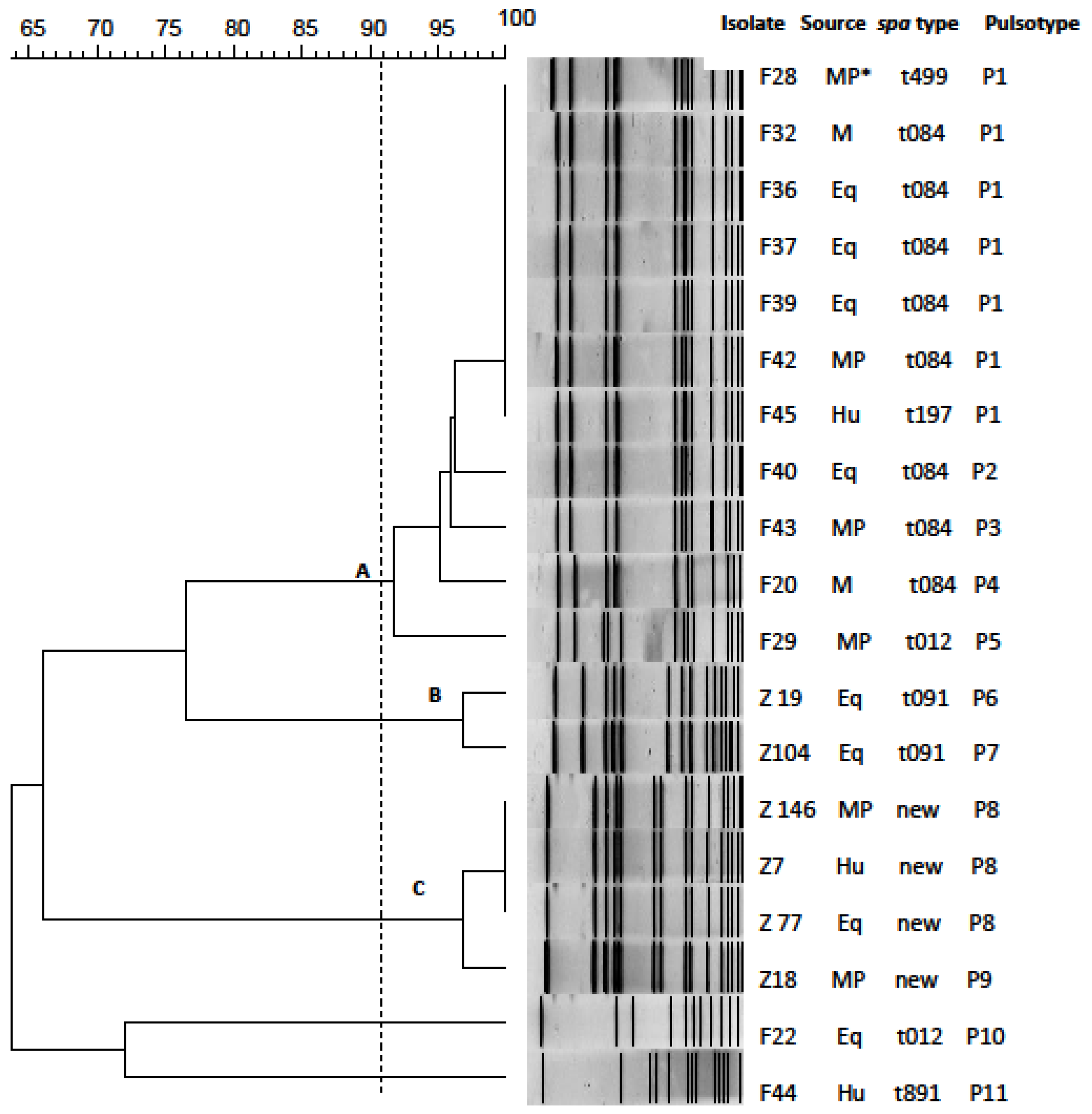
2.6. Pulsed Field Gel Electrophoresis Typing
2.7. Next-Generation Sequencing
3. Discussion
3.1. Isolation, Antimicrobial Susceptibly and Biofilm Formation Ability of S. aureus and MRSA Isolates
3.2. Enterotoxin Gene Carriage of S. aureus Isolates
3.3. Carriage of Genes Encoding for the Toxic Shock Syndrome and Panton–Valentine Leukocidin Toxins
3.4. Genetic Diversity of S. aureus Isolates
3.5. Next-Generation Sequencing
4. Materials and Methods
4.1. Sample Collection
4.2. Isolation and Identification of Staphylococcus aureus
4.3. Molecular Characterization of Staphylococcus aureus
4.4. Detection of Virulence Genes and Methicillin Resistance Genes
4.5. Antimicrobial Susceptibility Testing
4.6. Assessment of Biofilm-Formation Ability
4.7. Pulsed Field Gel Electrophoresis Analysis
4.8. Spa Typing
4.9. Next-Generation Sequencing and Analysis of the Whole Genome
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kaspar, U.; Kriegeskorte, A.; Schubert, T.; Peters, G.; Rudack, C.; Pieper, D.H.; Wos-Oxley, M.; Becker, K. The culturome of the human nose habitats reveals individual bacterial fingerprint patterns. Environ. Microbiol. 2016, 18, 2130–2142. [Google Scholar] [CrossRef] [PubMed]
- Lowy, F.D. Staphylococcus aureus Infections. N. Engl. J. Med. 1998, 339, 520–532. [Google Scholar] [CrossRef] [PubMed]
- Cuny, C.; Köck, R.; Witte, W. Livestock associated MRSA (LA-MRSA) and its relevance for humans in Germany. Int. J. Med. Microbiol. 2013, 303, 331–337. [Google Scholar] [CrossRef]
- Fitzgerald, J.R. Livestock-associated Staphylococcus aureus: Origin, evolution and public health threat. Trends Microbiol. 2012, 20, 192–198. [Google Scholar] [CrossRef]
- Verkade, E.; Kluytmans, J. Livestock-associated Staphylococcus aureus CC398: Animal reservoirs and human infections. Infect. Genet. Evol. 2014, 21, 523–530. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority. Assessment of the Public Health significance of meticillin resistant Staphylococcus aureus (MRSA) in animals and foods. EFSA J. 2009, 7, 1–73. [Google Scholar] [CrossRef]
- De Jonge, R.; E Verdier, J.; Havelaar, A.H. Prevalence of meticillin-resistant Staphylococcus aureus amongst professional meat handlers in the Netherlands, March–July 2008. Eurosurveillance 2010, 15, 19712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drougka, E.; Foka, A.; Giormezis, N.; Sergelidis, D.; Militsopoulou, M.; Jelastopulu, E.; Komodromos, D.; Sarrou, S.; Anastassiou, E.D.; Petinaki, E.; et al. Multidrug-resistant enterotoxigenic Staphylococcus aureus lineages isolated from animals, their carcasses, the personnel, and the environment of an abattoir in Greece. J. Food Process. Preserv. 2019, 43, e13961. [Google Scholar] [CrossRef]
- Huber, H.; Koller, S.; Giezendanner, N.; Stephan, R.; Zweifel, C. Prevalence and characteristics of meticillin-resistant Staphylococcus aureus in humans in contact with farm animals, in livestock, and in food of animal origin, Switzerland, 2009. Eurosurveillance 2010, 15, 19542. [Google Scholar] [CrossRef]
- Klimešová, M.; Manga, I.; Nejeschlebová, L.; Horáček, J.; Ponížil, A.; Vondrušková, E. Occurrence of Staphylococcus aureus in cattle, sheep, goat, and pig rearing in the Czech Republic. Acta Vet. Brno 2017, 86, 3–10. [Google Scholar] [CrossRef]
- Van Cleef, B.A.G.L.; Broens, E.M.; Voss, A.; Huijsdens, X.W.; Züchner, L.; VAN Benthem, B.H.B.; Kluytmans, J.A.J.W.; Mulders, M.N.; VAN DE Giessen, A.W. High prevalence of nasal MRSA carriage in slaughterhouse workers in contact with live pigs in The Netherlands. Epidemiol. Infect. 2010, 138, 756–763. [Google Scholar] [CrossRef]
- Bridier, A.; Sanchez-Vizuete, P.; Guilbaud, M.; Piard, J.-C.; Naïtali, M.; Briandet, R. Biofilm-associated persistence of food-borne pathogens. Food Microbiol. 2015, 45, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Møretrø, T.; Langsrud, S. Residential Bacteria on Surfaces in the Food Industry and Their Implications for Food Safety and Quality. Compr. Rev. Food Sci. Food Saf. 2017, 16, 1022–1041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donlan, R.M. Biofilms: Microbial Life on Surfaces. Emerg. Infect. Dis. 2002, 8, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Dass, S.C.; Wang, R. Biofilm through the Looking Glass: A Microbial Food Safety Perspective. Pathogens 2022, 11, 346. [Google Scholar] [CrossRef]
- Lianou, A.; Nychas, G.-J.E.; Koutsoumanis, K.P. Strain variability in biofilm formation: A food safety and quality perspective. Food Res. Int. 2020, 137, 109424. [Google Scholar] [CrossRef]
- Chen, C.-J.; Huang, Y.-C. New epidemiology of Staphylococcus aureus infection in Asia. Clin. Microbiol. Infect. 2014, 20, 605–623. [Google Scholar] [CrossRef] [Green Version]
- Stryjewski, M.; Corey, G.R. Methicillin-Resistant Staphylococcus aureus: An Evolving Pathogen. Clin. Infect. Dis. 2014, 58, S10–S19. [Google Scholar] [CrossRef] [Green Version]
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2017/2018. EFSA J. 2020, 18, e06007. [Google Scholar] [CrossRef] [Green Version]
- Ou, Q.; Peng, Y.; Lin, D.; Bai, C.; Zhang, T.; Lin, J.; Ye, X.; Yao, Z. A Meta-Analysis of the Global Prevalence Rates of Staphylococcus aureus and Methicillin-Resistant S. aureus Contamination of Different Raw Meat Products. J. Food Prot. 2017, 80, 763–774. [Google Scholar] [CrossRef]
- Palá, T.R.; Sevilla, A. Microbial Contamination of Carcasses, Meat, and Equipment from an Iberian Pork Cutting Plant. J. Food Prot. 2004, 67, 1624–1629. [Google Scholar] [CrossRef] [PubMed]
- Kluytmans, J.A.J.W. Methicillin-resistant Staphylococcus aureus in food products: Cause for concern or case for complacency? Clin. Microbiol. Infect. 2010, 16, 11–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, C.; Perry, M.J.; Sorock, G.S.; Hauser, R.; Spanjer, K.J.; Mittleman, M.; Stentz, T.L. Laceration injuries among workers at meat packing plants. Am. J. Ind. Med. 2005, 47, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Kyeremateng-Amoah, E.; Nowell, J.; Lutty, A.; Lees, P.S.; Ba, E.K.S. Laceration injuries and infections among workers in the poultry processing and pork meatpacking industries. Am. J. Ind. Med. 2014, 57, 669–682. [Google Scholar] [CrossRef]
- Neyra, R.C.; Frisancho, J.A.; Rinsky, J.L.; Resnick, C.; Carroll, K.C.; Rule, A.M.; Ross, T.; You, Y.; Price, L.B.; Silbergeld, E.K. Multidrug-Resistant and Methicillin-Resistant Staphylococcus aureus (MRSA) in Hog Slaughter and Processing Plant Workers and Their Community in North Carolina (USA). Environ. Health Perspect. 2014, 122, 471–477. [Google Scholar] [CrossRef] [Green Version]
- Karapsias, S.; Piperaki, E.T.; Spiliopoulou, I.; Katsanis, G.; Kotsovili, A.T. Methicillin-resistant Staphylococcus aureus nasal carriage among healthy employees of the Hellenic Air Force. Eurosurveillance 2008, 13, 18999. [Google Scholar] [CrossRef]
- Beneke, B.; Klees, S.; Stührenberg, B.; Fetsch, A.; Kraushaar, B.; Tenhagen, B.-A. Prevalence of Methicillin-Resistant Staphylococcus aureus in a Fresh Meat Pork Production Chain. J. Food Prot. 2011, 74, 126–129. [Google Scholar] [CrossRef]
- Jackson, C.R.; Davis, J.A.; Barrett, J.B. Prevalence and Characterization of Methicillin-Resistant Staphylococcus aureus Isolates from Retail Meat and Humans in Georgia. J. Clin. Microbiol. 2013, 51, 1199–1207. [Google Scholar] [CrossRef] [Green Version]
- Leibler, J.H.; Jordan, J.A.; Brownstein, K.; Lander, L.; Price, L.B.; Perry, M.J. Staphylococcus aureus Nasal Carriage among Beefpacking Workers in a Midwestern United States Slaughterhouse. PLoS ONE 2016, 11, e0148789. [Google Scholar] [CrossRef] [Green Version]
- Robinson, D.A.; Enright, M.C. Evolutionary Models of the Emergence of Methicillin-Resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2003, 47, 3926–3934. [Google Scholar] [CrossRef]
- Ivbule, M.; Miklaševičs, E.; Čupāne, L.; Bērziņa, L.; Bālinš, A.; Valdovska, A. Presence of methicillin-resistant Staphylococcus aureus in slaughterhouse environment, pigs, carcasses, and workers. J. Vet. Res. 2017, 61, 267–277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papadopoulos, P.; Papadopoulos, T.; Angelidis, A.S.; Boukouvala, E.; Zdragas, A.; Papa, A.; Hadjichristodoulou, C.; Sergelidis, D. Prevalence of Staphylococcus aureus and of methicillin-resistant S. aureus (MRSA) along the production chain of dairy products in north-western Greece. Food Microbiol. 2018, 69, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Parisi, A.; Caruso, M.; Normanno, G.; Latorre, L.; Miccolupo, A.; Fraccalvieri, R.; Intini, F.; Manginelli, T.; Santagada, G. MRSA in swine, farmers and abattoir workers in Southern Italy. Food Microbiol. 2019, 82, 287–293. [Google Scholar] [CrossRef]
- Okorie-Kanu, O.J.; Anyanwu, M.U.; Ezenduka, E.V.; Mgbeahuruike, A.C.; Thapaliya, D.; Gerbig, G.; Ugwuijem, E.E.; Okorie-Kanu, C.O.; Agbowo, P.; Olorunleke, S.; et al. Molecular epidemiology, genetic diversity and antimicrobial resistance of Staphylococcus aureus isolated from chicken and pig carcasses, and carcass handlers. PLoS ONE 2020, 15, e0232913. [Google Scholar] [CrossRef] [PubMed]
- Ou, C.; Shang, D.; Yang, J.; Chen, B.; Chang, J.; Jin, F.; Shi, C. Prevalence of multidrug-resistant Staphylococcus aureus isolates with strong biofilm formation ability among animal-based food in Shanghai. Food Control 2020, 112, 107106. [Google Scholar] [CrossRef]
- Mechesso, A.F.; Moon, D.C.; Ryoo, G.-S.; Song, H.-J.; Chung, H.Y.; Kim, S.U.; Choi, J.-H.; Kang, H.Y.; Na, S.H.; Yoon, S.-S.; et al. Resistance profiling and molecular characterization of Staphylococcus aureus isolated from goats in Korea. Int. J. Food Microbiol. 2021, 336, 108901. [Google Scholar] [CrossRef] [PubMed]
- Acco, M.; Ferreira, F.; Henriques, J.; Tondo, E. Identification of multiple strains of Staphylococcus aureus colonizing nasal mucosa of food handlers. Food Microbiol. 2003, 20, 489–493. [Google Scholar] [CrossRef]
- Castro, A.; Santos, C.; Meireles, H.; Silva, J.; Teixeira, P. Food handlers as potential sources of dissemination of virulent strains of Staphylococcus aureus in the community. J. Infect. Public Health 2016, 9, 153–160. [Google Scholar] [CrossRef] [Green Version]
- Seow, W.-L.; Mahyudin, N.A.; Amin-Nordin, S.; Radu, S.; Abdul-Mutalib, N.A. Antimicrobial resistance of Staphylococcus aureus among cooked food and food handlers associated with their occupational information in Klang Valley, Malaysia. Food Control 2021, 124, 107872. [Google Scholar] [CrossRef]
- di Cerbo, A.; Pezzuto, F.; Guidetti, G.; Canello, S.; Corsi, L. Tetracyclines: Insights and Updates of their Use in Human and Animal Pathology and their Potential Toxicity. Open Biochem. J. 2019, 13, 1–12. [Google Scholar] [CrossRef]
- Ge, B.; Mukherjee, S.; Hsu, C.-H.; Davis, J.A.; Tran, T.T.T.; Yang, Q.; Abbott, J.W.; Ayers, S.L.; Young, S.R.; Crarey, E.T.; et al. MRSA and multidrug-resistant Staphylococcus aureus in U.S. retail meats, 2010–2011. Food Microbiol. 2017, 62, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Jones, T.F.; E Kellum, M.; Porter, S.S.; Bell, M.; Schaffner, W. An outbreak of community-acquired foodborne illness caused by methicillin-resistant Staphylococcus aureus. Emerg. Infect. Dis. 2002, 8, 82–84. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, H.; Liang, L.; Xu, X.; Zhou, G. Prevalence, genetic characterization and biofilm formation in vitro of Staphylococcus aureus isolated from raw chicken meat at retail level in Nanjing, China. Food Control. 2018, 86, 11–18. [Google Scholar] [CrossRef]
- Di Ciccio, P.; Vergara, A.; Festino, A.; Paludi, D.; Zanardi, E.; Ghidini, S.; Ianieri, A. Biofilm formation by Staphylococcus aureus on food contact surfaces: Relationship with temperature and cell surface hydrophobicity. Food Control. 2015, 50, 930–936. [Google Scholar] [CrossRef]
- Gutiérrez, D.; Delgado, S.; Vázquez-Sánchez, D.; Martínez, B.; Cabo, M.L.; Rodríguez, A.; Herrera, J.J.; García, P. Incidence of Staphylococcus aureus and Analysis of Associated Bacterial Communities on Food Industry Surfaces. Appl. Environ. Microbiol. 2012, 78, 8547–8554. [Google Scholar] [CrossRef] [Green Version]
- Sofos, J.N.; Geornaras, I. Overview of current meat hygiene and safety risks and summary of recent studies on biofilms, and control of Escherichia coli O157:H7 in nonintact, and Listeria monocytogenes in ready-to-eat, meat products. Meat Sci. 2010, 86, 2–14. [Google Scholar] [CrossRef]
- Hao, D.; Xing, X.; Li, G.; Wang, X.; Zhang, M.; Zhang, W.; Xia, X.; Meng, J. Prevalence, Toxin Gene Profiles, and Antimicrobial Resistance of Staphylococcus aureus Isolated from Quick-Frozen Dumplings. J. Food Prot. 2015, 78, 218–223. [Google Scholar] [CrossRef]
- Normanno, G.; Firinu, A.; Virgilio, S.; Mula, G.; Dambrosio, A.; Poggiu, A.; Decastelli, L.; Mioni, R.; Scuota, S.; Bolzoni, G. Coagulase-positive Staphylococci and Staphylococcus aureus in food products marketed in Italy. Int. J. Food Microbiol. 2005, 98, 73–79. [Google Scholar] [CrossRef]
- Ho, J.; O’Donoghue, M.; Boost, M. Occupational exposure to raw meat: A newly-recognized risk factor for Staphylococcus aureus nasal colonization amongst food handlers. Int. J. Hyg. Environ. Health 2014, 217, 347–353. [Google Scholar] [CrossRef]
- Argudín, M.Á.; Mendoza, M.C.; Rodicio, M.R. Food Poisoning and Staphylococcus aureus Enterotoxins. Toxins 2010, 2, 1751–1773. [Google Scholar] [CrossRef]
- International Commission on Microbiological Specifications for Foods. Microorganisms in Foods 6: Microbial Ecology of Food Commodities; Kluwer Academic/Plenum Publishers: New York, NY, USA, 2005. [Google Scholar]
- Osman, M.; Kamal-Dine, K.; El Omari, K.; Rafei, R.; Dabboussi, F.; Hamze, M. Prevalence of Staphylococcus aureus methicillin-sensitive and methicillin-resistant nasal carriage in food handlers in Lebanon: A potential source of transmission of virulent strains in the community. Access Microbiol. 2019, 1, e000043. [Google Scholar] [CrossRef] [PubMed]
- Sospedra, I.; Mañes, J.; Soriano, J. Report of toxic shock syndrome toxin 1 (TSST-1) from Staphylococcus aureus isolated in food handlers and surfaces from foodservice establishments. Ecotoxicol. Environ. Saf. 2012, 80, 288–290. [Google Scholar] [CrossRef] [PubMed]
- Danelli, T.; Duarte, F.C.; De Oliveira, T.A.; Da Silva, R.S.; Alfieri, D.F.; Gonçalves, G.B.; De Oliveira, C.F.; Tavares, E.R.; Yamauchi, L.M.; Perugini, M.R.E.; et al. Nasal Carriage by Staphylococcus aureus among Healthcare Workers and Students Attending a University Hospital in Southern Brazil: Prevalence, Phenotypic, and Molecular Characteristics. Interdiscip. Perspect. Infect. Dis. 2020, 2020, 3808036. [Google Scholar] [CrossRef] [PubMed]
- Hogan, B.; Rakotozandrindrainy, R.; Al-Emran, H.; Dekker, D.; Hahn, A.; Jaeger, A.; Poppert, S.; Frickmann, H.; Hagen, R.M.; Micheel, V.; et al. Prevalence of nasal colonisation by methicillin-sensitive and methicillin-resistant Staphylococcus aureus among healthcare workers and students in Madagascar. BMC Infect. Dis. 2016, 16, 420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pakbaz, Z.; Sahraian, M.A.; Sabzi, S.; Mahmoodi, M.; Pourmand, M.R. Prevalence of sea, seb, sec, sed, and tsst-1 genes of Staphylococcus aureus in nasal carriage and their association with multiple sclerosis. Germs 2017, 7, 171–177. [Google Scholar] [CrossRef] [Green Version]
- Lozano, C.; Gómez-Sanz, E.; Benito, D.; Aspiroz, C.; Zarazaga, M.; Torres, C. Staphylococcus aureus nasal carriage, virulence traits, antibiotic resistance mechanisms, and genetic lineages in healthy humans in Spain, with detection of CC398 and CC97 strains. Int. J. Med. Microbiol. 2011, 301, 500–505. [Google Scholar] [CrossRef]
- Elbargisy, R.M.; Rizk, D.E.; Abdel-Rhman, S.H. Toxin gene profile and antibiotic resistance of Staphylococcus aureus isolated from clinical and food samples in Egypt. Afr. J. Microbiol. Res. 2016, 10, 428–437. [Google Scholar]
- Şahin, S.; Moğulkoç, M.; Kalin, R.; Karahan, M. Determination of the Important Toxin Genes of Staphylococcus aureus Isolated from Meat Samples, Food Handlers and Food Processing Surfaces in Turkey. Isr. J. Vet. Med. 2020, 75, 42–49. [Google Scholar]
- Weese, J.; Avery, B.; Reid-Smith, R. Detection and quantification of methicillin-resistant Staphylococcus aureus (MRSA) clones in retail meat products. Lett. Appl. Microbiol. 2010, 51, 338–342. [Google Scholar] [CrossRef]
- Odetokun, I.; Ballhausen, B.; Adetunji, V.O.; Ghali-Mohammed, I.; Adelowo, M.T.; Adetunji, S.; Fetsch, A. Staphylococcus aureus in two municipal abattoirs in Nigeria: Risk perception, spread and public health implications. Vet. Microbiol. 2018, 216, 52–59. [Google Scholar] [CrossRef]
- Strommenger, B.; Braulke, C.; Heuck, D.; Schmidt, C.; Pasemann, B.; Nübel, U.; Witte, W. spa Typing of Staphylococcus aureus as a Frontline Tool in Epidemiological Typing. J. Clin. Microbiol. 2008, 46, 574–581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krupa, P.; Bystroń, J.; Podkowik, M.; Empel, J.; Mroczkowska, A.; Bania, J. Population Structure and Oxacillin Resistance of Staphylococcus aureus from Pigs and Pork Meat in South-West of Poland. BioMed Res. Int. 2015, 2015, 141475. [Google Scholar] [CrossRef] [PubMed]
- Nulens, E.; Stobberingh, E.E.; van Dessel, H.; Sebastian, S.; van Tiel, F.H.; Beisser, P.S.; Deurenberg, R.H. Molecular Characterization of Staphylococcus aureus Bloodstream Isolates Collected in a Dutch University Hospital between 1999 and 2006. J. Clin. Microbiol. 2008, 46, 2438–2441. [Google Scholar] [CrossRef] [Green Version]
- Omuse, G.; Van Zyl, K.N.; Hoek, K.; Abdulgader, S.; Kariuki, S.; Whitelaw, A.; Revathi, G. Molecular characterization of Staphylococcus aureus isolates from various healthcare institutions in Nairobi, Kenya: A cross sectional study. Ann. Clin. Microbiol. Antimicrob. 2016, 15, 51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papadopoulos, P.; Papadopoulos, T.; Angelidis, A.S.; Kotzamanidis, C.; Zdragas, A.; Papa, A.; Filioussis, G.; Sergelidis, D. Prevalence, antimicrobial susceptibility and characterization of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus isolated from dairy industries in north-central and north-eastern Greece. Int. J. Food Microbiol. 2019, 291, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Nurjadi, D.; Fleck, R.; Lindner, A.; Schäfer, J.; Gertler, M.; Mueller, A.; Lagler, H.; Van Genderen, P.J.; Caumes, E.; Boutin, S.; et al. Import of community-associated, methicillin-resistant Staphylococcus aureus to Europe through skin and soft-tissue infection in intercontinental travellers, 2011–2016. Clin. Microbiol. Infect. 2019, 25, 739–746. [Google Scholar] [CrossRef] [Green Version]
- Singh-Moodley, A.; Lowe, M.; Mogokotleng, R.; Perovic, O. Diversity of SCCmec elements and spa types in South African Staphylococcus aureus mecA-positive blood culture isolates. BMC Infect. Dis. 2020, 20, 816. [Google Scholar] [CrossRef]
- Tenover, F.C.; Tickler, I.A.; Goering, R.V.; Kreiswirth, B.N.; Mediavilla, J.R.; Persing, D.H. Characterization of Nasal and Blood Culture Isolates of Methicillin-Resistant Staphylococcus aureus from Patients in United States Hospitals. Antimicrob. Agents Chemother. 2012, 56, 1324–1330. [Google Scholar] [CrossRef] [Green Version]
- Tegegne, H.A.; Koláčková, I.; Florianová, M.; Wattiau, P.; Gelbíčová, T.; Boland, C.; Madec, J.-Y.; Haenni, M.; Karpíšková, R. Genomic Insights into Methicillin-Resistant Staphylococcus aureus spa Type t899 Isolates Belonging to Different Sequence Types. Appl. Environ. Microbiol. 2021, 87, e01994-20. [Google Scholar] [CrossRef]
- Battisti, A.; Franco, A.; Merialdi, G.; Hasman, H.; Iurescia, M.; Lorenzetti, R.; Feltrin, F.; Zini, M.; Aarestrup, F. Heterogeneity among methicillin-resistant Staphylococcus aureus from Italian pig finishing holdings. Vet. Microbiol. 2010, 142, 361–366. [Google Scholar] [CrossRef]
- Köck, R.; Schaumburg, F.; Mellmann, A.; Köksal, M.; Jurke, A.; Becker, K.; Friedrich, A.W. Livestock-Associated Methicillin-Resistant Staphylococcus aureus (MRSA) as Causes of Human Infection and Colonization in Germany. PLoS ONE 2013, 8, e55040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pirolo, M.; Gioffrè, A.; Visaggio, D.; Gherardi, M.; Pavia, G.; Samele, P.; Ciambrone, L.; Di Natale, R.; Spatari, G.; Casalinuovo, F.; et al. Prevalence, molecular epidemiology, and antimicrobial resistance of methicillin-resistant Staphylococcus aureus from swine in southern Italy. BMC Microbiol. 2019, 19, 51. [Google Scholar] [CrossRef] [PubMed]
- Schulz, J.; Boklund, A.; Toft, N.; Halasa, T. Drivers for Livestock-Associated Methicillin-Resistant Staphylococcus aureus Spread Among Danish Pig Herds—A Simulation Study. Sci. Rep. 2018, 8, 16962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tegegne, H.A.; Koláčková, I.; Karpíšková, R. Diversity of livestock associated methicillin-resistant Staphylococcus aureus. Asian Pac. J. Trop. Med. 2017, 10, 929–931. [Google Scholar] [CrossRef] [PubMed]
- CARD. The Comprehensive Antibiotic Resistance Database. Available online: https://card.mcmaster.ca/home (accessed on 20 March 2021).
- Zhang, L.; Gao, J.; Barkema, H.W.; Ali, T.; Liu, G.; Deng, Y.; Naushad, S.; Kastelic, J.P.; Han, B. Virulence gene profiles: Alpha-hemolysin and clonal diversity in Staphylococcus aureus isolates from bovine clinical mastitis in China. BMC Vet. Res. 2018, 14, 63. [Google Scholar] [CrossRef] [Green Version]
- Pomorska, K.; Jakubu, V.; Malisova, L.; Fridrichova, M.; Musilek, M.; Zemlickova, H. Antibiotic Resistance, spa Typing and Clonal Analysis of Methicillin-Resistant Staphylococcus aureus (MRSA) Isolates from Blood of Patients Hospitalized in the Czech Republic. Antibiotics 2021, 10, 395. [Google Scholar] [CrossRef]
- Sato, T.; Usui, M.; Motoya, T.; Sugiyama, T.; Tamura, Y. Characterisation of meticillin-resistant Staphylococcus aureus ST97 and ST5 isolated from pigs in Japan. J. Glob. Antimicrob. Resist. 2015, 3, 283–285. [Google Scholar] [CrossRef]
- O’Brien, A.M.; Hanson, B.M.; Farina, S.A.; Wu, J.Y.; Simmering, J.; Wardyn, S.E.; Forshey, B.M.; Kulick, M.E.; Wallinga, D.B.; Smith, T.C. MRSA in Conventional and Alternative Retail Pork Products. PLoS ONE 2012, 7, e30092. [Google Scholar] [CrossRef] [Green Version]
- Antonios, Z.; Theofilos, P.; Ioannis, M.; Georgios, S.; Georgios, V.; Evridiki, B.; Loukia, E.; Kyriaki, M.; Athanasios, A.; Vasiliki, L. Prevalence, genetic diversity, and antimicrobial susceptibility profiles of Staphylococcus aureus isolated from bulk tank milk from Greek traditional ovine farms. Small Rumin. Res. 2015, 125, 120–126. [Google Scholar] [CrossRef]
- Hookey, J.V.; Richardson, J.F.; Cookson, B.D. Molecular Typing of Staphylococcus aureus Based on PCR Restriction Fragment Length Polymorphism and DNA Sequence Analysis of the Coagulase Gene. J. Clin. Microbiol. 1998, 36, 1083–1089. [Google Scholar] [CrossRef] [Green Version]
- Sudagidan, M.; Aydin, A. Screening virulence properties of staphylococci isolated from meat and meat products. Wien. Tierarztl. Mon. 2009, 96, 128–134. [Google Scholar]
- Løvseth, A.; Loncarevic, S.; Berdal, K.G. Modified Multiplex PCR Method for Detection of Pyrogenic Exotoxin Genes in Staphylococcal Isolates. J. Clin. Microbiol. 2004, 42, 3869–3872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stegger, M.; Andersen, P.; Kearns, A.; Pichon, B.; Holmes, M.; Edwards, G.; Laurent, F.; Teale, C.; Skov, R.; Larsen, A. Rapid detection, differentiation and typing of methicillin-resistant Staphylococcus aureus harbouring either mecA or the new mecA homologue mecALGA251. Clin. Microbiol. Infect. 2012, 18, 395–400. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.B. Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; CLSI Supplement M100: Berwyn, PA, USA, 2020. [Google Scholar]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Liu, F.; Baloch, Z.; Zhang, C.S.; Ma, K.; Peng, Z.X.; Yan, S.F.; Hu, Y.J.; Gan, X.; Dong, Y.P.; et al. Genotypic Characterization of Methicillin-resistant Staphylococcus aureus Isolated from Pigs and Retail Foods in China. Biomed. Environ. Sci. 2017, 30, 570–580. [Google Scholar] [CrossRef]
- Borges, S.; Silva, J.; Teixeira, P.M.L. Survival and biofilm formation by Group B streptococci in simulated vaginal fluid at different pHs. Antonie Van Leeuwenhoek 2011, 101, 677–682. [Google Scholar] [CrossRef]
- McDougal, L.K.; Steward, C.D.; Killgore, G.E.; Chaitram, J.M.; McAllister, S.K.; Tenover, F.C. Pulsed-Field Gel Electrophoresis Typing of Oxacillin-Resistant Staphylococcus aureus Isolates from the United States: Establishing a National Database. J. Clin. Microbiol. 2003, 41, 5113–5120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aires-De-Sousa, M.; Boye, K.; De Lencastre, H.; Deplano, A.; Enright, M.C.; Etienne, J.; Friedrich, A.; Harmsen, D.; Holmes, A.; Huijsdens, X.; et al. High Interlaboratory Reproducibility of DNA Sequence-Based Typing of Bacteria in a Multicenter Study. J. Clin. Microbiol. 2006, 44, 619–621. [Google Scholar] [CrossRef] [Green Version]
- Harmsen, D.; Claus, H.; Witte, W.; Rothgänger, J.; Claus, H.; Turnwald, D.; Vogel, U. Typing of Methicillin-Resistant Staphylococcus aureus in a University Hospital Setting by Using Novel Software for spa Repeat Determination and Database Management. J. Clin. Microbiol. 2003, 41, 5442–5448. [Google Scholar] [CrossRef] [Green Version]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In Silico Detection and Typing of Plasmids. Antimicrob using PlasmidFinder and plasmid multilocus sequence typing. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
| Sample Origin | Plant F | Plant Z | Total | ||||||
|---|---|---|---|---|---|---|---|---|---|
| n * | S. aureus ± (%) | MRSA (%) | n * | S. aureus ± (%) | MRSA (%) | n * | S. aureus ± (%) | MRSA (%) | |
| Incoming bovine meat | 6 | 1 (16.7) | - | 6 | - | - | 12 | 1 (8.3) | - |
| Incoming porcine meat | 6 | 1 (16.7) | - | 14 | 1 (7.1) | - | 20 | 2 (10.0) | - |
| Incoming ovine meat | 4 | - | - | 1 | - | - | 5 | - | - |
| Non-ground meat products | 6 | - | - | 2 | - | - | 8 | - | - |
| Ground meat products | 8 | 4 (50.0) | - | 8 | 2 (25.0) | 1 (12.5) | 16 | 6 (37.5) | 1 (6.3) |
| Nasal cavity of workers | 9 | 3 (33.3) | - | 11 | 2 (18.2) | - | 20 | 5 (25.0) | - |
| Infrastructure | 5 | - | - | 17 | 1 (5.9) | - | 22 | 1 (4.5) | - |
| Equipment | 16 | 5 (31.3) | - | 41 | 2 (4.9) | - | 57 | 7 (12.3) | - |
| Total | 60 | 14 (23.3) | - | 100 | 8 (8.0) | 1 (1.0) | 160 | 22 (13.8) | 1 (0.6) |
| Sample Code * | Sample Origin | Antimicrobial Resistance Profile ** | mecA | mecC | lukF-PV/tst | SE Genes | Biofilm Formation Ability | Spa Type | PFGE Electrophoretic Cluster |
|---|---|---|---|---|---|---|---|---|---|
| F20 | Meat | P, TET, TOB | - | - | -/- | - | moderate | t084 | A |
| F22 | Equipment | - | - | - | -/+ | sed, sei | moderate | t012 | NT *** |
| F28 | Meat product | P, A/C, TET | - | - | -/- | - | moderate | t499 | A |
| F29 | Meat product | P, TET | - | - | -/- | - | moderate | t774 | A |
| F32 | Meat product | P | - | - | -/- | seb | moderate | t084 | A |
| F36 | Equipment | P, A/C | - | - | -/- | seb | moderate | t084 | A |
| F37 | Equipment | P | - | - | -/- | seb | moderate | t084 | A |
| F39 | Equipment | P, A/C | - | - | -/- | seb | moderate | t084 | A |
| F40 | Equipment | P, AMP, A/C, TOB | - | - | -/- | seb | moderate | t084 | A |
| F42 | Meat product | P, A/C | - | - | -/- | seb | moderate | t084 | A |
| F43 | Meat product | P, A/C | - | - | -/- | seb | moderate | t084 | A |
| F44 | Human nasal cavity | P | - | - | -/- | sed, sei | moderate | t891 | NT |
| F45 | Human nasal cavity | P | - | - | -/- | sed | moderate | t197 | A |
| F46 | Human nasal cavity | P | - | - | -/- | seb | moderate | t084 | NT |
| Ζ5 | Human nasal cavity | P, A/C | - | - | -/+ | seb, sed, sei | moderate | t1510 | NT |
| Ζ7 | Human nasal cavity | - | - | - | -/- | - | moderate | new | C |
| Ζ18 | Infrastructure | - | - | - | -/- | - | moderate | new | C |
| Ζ19 | Equipment | - | - | - | -/- | - | moderate | t091 | B |
| Ζ77 | Meat product | - | - | - | -/- | - | moderate | new | C |
| Ζ104 | Equipment | - | - | - | -/- | - | strong | t091 | B |
| Ζ112 | Meat product | P, AMP, A/C, FOX, TET, TOB, C, SXT | + | - | -/- | - | moderate | t899 | NT *** |
| Ζ146 | Meat product | - | - | - | -/- | - | moderate | new | C |
| Feature | Isolate Z77 | Isolate Z112 |
|---|---|---|
| Number of contigs | 85 | 309 |
| N50 (bp) | 169,989 | 134,669 |
| GC (%), | 33.0 | 33.1 |
| Length (bp) | 2,896,939 | 2,840,021 |
| Sequence type (ST) | 97 | 398 |
| Spa type | Novel type (repeats 07-23-12-12-21-17-13-34-33-34) | t899 |
| Resistance genes | - | mecA, blaZ, tetM, fexA, dfrC |
| Antibiotic efflux genes | mgrA, arlR, mepR, norC, sdrM, sepA | mgrA, arlR, mepR, norA, norC, sdrM, sepA |
| Toxin genes | hlgA, hlgB, hlgC, lukD, lukE | hlgA, hlgB, hlgC |
| Exoenzyme genes | aur, splA, splB, splE | aur |
| Host immune defense genes | sak, scn | - |
| Plasmids | rep20 | rep21, repUS43 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Komodromos, D.; Kotzamanidis, C.; Giantzi, V.; Pappa, S.; Papa, A.; Zdragas, A.; Angelidis, A.; Sergelidis, D. Prevalence, Infectious Characteristics and Genetic Diversity of Staphylococcus aureus and Methicillin-Resistant Staphylococcus aureus (MRSA) in Two Raw-Meat Processing Establishments in Northern Greece. Pathogens 2022, 11, 1370. https://doi.org/10.3390/pathogens11111370
Komodromos D, Kotzamanidis C, Giantzi V, Pappa S, Papa A, Zdragas A, Angelidis A, Sergelidis D. Prevalence, Infectious Characteristics and Genetic Diversity of Staphylococcus aureus and Methicillin-Resistant Staphylococcus aureus (MRSA) in Two Raw-Meat Processing Establishments in Northern Greece. Pathogens. 2022; 11(11):1370. https://doi.org/10.3390/pathogens11111370
Chicago/Turabian StyleKomodromos, Dimitrios, Charalampos Kotzamanidis, Virginia Giantzi, Styliani Pappa, Anna Papa, Antonios Zdragas, Apostolos Angelidis, and Daniel Sergelidis. 2022. "Prevalence, Infectious Characteristics and Genetic Diversity of Staphylococcus aureus and Methicillin-Resistant Staphylococcus aureus (MRSA) in Two Raw-Meat Processing Establishments in Northern Greece" Pathogens 11, no. 11: 1370. https://doi.org/10.3390/pathogens11111370
APA StyleKomodromos, D., Kotzamanidis, C., Giantzi, V., Pappa, S., Papa, A., Zdragas, A., Angelidis, A., & Sergelidis, D. (2022). Prevalence, Infectious Characteristics and Genetic Diversity of Staphylococcus aureus and Methicillin-Resistant Staphylococcus aureus (MRSA) in Two Raw-Meat Processing Establishments in Northern Greece. Pathogens, 11(11), 1370. https://doi.org/10.3390/pathogens11111370







