Characterizing the Virome of Apple Orchards Affected by Rapid Decline in the Okanagan and Similkameen Valleys of British Columbia (Canada)
Abstract
:1. Introduction
2. Results
2.1. Detection of 21 Plant Viruses and One Plant Viroid by NGS in Apple Orchards Affected by RAD
2.2. Virus Prevalence in Apple Orchards Affected by RAD
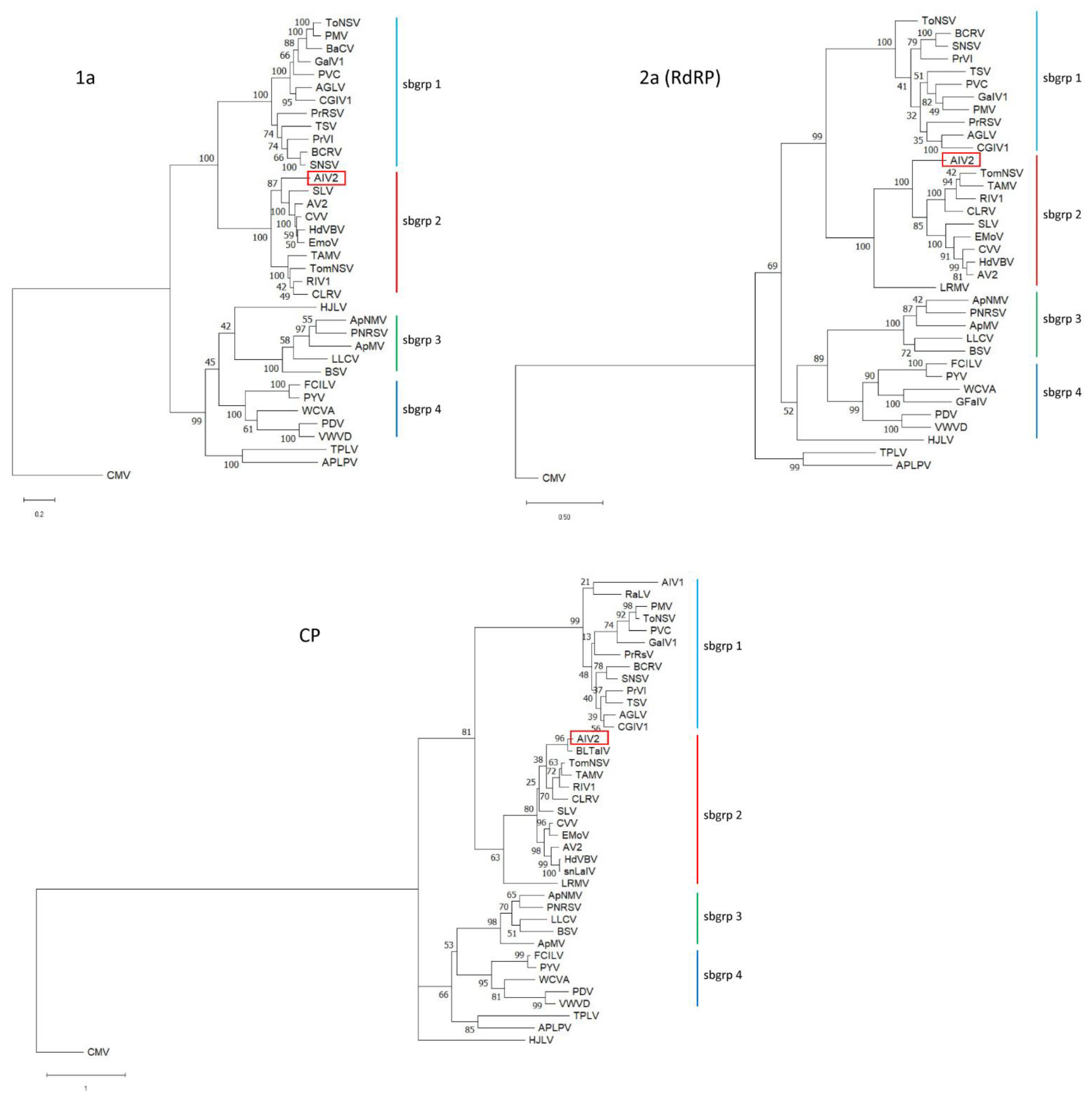
2.3. Complete Genome Sequence and Phylogenetic Analysis of Apple Ilarvirus 2
3. Discussion
4. Materials and Methods
4.1. Sample Collection and Preparation
4.2. High Throughput Sequencing and Analyses
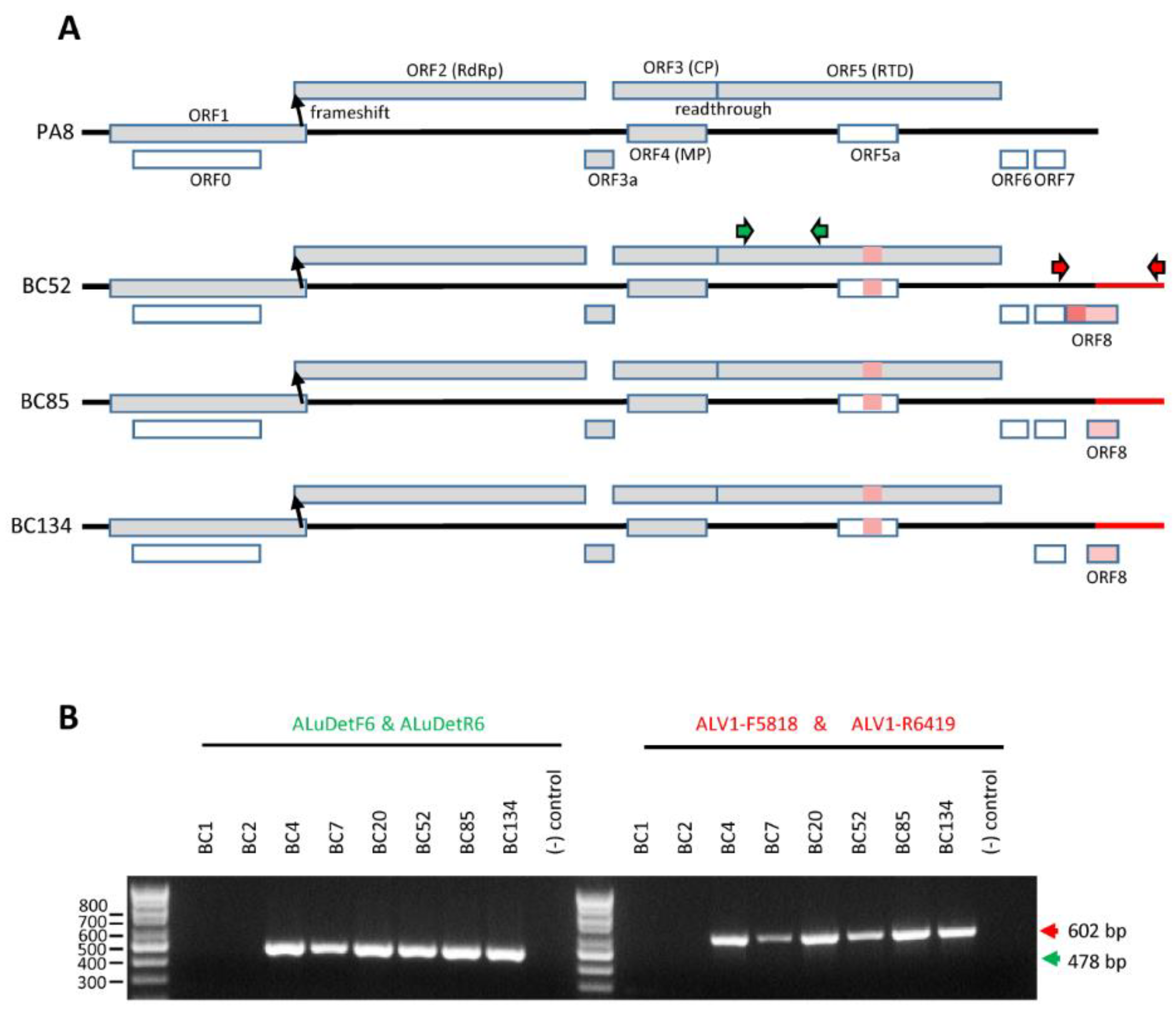
4.3. Analysis of Virus Prevalence Using RT-PCR
4.4. Determination of the Complete Genome Sequence of AIV2
4.5. Phylogenetic Analyses
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wright, A.A.; Cross, A.R.; Harper, S.J. A bushel of viruses: Identification of seventeen novel putative viruses by RNA-seq in six apple trees. PLoS ONE 2020, 15, e0227669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stokstad, E. Rapid apple decline has researchers stumped. Science 2019, 363, 1259. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.; Silva, K.J.P.; Fuchs, M.; Khan, A. Potential role of weather, soil and plant microbial communities in rapid decline of apple trees. PLoS ONE 2019, 14, e0213293. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wu, L.; Nikolaeva, E.; Peter, K.; Liu, Z.; Mollov, D.; Cao, M.; Li, R. Characterization of a new apple luteovirus identified by high-throughput sequencing. Virol. J. 2018, 15, 85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wright, A.A.; Szostek, S.A.; Beaver-Kanuya, E.; Harper, S.J. Diversity of three bunya-like viruses infecting apple. Arch. Virol. 2018, 163, 3339–3343. [Google Scholar] [CrossRef]
- Ilyukhin, E.; Schneider, K.; Ellouze, W. First Report of Botryosphaeria dothidea Causing Stem Canker and Dieback of Apple Trees in Ontario, Canada. Plant Dis. 2022. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Renderos, W.; Bevis, E.; Hebb, J.; Abbasi, P.A. Diaporthe eres causes stem cankers and death of young apple rootstocks in Canada. Can. J. Plant Pathol. 2020, 42, 218–227. [Google Scholar] [CrossRef]
- MacDonald, J.; O’Gorman, D.T.; Moffat, C.; Judd, G.R.; Xu, H.; Sanfacon, H.; Hannam, K.; Forge, T.A. Recognizing apple decline in your orchard. In Proceedings of the Apple Decline Regional Research Users’ Meeting, Summerland, BC, Canada, 6 June 2019. [Google Scholar] [CrossRef]
- Umer, M.; Liu, J.; You, H.; Xu, C.; Dong, K.; Luo, N.; Kong, L.; Li, X.; Hong, N.; Wang, G.; et al. Genomic, Morphological and Biological Traits of the Viruses Infecting Major Fruit Trees. Viruses 2019, 11, 515. [Google Scholar] [CrossRef] [Green Version]
- Fuchs, M.; Kahke, C.; Donahue, D.; Wallis, A.; Basedow, M. Distribution of Viruses in New York Apple Orchards. Fruit Quaterly 2018, 26, 5–8. [Google Scholar]
- James, D.; Varga, A.; Jesperson, G.D.; Navratil, M.; Safarova, D.; Constable, F.; Horner, M.; Eastwell, K.; Jelkmann, W. Identification and complete genome analysis of a virus variant or putative new foveavirus associated with apple green crinkle disease. Arch. Virol. 2013, 158, 1877–1887. [Google Scholar] [CrossRef]
- Kutnjak, D.; Tamisier, L.; Adams, I.; Boonham, N.; Candresse, T.; Chiumenti, M.; De Jonghe, K.; Kreuze, J.F.; Lefebvre, M.; Silva, G.; et al. A Primer on the Analysis of High-Throughput Sequencing Data for Detection of Plant Viruses. Microorganisms 2021, 9, 841. [Google Scholar] [CrossRef] [PubMed]
- Roossinck, M.J. Deep sequencing for discovery and evolutionary analysis of plant viruses. Virus Res. 2017, 239, 82–86. [Google Scholar] [CrossRef] [PubMed]
- Pallas, V.; Aparicio, F.; Herranz, M.C.; Amari, K.; Sanchez-Pina, M.A.; Myrta, A.; Sanchez-Navarro, J.A. Ilarviruses of Prunus spp.: A continued concern for fruit trees. Phytopathology 2012, 102, 1108–1120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tokarz, R.; Sameroff, S.; Tagliafierro, T.; Jain, K.; Williams, S.H.; Cucura, D.M.; Rochlin, I.; Monzon, J.; Carpi, G.; Tufts, D.; et al. Identification of Novel Viruses in Amblyomma americanum, Dermacentor variabilis, and Ixodes scapularis Ticks. mSphere 2018, 3, e00614-17. [Google Scholar] [CrossRef] [Green Version]
- Pallas, V.; Aparicio, F.; Herranz, M.C.; Sanchez-Navarro, J.A.; Scott, S.W. The molecular biology of ilarviruses. Adv. Virus Res. 2013, 87, 139–181. [Google Scholar] [CrossRef]
- Shimura, H.; Masuta, C.; Yoshida, N.; Sueda, K.; Suzuki, M. The 2b protein of Asparagus virus 2 functions as an RNA silencing suppressor against systemic silencing to prove functional synteny with related cucumoviruses. Virology 2013, 442, 180–188. [Google Scholar] [CrossRef] [Green Version]
- Bujarski, J.; Gallitelli, D.; García-Arenal, F.; Pallás, V.; Palukaitis, P.; Reddy, M.K.; Wang, A.; ICTV Report Consortium. ICTV virus taxonomy profile: Bromoviridae. J. Gen. Virol. 2019, 100, 1206–1207. [Google Scholar] [CrossRef]
- Paudel, D.B.; Sanfaçon, H. Exploring the diversity of mechanisms associated with plant tolerance to virus infection. Front. Plant Sci. 2018, 9, 1575. [Google Scholar] [CrossRef]
- Ghoshal, B.; Sanfaçon, H. Symptom recovery in virus-infected plants: Revisiting the role of RNA silencing mechanisms. Virology 2015, 479–480, 167–179. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, R.; Butkovic, A.; Escaray, F.J.; Martinez-Latorre, J.; Melero, I.; Perez-Parets, E.; Gomez-Cadenas, A.; Carrasco, P.; Elena, S.F. Plant virus evolution under strong drought conditions results in a transition from parasitism to mutualism. Proc. Natl. Acad. Sci. USA 2021, 118, e2020990118. [Google Scholar] [CrossRef]
- van Munster, M. Impact of Abiotic Stresses on Plant Virus Transmission by Aphids. Viruses 2020, 12, 216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreno, A.B.; López-Moya, J.J. When viruses play team sports: Mixed infections in plants. Phytopathology 2020, 110, 29–48. [Google Scholar] [CrossRef]
- Syller, J.; Grupa, A. Antagonistic within-host interactions between plant viruses: Molecular basis and impact on viral and host fitness. Mol. Plant Pathol. 2016, 17, 769–782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mascia, T.; Gallitelli, D. Synergies and antagonisms in virus interactions. Plant Sci. 2016, 252, 176–192. [Google Scholar] [CrossRef]
- Xiao, H.; Kim, W.S.; Meng, B. A highly effective and versatile technology for the isolation of RNAs from grapevines and other woody perennials for use in virus diagnostics. Virol. J. 2015, 12, 171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rott, M.E.; Kesanakurti, P.; Berwarth, C.; Rast, H.; Boyes, I.; Phelan, J.; Jelkmann, W. Discovery of negative-sense RNA viruses in trees infected with apple rubbery wood disease by next-generation sequencing. Plant Dis. 2018, 102, 1254–1263. [Google Scholar] [CrossRef] [Green Version]
- Xie, W.; Marty, D.M.; Xu, J.; Khatri, N.; Willie, K.; Moraes, W.B.; Stewart, L.R. Simultaneous gene expression and multi-gene silencing in Zea mays using maize dwarf mosaic virus. BMC Plant Biol. 2021, 21, 208. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]


| Orchards * | Number of Samples | Scions | Number of Samples | Rootstocks | Number of Samples | RAD Symptoms | Number of Samples |
|---|---|---|---|---|---|---|---|
| A | 14 | Ambrosia | 78 | M9 | 104 | Yes | 80 |
| B | 14 | Honey Crisp | 26 | Nic29 | 19 | No | 68 |
| C | 30 | Salish | 10 | Others (17) | 25 | ||
| D | 19 | Gala | 12 | ||||
| E | 13 | Others (14) | 19 | ||||
| F | 15 | Unknown | 3 | ||||
| G | 10 | ||||||
| H | 10 | ||||||
| I | 20 | ||||||
| J | 3 |
| NGS Samples * | Orchards | Scions | Rootstocks | RAD Symptoms ** |
|---|---|---|---|---|
| HX1 | B | Fuji, Honey Crispy, 8S-54-28 | M9 | Yes (3) |
| HX2 | A | Ambrosia, Spartan | M9 | Yes (3) |
| HX3 | D, E, F | Ambrosia, Honey Crisp | M9, Nic29 | Yes (2), No (1) |
| HX4 | I, J | Ambrosia, Pink Lady | M9, G11 | Yes (3) |
| HX5 | C | Honey Crisp | G935N, Bud9, Bud 71-7-22 | No (2), Yes (1) |
| HX6 | C | Honey Crisp | G4124, G2034, Bud9 | No (3) |
| HX7 | C | G. Reinette, Mcintosh, Rein Russ | M9 | No (2), Yes (1) |
| HX8 | C | Winter banana, Splendour, Bisbee RD | M9 | No (3) |
| HX9 | B | 8S-06-28, 8S-56-50, 9C-06-140 | M9 | Yes (3) |
| HX10 | A, E | Ambrosia | M9 | Yes (2), No (1) |
| Acronym | Virus or Viroid Name | Species Name | GenBank Accession Number of Reference Isolate | Genus | Family |
|---|---|---|---|---|---|
| AHVd | Apple hammerhead viroid | Apple hammerhead viroid | NC_028132.1 | Pelamoviroid | Avsunviroidae |
| ACLSV | Apple chlorotic leaf spot virus | Apple chlorotic leaf spot virus | NC_001409.1 | Trichovirus | Betaflexiviridae |
| ASGV | Apple stem grooving virus | Apple stem grooving virus | NC_001749.2 | Capillovirus | |
| CVA | Cherry virus A | Cherry virus A | NC_003689.1 | ||
| ASPV | Apple stem pitting virus | Apple stem pitting virus | NC_003462.2 | Foveavirus | |
| ALV1 | Apple luteovirus 1 | Apple luteovirus 1 | NC_040680.1 | Luteovirus | Tombusviridae |
| TCV | Turnip crinkle virus | Turnip crinkle virus | NC_003821.3 | Betacarmovirus | |
| ARWaV-1 | Apple rubbery wood virus 1 | Apple rubodvirus 1 | NC_055390.1, NC_055391.1, NC_055392.1 | Rubodvirus | Phenuiviridae |
| ARWaV-2 | Apple rubbery wood virus 2 | Apple rubodvirus 2 | NC_055533.1, NC_055534.1, NC_055535.1, NC_055536.1, NC_055537.1 | ||
| CCGaV | Citrus concave gum-associated virus | Citrus coguvirus | NC_035759.1, NC_035454.1 | Coguvirus | |
| AIV2 | Apple ilarvirus 2 | Identified in this study, not classified | ON932434, ON932435, ON932436 | Ilarvirus | Bromoviridae |
| ApMV | Apple mosaic virus | Apple mosaic virus | NC_003464.1, NC_003465.1, NC_003480.1 | ||
| SNIV | Solanum nigrum ilarvirus | Not classified | MN216370.1, MN216373.1, MN216376.1 | ||
| TSV | Tobacco streak virus | Tobacco streak virus | NC_003844.1, NC_003842.1, NC_003845.1 | ||
| AIV1 | Apple ilarvirus 1 | Not classified | MN386957.1, MN386958.1 | ||
| PNRSV | Prunus necrotic ringspot virus | Prunus necrotic ringspot virus | NC_004362.1, NC_004363.1, NC_004364.1 | ||
| AlMV | Alfalfa mosaic virus | Alfalfa mosaic virus | NC_001495.1, NC_002024.2, NC_002025.1 | Alfamovirus | |
| CYMV | Clover yellow mosaic virus | Clover yellow mosaic virus | NC_001753.1 | Potexvirus | Alphaflexiviridae |
| WClMV | White clover mosaic virus | White clover mosaic virus | NC_003820.1 | ||
| TVCV | Turnip vein clearing virus | Turnip vein-clearing virus | NC_001873.1 | Tobamovirus | Virgaviridae |
| TuMV | Turnip mosaic virus | Turnip mosaic virus | NC_002509.2 | Potyvirus | Potyviridae |
| SoMV | Sowbane mosaic virus | Sowbane mosaic virus | NC_011187.1 | Sobemovirus | Solemoviridae |
| NGS Samples | HX1 | HX2 | HX3 | HX4 | HX5 | HX6 | HX7 | HX8 | HX9 | HX10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Total reads | 103,066,652 | 81,797,780 | 101,947,128 | 86,362,078 | 81,880,242 | 102,784,288 | 71,349,254 | 94,601,674 | 85,908,874 | 77,797,014 |
| Reads Mapped to Malus Domestica | 97,549,197 | 75,364,116 | 95,651,348 | 80,689,725 | 72,958,929 | 92,960,006 | 65,850,694 | 87,214,426 | 79,122,763 | 71,389,255 |
| Reads not Mapped to M. domestica | 5,517,455 | 6,433,664 | 6,295,780 | 5,672,353 | 8,921,313 | 9,824,282 | 5,498,560 | 7,387,248 | 6,786,111 | 6,407,759 |
| Reads Mapped to Plant Viruses | 40,286 | 161 | 132,732 | 52,869 | 1994 | 986 | 148,093 | 357,639 | 4838 | 76,969 |
| AHVd | 8699 | 3435 | 704 | 54 | 63,949 | 180,962 | 200 | |||
| ACLSV | 12,174 | 20,423 | 8 | 12,801 | 10,955 | 677 | ||||
| ASGV | 9785 | 4467 | 2643 | |||||||
| CVA | 187 | |||||||||
| ASPV | 5 | 35,542 | 11,105 | 8 | 12 | 43,759 | 151,192 | 6532 | ||
| ALV1 | 39,292 | 12,494 | 1978 | 10,409 | 7413 | |||||
| ARWaV-1 | 3840 | 15,168 | 3970 | |||||||
| ARWaV-2 | 41,807 | 6141 | 232 | 874 | ||||||
| CCGaV | 10,782 | 9 | 12,967 | 56,775 | ||||||
| AIV2 * | 114 | 156 | 1389 | 120 | 1257 | 882 | 1656 | 1521 | 848 | 1652 |
| ApMV | 931 | |||||||||
| SNIV | 110 | 211 | 30 | 36 | 14 | 22 | ||||
| TSV | 202 | |||||||||
| AIV1 | 81 | 10 | ||||||||
| PNRSV | 250 | |||||||||
| AlMV | 16 | 14 | 10 | 14 | ||||||
| CYMV | 149 | 10 | 7 | 7 | 71 | |||||
| WClMV | 116 | 8 | 18 | 6 | ||||||
| TVCV | 40 | 6 | 149 | |||||||
| TuMV | 12 | 46 | 6 | |||||||
| TCV | 10 | |||||||||
| SoMV | 22 |
| Number of Viruses and/or Viroid | Number of Samples | Percentage of Samples (%) |
|---|---|---|
| 0 | 1 | 0.7 |
| 1 | 26 | 17.6 |
| 2 | 26 | 17.6 |
| 3 | 24 | 16.2 |
| 4 | 15 | 10.1 |
| 5 | 29 | 19.6 |
| 6 | 15 | 10.1 |
| 7 | 9 | 6.1 |
| 8 | 3 | 2.0 |
| Viruses Tested | RAD Symptoms (80) * | No RAD Symptoms (68) | All Samples (148) | |||
|---|---|---|---|---|---|---|
| No. of Positive Samples | Percentage of Positive Samples (%) | No. of Positive Samples | Percentage of Positive Samples (%) | No. of Positive Samples | Percentage of Positive Samples (%) | |
| AIV2 | 46 | 57.5 | 48 | 70.6 | 94 | 63.5 |
| CCGaV | 49 | 61.3 | 43 | 63.2 | 92 | 62.2 |
| ASPV | 43 | 53.8 | 38 | 55.9 | 81 | 54.7 |
| ACLSV | 36 | 45.0 | 38 | 55.9 | 74 | 50.0 |
| ASGV | 34 | 42.5 | 28 | 41.2 | 62 | 41.9 |
| AHVd | 15 | 18.8 | 19 | 27.9 | 34 | 23.0 |
| ALV1 | 15 | 18.8 | 12 | 17.6 | 27 | 18.2 |
| ARWaV2 | 14 | 17.5 | 9 | 13.2 | 23 | 15.5 |
| TSV | 6 | 7.5 | 8 | 11.8 | 14 | 9.5 |
| ARWaV1 | 8 | 10.0 | 5 | 7.4 | 13 | 8.8 |
| SNIV | 7 | 8.8 | 4 | 5.9 | 11 | 7.4 |
| ApMV | 5 | 6.3 | 1 | 1.5 | 6 | 4.1 |
| PNRSV | 1 | 1.3 | 0 | 0.0 | 1 | 0.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022, His Majesty the King in Right of Canada. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, H.; Hao, W.; Storoschuk, G.; MacDonald, J.L.; Sanfaçon, H. Characterizing the Virome of Apple Orchards Affected by Rapid Decline in the Okanagan and Similkameen Valleys of British Columbia (Canada). Pathogens 2022, 11, 1231. https://doi.org/10.3390/pathogens11111231
Xiao H, Hao W, Storoschuk G, MacDonald JL, Sanfaçon H. Characterizing the Virome of Apple Orchards Affected by Rapid Decline in the Okanagan and Similkameen Valleys of British Columbia (Canada). Pathogens. 2022; 11(11):1231. https://doi.org/10.3390/pathogens11111231
Chicago/Turabian StyleXiao, Huogen, Wenjia Hao, Gavin Storoschuk, Jesse L. MacDonald, and Hélène Sanfaçon. 2022. "Characterizing the Virome of Apple Orchards Affected by Rapid Decline in the Okanagan and Similkameen Valleys of British Columbia (Canada)" Pathogens 11, no. 11: 1231. https://doi.org/10.3390/pathogens11111231
APA StyleXiao, H., Hao, W., Storoschuk, G., MacDonald, J. L., & Sanfaçon, H. (2022). Characterizing the Virome of Apple Orchards Affected by Rapid Decline in the Okanagan and Similkameen Valleys of British Columbia (Canada). Pathogens, 11(11), 1231. https://doi.org/10.3390/pathogens11111231








