Biomolecular Investigation of Bartonella spp. in Wild Rodents of Two Swiss Regions
Abstract
:1. Introduction
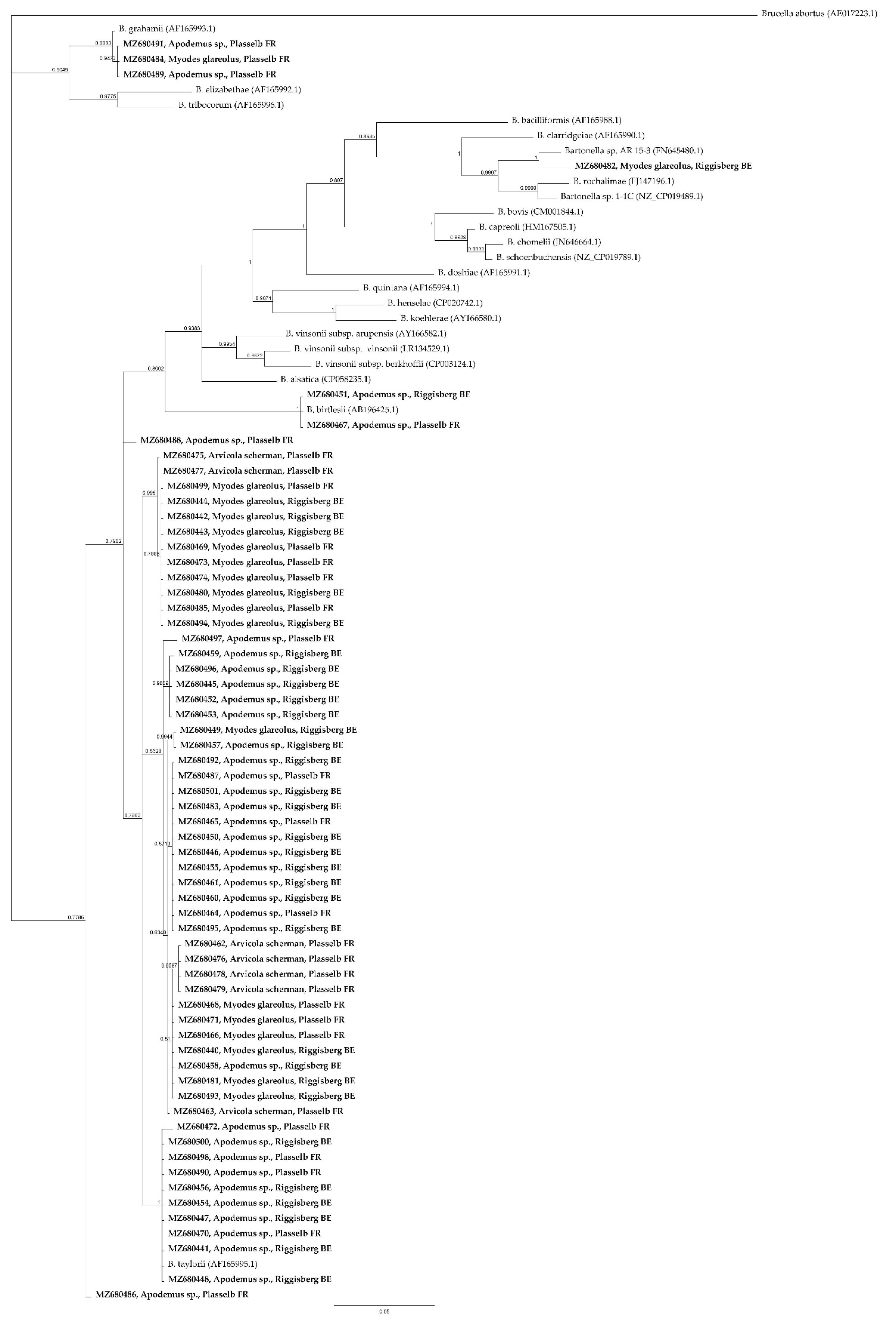
2. Results
3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Deng, H.; Le Rhun, D.; Buffet, J.P.R.; Cotté, V.; Read, A.; Birtles, R.J.; Vayssier-Taussat, M. Strategies of exploitation of mammalian reservoirs by Bartonella species. Vet. Res. 2012, 43, 1–14. [Google Scholar] [CrossRef]
- Regier, Y.; Órourke, F.; Kempf, V.A.J. Bartonella spp.—A chance to establish One Health concepts in veterinary and human medicine. Parasites Vectors 2016, 9, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guptill, L. Bartonellosis. Vet. Microbiol. 2010, 140, 347–359. [Google Scholar] [CrossRef] [PubMed]
- Cheslock, M.A.; Embers, M.E. Human bartonellosis: An underappreciated public health problem? Trop. Med. Infect. Dis. 2019, 4, 69. [Google Scholar] [CrossRef] [Green Version]
- Angelakis, E.; Raoult, D. Pathogenicity and treatment of Bartonella infections. Int. J. Antimicrob. Agents 2014, 44, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Breitschwerdt, E.B. Bartonellosis: One health perspectives for an emerging infectious disease. ILAR J. 2014, 55, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Breitschwerdt, E.B. Bartonellosis, One Health and all creatures great and small. Vet. Dermatol. 2017, 28, 96-e21. [Google Scholar] [CrossRef] [PubMed]
- Maggi, R.G.; Mozayeni, B.R.; Pultorak, E.L.; Hegarty, B.C.; Bradley, J.M.; Correa, M.; Breitschwerdt, E.B. Bartonella spp. Bacteremia and rheumatic symptoms in patients from Lyme disease-endemic region. Emerg. Infect. Dis. 2012, 18, 783–791. [Google Scholar] [CrossRef]
- Okaro, U.; Addisu, A.; Casanas, B.; Anderson, B. Bartonella Species, an Emerging Cause of Blood-Culture-Negative Endocarditis. Clin. Microbiol. Rev. 2017, 30, 709–746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Álvarez-Fernández, A.; Breitschwerdt, E.B.; Solano-Gallego, L. Bartonella infections in cats and dogs including zoonotic aspects. Parasites Vectors 2018, 11, 1–21. [Google Scholar] [CrossRef]
- Boularias, G.; Azzag, N.; Gandoin, C.; Bouillin, C.; Chomel, B.; Haddad, N.; Boulouis, H.J. Bartonella bovis and Bartonella chomelii infection in dairy cattle and their ectoparasites in Algeria. Comp. Immunol. Microbiol. Infect. Dis. 2020, 70, 101450. [Google Scholar] [CrossRef] [PubMed]
- Magni, E.; Bertelloni, F.; Sgorbini, M.; Ebani, V.V. Bartonella infection in asymptomatic horses and donkeys from Tuscany, Central Italy. Asian Pac. J. Trop. Med. 2017, 10, 1077–1079. [Google Scholar] [CrossRef]
- Stuckey, M.J.; Chomel, B.B.; de Fleurieu, E.C.; Aguilar-Setién, A.; Boulouis, H.-J.; Chang, C. Bartonella, bats and bugs: A review. Comp. Immunol. Microbiol. Infect. Dis. 2017, 55, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Greco, G.; Zarea, A.A.K.; Sgroi, G.; Tempesta, M.; D’Alessio, N.; Lanave, G.; Bezerra-Santos, M.A.; Iatta, R.; Veneziano, V.; Otranto, D.; et al. Zoonotic Bartonella species in Eurasian wolves and other free-ranging wild mammals from Italy. Zoonoses Public Health 2021, 68, 316–326. [Google Scholar] [CrossRef]
- Harms, C.A.; Maggi, R.G.; Breitschwerdt, E.B.; Clemons-Chevis, C.L.; Solangi, M.; Rotstein, D.S.; Fair, P.A.; Hansen, L.J.; Hohn, A.A.; Lovewell, G.N.; et al. Bartonella species detection in captive, stranded and free-ranging cetaceans. Vet. Res. 2008, 39, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Dahmana, H.; Granjon, L.; Diagne, C.; Davoust, B.; Fenollar, F.; Mediannikov, O. Rodents as hosts of pathogens and related zoonotic disease risk. Pathogens 2020, 9, 202. [Google Scholar] [CrossRef] [Green Version]
- Kosoy, M.; Bai, Y.; Enscore, R.; Rizzo, M.R.; Bender, S.; Popov, V.; Albayrak, L.; Fofanov, Y.; Chomel, B. Bartonella melophagi in blood of domestic sheep (Ovis aries) and sheep keds (Melophagus ovinus) from the southwestern US: Cultures, genetic characterization, and ecological connections. Vet. Microbiol. 2016, 190, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Williams, H.M.; Dittmar, K. Expanding our view of Bartonella and its hosts: Bartonella in nest ectoparasites and their migratory avian hosts. Parasites and Vectors 2020, 13, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valentine, K.H.; Harms, C.A.; Cadenas, M.B.; Birkenheuer, A.J.; Marr, H.S.; Braun-McNeill, J.; Maggi, R.G.; Breitschwerdt, E.B. Bartonella DNA in Loggerhead Sea Turtles. Emerg. Infect. Dis. 2007, 13, 949–950. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, R.; Krasnov, B.; Morick, D.; Gottlieb, Y.; Khokhlova, I.S.; Harrus, S. Bartonella Infection in Rodents and Their Flea Ectoparasites: An Overview. Vector Borne Zoonotic Dis. 2015, 15, 27–39. [Google Scholar] [CrossRef] [Green Version]
- Gundi, V.A.K.B.; Billeter, S.A.; Rood, M.P.; Kosoy, M.Y. Bartonella spp. in Rats and Zoonoses, Los Angeles, California, USA. Emerg. Infect. Dis. 2012, 18, 631–633. [Google Scholar] [CrossRef] [PubMed]
- Kandelaki, G.; Malania, L.; Bai, Y.; Chakvetadze, N.; Katsitadze, G.; Imnadze, P.; Nelson, C.; Harrus, S.; Kosoy, M. Human Lymphadenopathy Caused by Ratborne Bartonella, Tbilisi, Georgia. Emerg. Infect. Dis. 2016, 22, 544–546. [Google Scholar] [CrossRef]
- Helan, J.V.G.; Grinberg, A.; Gedye, K.; Potter, M.A.; Harrus, S. Molecular detection of Bartonella coopersplainsensis and B. henselae in rats from New Zealand. N. Z. Vet. J. 2018, 66, 257–260. [Google Scholar] [CrossRef]
- Engbæk, K.; Lawson, P.A. Identification of Bartonella species in rodents, shrews and cats, in Denmark: Detection of two B. henselae variants, one in cafe and the other to the long-tailed field mouse. Apmis 2004, 112, 336–341. [Google Scholar] [CrossRef] [PubMed]
- Divari, S.; Pregel, P.; Zanet, S.; Ferroglio, E.; Giannini, F.; Scaglione, F.E.; Grinberg, A.; Biolatti, B.; Bollo, E. Molecular Evidence of Bartonella spp. in Rodents: A Study in Pianosa Island, Italy. Animals 2020, 10, 2070. [Google Scholar] [CrossRef] [PubMed]
- Maggi, R.G.; Duncan, A.W.; Breitschwerdt, E.B. Novel chemically modified liquid medium that will support the growth of seven Bartonella species. J. Clin. Microbiol. 2005, 43, 2651–2655. [Google Scholar] [CrossRef] [Green Version]
- Diddi, K.; Chaudhry, R.; Sharma, N.; Dhawan, B. Strategy for identification & characterization of Bartonella henselae with conventional & molecular methods. Indian J. Med. Res. 2013, 137, 380–387. [Google Scholar]
- Gutiérrez, R.; Vayssier-Taussat, M.; Buffet, J.-P.; Harrus, S. Guidelines for the Isolation, Molecular Detection, and Characterization of Bartonella Species. Vector Borne Zoonotic Dis. 2017, 17, 42–50. [Google Scholar] [CrossRef] [Green Version]
- Breitschwerdt, E.B.; Kordick, D.L. Bartonella Infection in Animals: Carriership, Reservoir Potential, Pathogenicity, and Zoonotic Potential for Human Infection. Clin. Microbiol. Rev. 2000, 13, 428–438. [Google Scholar] [CrossRef]
- La Scola, B.; Zeaiter, Z.; Khamis, A.; Raoult, D. Gene-sequence-based criteria for species definition in bacteriology: The Bartonella paradigm. Trends Microbiol. 2003, 11, 318–321. [Google Scholar] [CrossRef]
- Gonçalves, L.R.; de Mendonça Favacho, A.R.; Roque, A.L.R.; Mendes, N.S.; Fidelis, O.L.; Benevenute, J.L.; Herrera, H.M.; D’Andrea, P.S.; de Lemos, E.R.S.; Machado, R.Z.; et al. Association of Bartonella species with wild and synanthropic rodents in different Brazilian biomes. Appl. Environ. Microbiol. 2016, 82, 7154–7164. [Google Scholar] [CrossRef] [Green Version]
- Buffet, J.P.; Kosoy, M.; Vayssier-Taussat, M. Natural history of Bartonella-infecting rodents in light of new knowledge on genomics, diversity and evolution. Future Microbiol. 2013, 8, 1117–1128. [Google Scholar] [CrossRef]
- Szewczyk, T.; Werszko, J.; Slivinska, K.; Laskowski, Z.; Karbowiak, G. Molecular Detection of Bartonella spp. in Rodents in Chernobyl Exclusion Zone, Ukraine. Acta Parasitol. 2020, 66, 222–227. [Google Scholar] [CrossRef]
- Lipatova, I.; Paulauskas, A.; Puraite, I.; Radzijevskaja, J.; Balciauskas, L.; Gedminas, V. Bartonella infection in small mammals and their ectoparasites in Lithuania. Microbes Infect. 2015, 17, 884–888. [Google Scholar] [CrossRef]
- Engel, P.; Salzburger, W.; Liesch, M.; Chang, C.-C.; Maruyama, S.; Lanz, C.; Calteau, A.; Lajus, A.; Médigue, C.; Schuster, S.C.; et al. Parallel Evolution of a Type IV Secretion System in Radiating Lineages of the Host-Restricted Bacterial Pathogen Bartonella. PLoS Genet. 2011, 7, e1001296. [Google Scholar] [CrossRef] [Green Version]
- Birtles, R.J.; Harrison, T.G.; Saunders, N.A.; Molyneux, D.H. Proposals To Unify the Genera Grahamella and Bartonella, with Descriptions of Bartonella talpae comb, nov., Bartonella peromysci comb. nov., and Three New Species, Bartonella grahamii sp. nov., Bartonella taylorii sp. nov., and Bartonella doshiae sp. nov. Int. J. Syst. Bacteriol. 1995, 45, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Mediannikov, O.; Ivanov, L.; Zdanovskaya, N.; Vysochina, N.; Fournier, P.E.; Tarasevich, I.; Raoult, D. Molecular screening of Bartonella species in rodents from the Russian Far East. Ann. N. Y. Acad. Sci. 2005, 1063, 308–311. [Google Scholar] [CrossRef] [PubMed]
- Buffet, J.-P.; Pisanu, B.; Brisse, S.; Roussel, S.; Félix, B.; Halos, L.; Chapuis, J.-L.; Vayssier-Taussat, M. Deciphering Bartonella Diversity, Recombination, and Host Specificity in a Rodent Community. PLoS ONE 2013, 8, e68956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kerkhoff, F.T.; Bergmans, A.M.C.; Van Der Zee, A.; Rothova, A. Demonstration of Bartonella grahamii DNA in ocular fluids of a patient with neuroretinitis. J. Clin. Microbiol. 1999, 37, 4034–4038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bermond, D.; Heller, R.; Barrat, F.; Delacour, G.; Dehio, C.; Alliot, A.; Monteil, H.; Chomel, B.; Boulouis, H.J.; Piémont, Y. Bartonella birtlesii sp. nov., isolated from small mammals (Apodemus spp.). Int. J. Syst. Evol. Microbiol. 2000, 50, 1973–1979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paziewska, A.; Harris, P.D.; Zwolińska, L.; Bajer, A.; Siński, E. Recombination Within and Between Species of the Alpha Proteobacterium Bartonella Infecting Rodents. Microb. Ecol. 2011, 61, 134–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abbot, P.; Aviles, A.E.; Eller, L.; Durden, L.A. Mixed infections, cryptic diversity, and vector-borne pathogens: Evidence from Polygenis fleas and Bartonella species. Appl. Environ. Microbiol. 2007, 73, 6045–6052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vayssier-Taussat, M.; Moutailler, S.; Féménia, F.; Raymond, P.; Croce, O.; La Scola, B.; Fournier, P.-E.; Raoult, D. Identification of Novel Zoonotic Activity of Bartonella spp., France. Emerg. Infect. Dis. 2016, 22, 457–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paziewska, A.; Harris, P.D.; Zwolińska, L.; Bajer, A.; Siński, E. Differences in the ecology of Bartonella infections of Apodemus flavicollis and Myodes glareolus in a boreal forest. Parasitology 2012, 139, 881–893. [Google Scholar] [CrossRef]
- Mardosaitė-Busaitienė, D.; Radzijevskaja, J.; Balčiauskas, L.; Bratchikov, M.; Jurgelevičius, V.; Paulauskas, A. Prevalence and diversity of Bartonella species in small rodents from coastal and continental areas. Sci. Rep. 2019, 9, 12349. [Google Scholar] [CrossRef]
- Krasnov, B.R.; Khokhlova, I.S.; Fielden, L.J.; Burdelova, N.V. Effect of Air Temperature and Humidity on the Survival of Pre-Imaginal Stages of Two Flea Species (Siphonaptera: Pulicidae). J. Med. Entomol. 2001, 38, 629–637. [Google Scholar] [CrossRef]
- Krasnov, B.R.; Poulin, R.; Shenbrot, G.I.; Mouillot, D.; Khokhlova, I.S. Ectoparasitic “jacks-of-all-trades”: Relationship between abundance and host specificity in fleas (Siphonaptera) parasitic on small mammals. Am. Nat. 2004, 164, 506–516. [Google Scholar] [CrossRef]
- Kim, B.J.; Kim, S.J.; Kang, J.G.; Ko, S.; Won, S.; Kim, H.; Kim, H.C.; Kim, M.S.; Chong, S.T.; Klein, T.A.; et al. First report for the seasonal and annual prevalence of Flea-Borne Bartonella from Rodents and soricomorphs in the Republic of Korea. Vector Borne Zoonotic Dis. 2013, 13, 457–467. [Google Scholar] [CrossRef] [Green Version]
- Peterhans, S.; Landolt, P.; Friedel, U.; Oberhänsli, F.; Dennler, M.; Willi, B.; Senn, M.; Hinden, S.; Kull, K.; Kipar, A.; et al. Mycobacterium microti: Not Just a Coincidental Pathogen for Cats. Front. Vet. Sci. 2020, 7, 1–15. [Google Scholar] [CrossRef]
- Burlet, P.; Deplazes, P.; Hegglin, D. Efficient age determination: How freezing affects eye lens weight of the small rodent species arvicola terrestris. Eur. J. Wildl. Res. 2010, 56, 685–688. [Google Scholar] [CrossRef] [Green Version]
- Burlet, P.; Deplazes, P.; Hegglin, D. Age, season and spatio-temporal factors affecting the prevalence of Echinococcus multilocularis and Taenia taeniaeformis in Arvicola terrestris. Parasites Vectors 2011, 4, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beerli, O.; Guerra, D.; Baltrunaite, L.; Deplazes, P.; Hegglin, D. Microtus arvalis and Arvicola scherman: Key players in the Echinococcus multilocularis life cycle. Front. Vet. Sci. 2017, 4, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutiérrez, R.; Morick, D.; Gross, I.; Winkler, R.; Abdeen, Z.; Harrus, S. Bartonellae in Domestic and Stray Cats from Israel: Comparison of Bacterial Cultures and High-Resolution Melt Real-Time PCR As Diagnostic Methods. Vector Borne Zoonotic Dis. 2013, 13, 857–864. [Google Scholar] [CrossRef]
- Gutiérrez, R.; Nachum-Biala, Y.; Harrus, S. Relationship between the presence of Bartonella species and bacterial loads in cats and cat fleas (Ctenocephalides felis) under natural conditions. Appl. Environ. Microbiol. 2015, 81, 5613–5621. [Google Scholar] [CrossRef] [Green Version]
- Renesto, P.; Gouvernet, J.; Drancourt, M.; Roux, V.; Raoult, D. Use of rpoB gene analysis for detection and identification of Bartonella species. J. Clin. Microbiol. 2001, 39, 430–437. [Google Scholar] [CrossRef] [Green Version]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Rodent Species | Site | Area | Gender | Age | Total | ||||
|---|---|---|---|---|---|---|---|---|---|
| BE | FR | WL | OF | Females | Males | Adult | Juvenile | ||
| Myodes glareolus | 55 | 74 | 128 | 1 | 66 | 63 | 84 | 45 | 129 |
| Microtus agrestis | 2 | 0 | 2 | 0 | 2 | 0 | 2 | 0 | 2 |
| Arvicola scherman | 0 | 86 | 0 | 86 | 47 | 39 | 69 | 17 | 86 |
| Apodemus sp. | 83 | 46 | 127 | 2 | 63 | 66 | 90 | 39 | 129 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Divari, S.; Danelli, M.; Pregel, P.; Ghielmetti, G.; Borel, N.; Bollo, E. Biomolecular Investigation of Bartonella spp. in Wild Rodents of Two Swiss Regions. Pathogens 2021, 10, 1331. https://doi.org/10.3390/pathogens10101331
Divari S, Danelli M, Pregel P, Ghielmetti G, Borel N, Bollo E. Biomolecular Investigation of Bartonella spp. in Wild Rodents of Two Swiss Regions. Pathogens. 2021; 10(10):1331. https://doi.org/10.3390/pathogens10101331
Chicago/Turabian StyleDivari, Sara, Marta Danelli, Paola Pregel, Giovanni Ghielmetti, Nicole Borel, and Enrico Bollo. 2021. "Biomolecular Investigation of Bartonella spp. in Wild Rodents of Two Swiss Regions" Pathogens 10, no. 10: 1331. https://doi.org/10.3390/pathogens10101331
APA StyleDivari, S., Danelli, M., Pregel, P., Ghielmetti, G., Borel, N., & Bollo, E. (2021). Biomolecular Investigation of Bartonella spp. in Wild Rodents of Two Swiss Regions. Pathogens, 10(10), 1331. https://doi.org/10.3390/pathogens10101331








