Kinetic Study of BLV Infectivity in BLV Susceptible and Resistant Cattle in Japan from 2017 to 2019
Abstract
:1. Introduction
2. Results
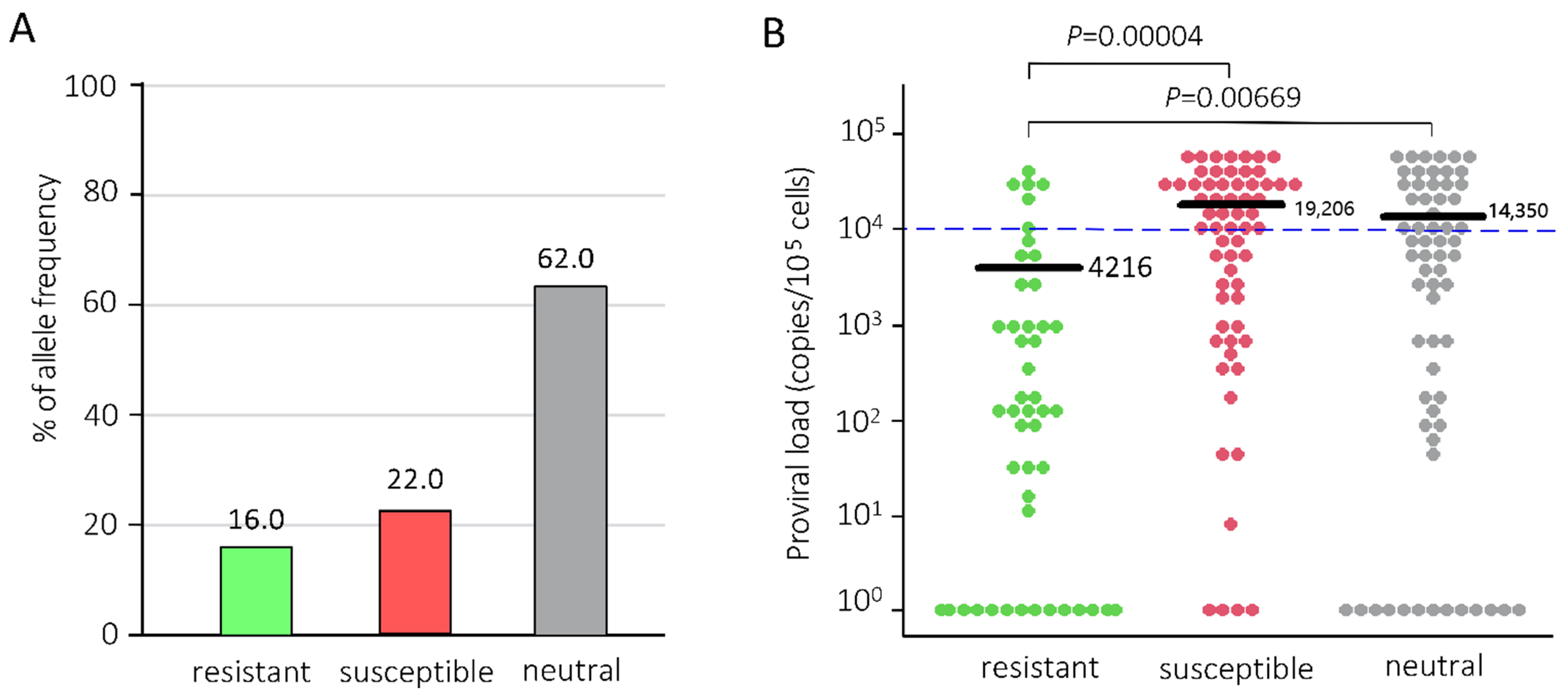
2.1. Determination of BoLA-DRB3 Alleles and BLV Infection
2.2. Syncytium Formation Abilities in Cattle with Different BoLA-DRB3 Alleles
2.3. Assessing BLV Infectivity in Seven Selected Cattle in Three-Year Follow-Up Study
2.4. Correlation of BLV Infectivity and PVL in Three-Year Follow-Up Study
3. Discussion
4. Materials and Methods
4.1. Blood Sample Collection
4.2. DNA Extraction
4.3. Detection of BLV Proviral Load
4.4. BoLA-DRB3 Allele Typing
4.5. Measurement of gp51 Antibodies
4.6. Detection of Syncytium Formation
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aida, Y.; Murakami, H.; Takahashi, M.; Takeshima, S.N. Mechanisms of pathogenesis induced by bovine leukemia virus as a model for human T-cell leukemia virus. Front. Microbiol. 2013, 4, 328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gillet, N.; Florins, A.; Boxus, M.; Burteau, C.; Nigro, A.; Vandermeers, F.; Balon, H.; Bouzar, A.B.; Defoiche, J.; Burny, A.; et al. Mechanisms of leukemogenesis induced by bovine leukemia virus: Prospects for novel anti-retroviral therapies in human. Retrovirology 2007, 4, 18. [Google Scholar] [CrossRef] [Green Version]
- Enzootic Bovine Leukosis. OIE World Animal Health Information Database (WAHID Interface). 2019, pp. 1113–1124. Available online: https://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/3.04.09_EBL.pdf (accessed on 10 August 2021).
- Murakami, K.; Kobayashi, S.; Konishi, M.; Kameyama, K.; Tsutsui, T. Nationwide survey of bovine leukemia virus infection among dairy and beef breeding cattle in Japan from 2009–2011. J. Vet. Med. Sci. 2013, 75, 1123–1126. [Google Scholar] [CrossRef] [Green Version]
- LaDronka, R.M.; Ainsworth, S.; Wilkins, M.J.; Norby, B.; Byrem, T.M.; Bartlett, P.C. Prevalence of Bovine Leukemia Virus Antibodies in US Dairy Cattle. Vet. Med. Int. 2018, 2018, 5831278. [Google Scholar] [CrossRef] [Green Version]
- Nekouei, O.; VanLeeuwen, J.; Sanchez, J.; Kelton, D.; Tiwari, A.; Keefe, G. Herd-level risk factors for infection with bovine leukemia virus in Canadian dairy herds. Prev. Vet. Med. 2015, 119, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, J.C.; Li, C.Y.; Hsu, W.L.; Chuang, S.T. Molecular Epidemiological and Serological Studies of Bovine Leukemia Virus in Taiwan Dairy Cattle. Front. Vet. Sci. 2019, 6, 427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suh, G.H.; Lee, J.C.; Lee, C.Y.; Hur, T.Y.; Son, D.S.; Ahn, B.S.; Kim, N.C.; Lee, C.G. Establishment of a bovine leukemia virus-free dairy herd in Korea. J. Vet. Sci. 2005, 6, 227–230. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Fan, W.; Mao, Y.; Yang, Z.; Lu, G.; Zhang, R.; Zhang, H.; Szeto, C.; Wang, C. Bovine leukemia virus infection in cattle of China: Association with reduced milk production and increased somatic cell score. J. Dairy Sci. 2016, 99, 3688–3697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, C.; Wang, X.; Zhou, Y.; Wang, Y.; Zhang, X.; Zheng, Y. Genotyping bovine leukemia virus in dairy cattle of Heilongjiang, northeastern China. BMC Vet. Res. 2019, 15, 179. [Google Scholar] [CrossRef]
- Ott, S.L.; Johnson, R.; Wells, S.J. Association between bovine-leukosis virus seroprevalence and herd-level productivity on US dairy farms. Prev. Vet. Med. 2003, 61, 249–262. [Google Scholar] [CrossRef]
- Erskine, R.J.; Bartlett, P.C.; Byrem, T.M.; Render, C.L.; Febvay, C.; Houseman, J.T. Association between bovine leukemia virus, production, and population age in Michigan dairy herds. J. Dairy Sci. 2012, 95, 727–734. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, J.K.; Pelzer, K.D.; Johnson, Y.J.; Russek-Cohen, E. Comparison of culling rates among dairy cows grouped on the basis of serologic status for bovine leukemia virus. J. Am. Vet. Med. Assoc. 2003, 223, 229–231. [Google Scholar] [CrossRef] [PubMed]
- Alessa, K.; Henk, H.; Karin, O.; Robert, W.; Jada, T.; Eldon, S.; Frank, V.D.M. Economic evaluation of 4 bovine leukemia virus control strategies for Alberta dairy farms. J. Dairy Sci. 2019, 102, 2578–2592. [Google Scholar] [CrossRef] [Green Version]
- Mekata, H.; Sekiguchi, S.; Konnai, S.; Kirino, Y.; Horii, Y.; Norimine, J. Horizontal transmission and phylogenetic analysis of bovine leukemia virus in two districts of Miyazaki, Japan. J. Vet. Med. Sci. 2015, 77, 1115–1120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dimmock, C.K.; Chung, Y.S.; MacKenzie, A.R. Factors affecting the natural transmission of bovine leukaemia virus infection in Queensland dairy herds. Aust. Vet. J. 1991, 68, 230–233. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, S.G.; DiGiacomo, R.F. Natural transmission of bovine leukemia virus in dairy and beef cattle. Vet. Clin. N. Am. Food Anim. Pract. 1997, 13, 107–128. [Google Scholar] [CrossRef]
- Kohara, J.; Konnai, S.; Onuma, M. Experimental transmission of Bovine leukemia virus in cattle via rectal palpation. Jpn. J. Vet. Res. 2006, 54, 25–30. [Google Scholar] [PubMed]
- Kohara, J.; Takeuchi, M.; Hirano, Y.; Sakurai, Y.; Takahashi, T. Vector control efficacy of fly nets on preventing bovine leukemia virus transmission. J. Vet. Med. Sci. 2018, 80, 1524–1527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manet, G.; Guilbert, X.; Roux, A.; Vuillaume, A.; Parodi, A.L. Natural mode of horizontal transmission of bovine leukemia virus (BLV): The potential role of tabanids (Tabanus spp.). Vet. Immunol. Immunopathol. 1989, 22, 255–263. [Google Scholar] [CrossRef]
- Meas, S.; Usui, T.; Ohashi, K.; Sugimoto, C.; Onuma, M. Vertical transmission of bovine leukemia virus and bovine immunodeficiency virus in dairy cattle herds. Vet. Microbiol. 2002, 84, 275–282. [Google Scholar] [CrossRef]
- Agresti, A.; Ponti, W.; Rocchi, M.; Meneveri, R.; Marozzi, A.; Cavalleri, D.; Peri, E.; Poli, G.; Ginelli, E. Use of polymerase chain reaction to diagnose bovine leukemia virus infection in calves at birth. Am. J. Vet. Res. 1993, 54, 373–378. [Google Scholar]
- Bai, L.; Sato, H.; Kubo, Y.; Wada, S.; Aida, Y. CAT1/SLC7A1 acts as a cellular receptor for bovine leukemia virus infection. FASEB J. 2019, fj201901528R. [Google Scholar] [CrossRef] [Green Version]
- Ohno, A.; Takeshima, S.N.; Matsumoto, Y.; Aida, Y. Risk factors associated with increased bovine leukemia virus proviral load in infected cattle in Japan from 2012 to 2014. Virus Res. 2015, 210, 283–290. [Google Scholar] [CrossRef]
- Jimba, M.; Takeshima, S.N.; Murakami, H.; Kohara, J.; Kobayashi, N.; Matsuhashi, T.; Ohmori, T.; Nunoya, T.; Aida, Y. BLV-CoCoMo-qPCR: A useful tool for evaluating bovine leukemia virus infection status. BMC Vet. Res. 2012, 8, 167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, H.; Watanuki, S.; Murakami, H.; Sato, R.; Ishizaki, H.; Aida, Y. Development of a luminescence syncytium induction assay (LuSIA) for easily detecting and quantitatively measuring bovine leukemia virus infection. Arch. Virol. 2018, 163, 1519–1530. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Kitamura-Muramatsu, Y.; Saito, S.; Ishizaki, H.; Nakano, M.; Haga, S.; Matoba, K.; Ohno, A.; Murakami, H.; Takeshima, S.N.; et al. Detection of the BLV provirus from nasal secretion and saliva samples using BLV-CoCoMo-qPCR-2: Comparison with blood samples from the same cattle. Virus Res. 2015, 210, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Watanuki, S.; Takeshima, S.N.; Borjigin, L.; Sato, H.; Bai, L.; Murakami, H.; Sato, R.; Ishizaki, H.; Matsumoto, Y.; Aida, Y. Visualizing bovine leukemia virus (BLV)-infected cells and measuring BLV proviral loads in the milk of BLV seropositive dams. Vet. Res. 2019, 50, 102. [Google Scholar] [CrossRef] [Green Version]
- Juliarena, M.A.; Barrios, C.N.; Ceriani, M.C.; Esteban, E.N. Hot topic: Bovine leukemia virus (BLV)-infected cows with low proviral load are not a source of infection for BLV-free cattle. J. Dairy Sci. 2016, 99, 4586–4589. [Google Scholar] [CrossRef] [Green Version]
- Juliarena, M.A.; Poli, M.; Sala, L.; Ceriani, C.; Gutierrez, S.; Dolcini, G.; Rodriguez, E.M.; Marino, B.; Rodriguez-Dubra, C.; Esteban, E.N. Association of BLV infection profiles with alleles of the BoLA-DRB3.2 gene. Anim. Genet. 2008, 39, 432–438. [Google Scholar] [CrossRef]
- Takeshima, S.N.; Ohno, A.; Aida, Y. Bovine leukemia virus proviral load is more strongly associated with bovine major histocompatibility complex class II DRB3 polymorphism than with DQA1 polymorphism in Holstein cow in Japan. Retrovirology 2019, 16, 14. [Google Scholar] [CrossRef]
- Takeshima, S.N.; Sasaki, S.; Meripet, P.; Sugimoto, Y.; Aida, Y. Single nucleotide polymorphisms in the bovine MHC region of Japanese Black cattle are associated with bovine leukemia virus proviral load. Retrovirology 2017, 14, 24. [Google Scholar] [CrossRef]
- Lo, C.W.; Borjigin, L.; Saito, S.; Fukunaga, K.; Saitou, E.; Okazaki, K.; Mizutani, T.; Wada, S.; Takeshima, S.N.; Aida, Y. BoLA-DRB3 Polymorphism is Associated with Differential Susceptibility to Bovine Leukemia Virus-Induced Lymphoma and Proviral Load. Viruses 2020, 12, 352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lutzelschwab, C.M.; Forletti, A.; Cepeda, R.; Esteban, E.N.; Confalonieri, O.; Gutierrez, S.E. Co-infection with Mycobacterium bovis does not alter the response to bovine leukemia virus in BoLA DRB3*0902, genetically resistant cattle. Res. Vet. Sci. 2016, 109, 10–16. [Google Scholar] [CrossRef]
- Takeshima, S.N.; Miyasaka, T.; Polat, M.; Kikuya, M.; Matsumoto, Y.; Mingala, C.N.; Villanueva, M.A.; Salces, A.J.; Onuma, M.; Aida, Y. The great diversity of major histocompatibility complex class II genes in Philippine native cattle. Meta Gene 2014, 2, 176–190. [Google Scholar] [CrossRef] [PubMed]
- Borjigin, L.; Yoneyama, S.; Saito, S.; Polat, M.; Inokuma, M.; Shinozaki, Y.; Tanaka, N.; Yamanaka, R.; Yasui, A.; Mimura, M.; et al. A novel real time PCR assay for bovine leukemia virus detection using mixed probes and degenerate primers targeting novel BLV strains. J. Virol. Methods 2021, 114264. [Google Scholar] [CrossRef] [PubMed]
- Jimba, M.; Takeshima, S.N.; Matoba, K.; Endoh, D.; Aida, Y. BLV-CoCoMo-qPCR: Quantitation of bovine leukemia virus proviral load using the CoCoMo algorithm. Retrovirology 2010, 7, 91. [Google Scholar] [CrossRef] [Green Version]
- Takeshima, S.N.; Kitamura-Muramatsu, Y.; Yuan, Y.; Polat, M.; Saito, S.; Aida, Y. BLV-CoCoMo-qPCR-2: Improvements to the BLV-CoCoMo-qPCR assay for bovine leukemia virus by reducing primer degeneracy and constructing an optimal standard curve. Arch. Virol. 2015, 160, 1325–1332. [Google Scholar] [CrossRef]
- Sato, H.; Watanuki, S.; Bai, L.; Borjigin, L.; Ishizaki, H.; Matsumoto, Y.; Hachiya, Y.; Sentsui, H.; Aida, Y. A sensitive luminescence syncytium induction assay (LuSIA) based on a reporter plasmid containing a mutation in the glucocorticoid response element in the long terminal repeat U3 region of bovine leukemia virus. Virol. J. 2019, 16, 66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, L.; Hirose, T.; Assi, W.; Wada, S.; Takeshima, S.N.; Aida, Y. Bovine Leukemia Virus Infection Affects Host Gene Expression Associated with DNA Mismatch Repair. Pathogens 2020, 9, 909. [Google Scholar] [CrossRef]
- Assi, W.; Hirose, T.; Wada, S.; Matsuura, R.; Takeshima, S.N.; Aida, Y. PRMT5 Is Required for Bovine Leukemia Virus Infection In Vivo and Regulates BLV Gene Expression, Syncytium Formation, and Glycosylation In Vitro. Viruses 2020, 12, 650. [Google Scholar] [CrossRef] [PubMed]
- Komori, H.; Ishiguro, N.; Horiuchi, M.; Shinagawa, M.; Aida, Y. Predominant p53 mutations in enzootic bovine leukemic cell lines. Vet. Immunol. Immunopathol. 1996, 52, 53–63. [Google Scholar] [CrossRef]
- Lewin, H.A.; Bernoco, D. Evidence for BoLA-linked resistance and susceptibility to subclinical progression of bovine leukaemia virus infection. Anim. Genet. 1986, 17, 197–207. [Google Scholar] [CrossRef]
- Zanotti, M.; Poli, G.; Ponti, W.; Polli, M.; Rocchi, M.; Bolzani, E.; Longeri, M.; Russo, S.; Lewin, H.A.; van Eijk, M.J. Association of BoLA class II haplotypes with subclinical progression of bovine leukaemia virus infection in Holstein-Friesian cattle. Anim. Genet. 1996, 27, 337–341. [Google Scholar] [PubMed]
- Miyasaka, T.; Takeshima, S.N.; Matsumoto, Y.; Kobayashi, N.; Matsuhashi, T.; Miyazaki, Y.; Tanabe, Y.; Ishibashi, K.; Sentsui, H.; Aida, Y. The diversity of bovine MHC class II DRB3 and DQA1 alleles in different herds of Japanese Black and Holstein cattle in Japan. Gene 2011, 472, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Xu, A.; van Eijk, M.J.; Park, C.; Lewin, H.A. Polymorphism in BoLA-DRB3 exon 2 correlates with resistance to persistent lymphocytosis caused by bovine leukemia virus. J. Immunol. 1993, 151, 6977–6985. [Google Scholar]
- Hayashi, T.; Mekata, H.; Sekiguchi, S.; Kirino, Y.; Mitoma, S.; Honkawa, K.; Horii, Y.; Norimine, J. Cattle with the BoLA class II DRB3*0902 allele have significantly lower bovine leukemia proviral loads. J. Vet. Med. Sci. 2017, 79, 1552–1555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agustina, F.; Claudia, M.L.; Rosana, C.; Eduardo, N.E.; Silvina, E.G. Early events following bovine leukaemia virus infection in calves with different alleles of the major histocompatibility complex DRB3 gene. Vet. Res. 2020, 51, 4. [Google Scholar] [CrossRef] [Green Version]
- Borjigin, L.; Lo, C.W.; Bai, L.; Hamada, R.; Sato, H.; Yoneyama, S.; Yasui, A.; Yasuda, S.; Yamanaka, R.; Mimura, M.; et al. Risk Assessment of Bovine Major Histocompatibility Complex Class II DRB3 Alleles for Perinatal Transmission of Bovine Leukemia Virus. Pathogens 2021, 10, 502. [Google Scholar] [CrossRef]
- Polat, M.; Moe, H.H.; Shimogiri, T.; Moe, K.K.; Takeshima, S.N.; Aida, Y. The molecular epidemiological study of bovine leukemia virus infection in Myanmar cattle. Arch. Virol. 2016. [Google Scholar] [CrossRef]
- Panei, C.J.; Takeshima, S.N.; Omori, T.; Nunoya, T.; Davis, W.C.; Ishizaki, H.; Matoba, K.; Aida, Y. Estimation of bovine leukemia virus (BLV) proviral load harbored by lymphocyte subpopulations in BLV-infected cattle at the subclinical stage of enzootic bovine leucosis using BLV-CoCoMo-qPCR. BMC Vet. Res. 2013, 9, 95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeshima, S.N.; Matsumoto, Y.; Miyasaka, T.; Arainga-Ramirez, M.; Saito, H.; Onuma, M.; Aida, Y. A new method for typing bovine major histocompatibility complex class II DRB3 alleles by combining two established PCR sequence-based techniques. Tissue Antigens 2011, 78, 208–213. [Google Scholar] [CrossRef] [PubMed]
- Tajima, S.; Aida, Y. Induction of expression of bovine leukemia virus (BLV) in blood taken from BLV-infected cows without removal of plasma. Microbes Infect. 2005, 7, 1211–1216. [Google Scholar] [CrossRef] [PubMed]
- Sato, H.; Bai, L.; Borjigin, L.; Aida, Y. Overexpression of bovine leukemia virus receptor SLC7A1/CAT1 enhances cellular susceptibility to BLV infection on luminescence syncytium induction assay (LuSIA). Virol. J. 2020, 17, 57. [Google Scholar] [CrossRef]
- Van Der Maaten, M.J.; Miller, J.M. Replication of bovine leukemia virus in monolayer cell cultures. Bibl. Haematol. 1975, 360–362. [Google Scholar]
- Onuma, M.; Koyama, H.; Aida, Y.; Okada, K.; Ogawa, Y.; Kirisawa, R.; Kawakami, Y. Establishment of B-cell lines from tumor of enzootic bovine leukosis. Leuk. Res. 1986, 10, 689–695. [Google Scholar] [CrossRef]



| Cattle Classification | Cattle No. | High PVL 1 Group | Low PVL Group | BLV-Free | ||
|---|---|---|---|---|---|---|
| BLV+ (%) | Mean PVL | BLV+ (%) | Mean PVL | BLV− (%) | ||
| Susceptible cattle 2 | 62 | 35 (56.5%) | 32,218 | 23 (37.1%) | 2746 | 4 (6.4%) |
| Neutral cattle 3 | 68 | 26 (38.2%) | 34,634 | 25 (36.8%) | 3014 | 17 (25.0%) |
| Resistant cattle 4 | 49 | 6 (12.2%) | 28,688 | 27 (55.1%) | 1275 | 16 (32.7%) |
| Total | 179 | 67 (37.4%) | 32,839 | 75 (41.9%) | 2306 | 37 (20.7%) |
| Cattle No. | Genotype | BoLA-DRB3 | PVL 1,2 | gp51 Abs 2 | Age 2 (Year) | Syncytium Formation Assay for Three-Year Follow-Up Study | Correlation Analysis of BLV Infectivity with PVL | |
|---|---|---|---|---|---|---|---|---|
| A | B | |||||||
| S1 | Susceptible | 012:01 | 015:01 | 79,519 | + 3 | 3.7 | NT 4 | NT |
| S2 | Susceptible | 001:01 | 015:01 | 35,758 | + | 7.8 | selected | Tested |
| S3 | Susceptible | 011:01 | 015:01 | 37,707 | + | 6.2 | selected | Tested |
| S4 | Susceptible | 011:01 | 015:01 | 10,780 | + | 3.9 | NT | NT |
| S5 | Susceptible | 001:01 | 015:01 | 27,642 | + | 10.7 | NT | Tested |
| S6 | Susceptible | 011:01 | 015:01 | 36,149 | + | 7.1 | NT | NT |
| S7 | Susceptible | 001:01 | 015:01 | 39,024 | + | 11.2 | selected | Tested |
| S8 | Susceptible | 001:01 | 015:01 | 17,981 | + | 5.8 | NT | NT |
| S9 | Susceptible | 011:01 | 015:01 | 40,534 | + | 5.1 | NT | NT |
| S10 | Susceptible | 015:01 | 015:01 | 18,062 | + | 6.8 | NT | Tested |
| S11 | Susceptible | 015:01 | 018:01 | 7733 | + | 4.8 | NT | Tested |
| S12 | Susceptible | 011:01 | 012:01 | 18,879 | + | 6.2 | NT | NT |
| S13 | Susceptible | 010:01 | 015:01 | 12,097 | + | 1.4 | NT | NT |
| S14 | Susceptible | 010:01 | 015:01 | 10,169 | + | 6.3 | NT | Tested |
| S15 | Susceptible | 015:01 | 015:01 | 16,337 | + | 6.1 | NT | NT |
| N1 | Neutral | 001:01 | 010:01 | 12,424 | NT | 0.5 | selected | Tested |
| R1 | Resistant | 011:01 | 014:01:01 | 8597 | + | 6.2 | NT | Tested |
| R2 | Resistant | 009:02 | 015:01 | 222 | + | 4.8 | selected | Tested |
| R3 | Resistant | 001:01 | 014:01:01 | 165 | + | 10.8 | NT | NT |
| R4 | Resistant | 014:01:01 | 014:01:01 | 0 | + | 6.2 | selected | Tested |
| R5 | Resistant | 001:01 | 009:02 | 0 | + | 7.8 | selected | Tested |
| R6 | Resistant | 011:01 | 014:01:01 | 54 | + | 5.4 | NT | NT |
| R7 | Resistant | 011:01 | 014:01:01 | 468 | + | 5.1 | NT | Tested |
| R8 | Resistant | 011:01 | 014:01:01 | 0 | + | 4.3 | NT | NT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bai, L.; Borjigin, L.; Sato, H.; Takeshima, S.-N.; Asaji, S.; Ishizaki, H.; Kawashima, K.; Obuchi, Y.; Sunaga, S.; Ando, A.; et al. Kinetic Study of BLV Infectivity in BLV Susceptible and Resistant Cattle in Japan from 2017 to 2019. Pathogens 2021, 10, 1281. https://doi.org/10.3390/pathogens10101281
Bai L, Borjigin L, Sato H, Takeshima S-N, Asaji S, Ishizaki H, Kawashima K, Obuchi Y, Sunaga S, Ando A, et al. Kinetic Study of BLV Infectivity in BLV Susceptible and Resistant Cattle in Japan from 2017 to 2019. Pathogens. 2021; 10(10):1281. https://doi.org/10.3390/pathogens10101281
Chicago/Turabian StyleBai, Lanlan, Liushiqi Borjigin, Hirotaka Sato, Shin-Nosuke Takeshima, Sakurako Asaji, Hiroshi Ishizaki, Keiji Kawashima, Yuko Obuchi, Shinji Sunaga, Asako Ando, and et al. 2021. "Kinetic Study of BLV Infectivity in BLV Susceptible and Resistant Cattle in Japan from 2017 to 2019" Pathogens 10, no. 10: 1281. https://doi.org/10.3390/pathogens10101281
APA StyleBai, L., Borjigin, L., Sato, H., Takeshima, S.-N., Asaji, S., Ishizaki, H., Kawashima, K., Obuchi, Y., Sunaga, S., Ando, A., Inoko, H., Wada, S., & Aida, Y. (2021). Kinetic Study of BLV Infectivity in BLV Susceptible and Resistant Cattle in Japan from 2017 to 2019. Pathogens, 10(10), 1281. https://doi.org/10.3390/pathogens10101281








