Plasmepsin-like Aspartyl Proteases in Babesia
Abstract
:1. Introduction
2. Results and Discussion
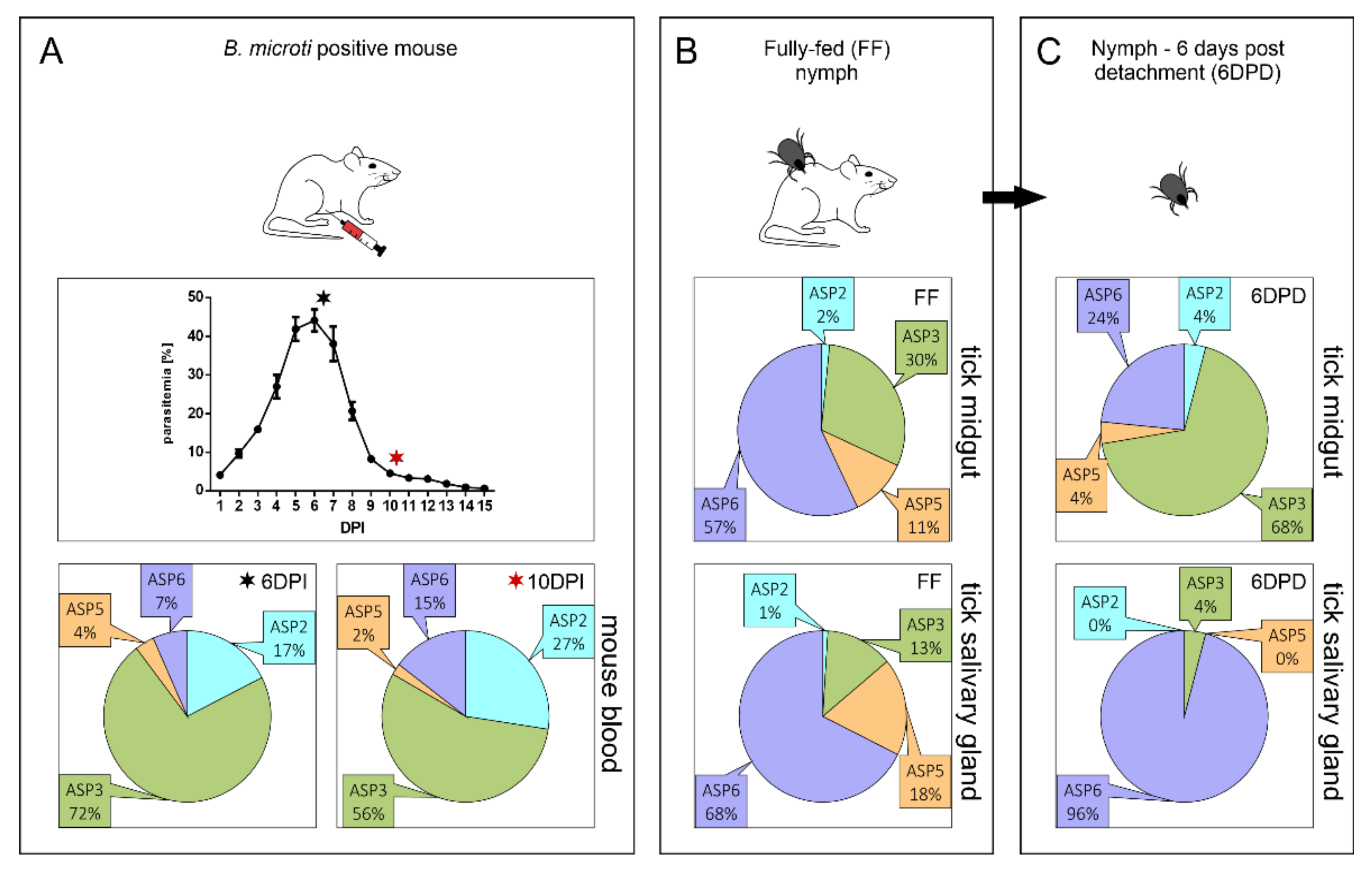
2.1. Expression Profiling of BmASPs Indicate Their Various Roles throughout the Life Cycle of Babesia microti
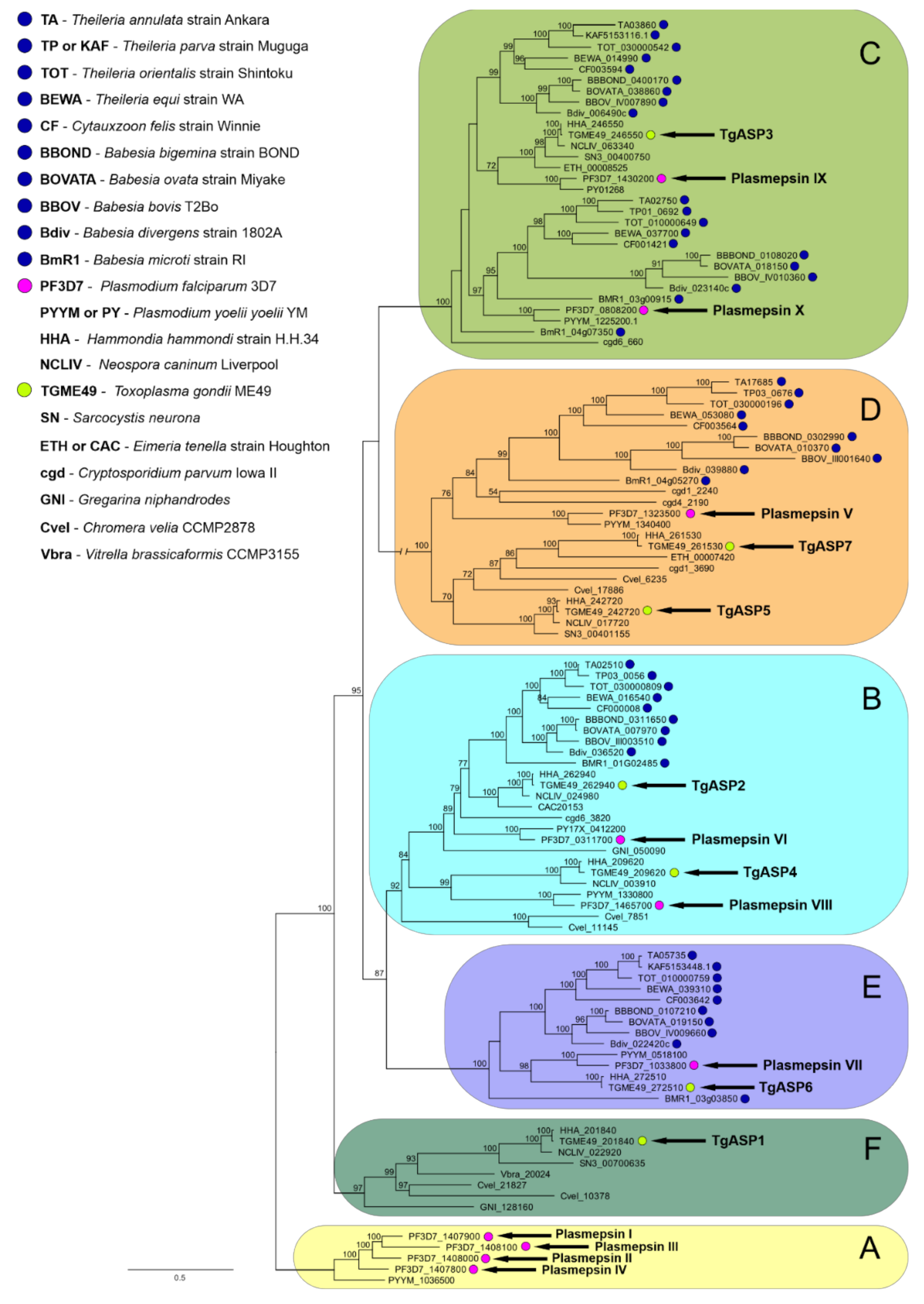
2.2. Data-Mining and Phylogenetic Analysis of Piroplasmid APs Reveals the Presence of Multiple AP Isoenzymes Clustering to Several Apicomplexan AP Clades
2.2.1. Clade A
2.2.2. Clade B
2.2.3. Clade C
2.2.4. Clade D
2.2.5. Clade E
2.2.6. Clade F
3. Conclusions
4. Materials and Methods
4.1. B. microti Propagation in Mice
4.2. RNA Isolation from Tick Tissues and Murine Blood Cells
4.3. Quantitative RT-PCR
4.4. Phylogenetic Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jalovecka, M.; Sojka, D.; Ascencio, M.; Schnittger, L. Babesia life cycle–when phylogeny meets biology. Trends Parasitol. 2019, 35, 356–368. [Google Scholar] [CrossRef]
- Vannier, E.; Krause, P.J. Babesiosis. In Hunter’s Tropical Medicine and Emerging Infectious Diseases; Elsevier: Amsterdam, The Netherlands, 2020; pp. 799–802. [Google Scholar]
- Schnittger, L.; Rodriguez, A.E.; Florin-Christensen, M.; Morrison, D.A. Babesia: A world emerging. Infect. Genet. Evol. 2012, 12, 1788–1809. [Google Scholar] [CrossRef]
- Florin-Christensen, M.; Suarez, C.E.; Rodriguez, A.E.; Flores, D.A.; Schnittger, L. Vaccines against bovine babesiosis: Where we are now and possible roads ahead. Parasitology 2014, 141, 1563–1592. [Google Scholar] [CrossRef]
- Rubel, F.; Brugger, K.; Pfeffer, M.; Chitimia-Dobler, L.; Didyk, Y.M.; Leverenz, S.; Dautel, H.; Kahl, O. Geographical distribution of Dermacentor marginatus and Dermacentor reticulatus in Europe. Ticks Tick-Borne Dis. 2016, 7, 224–233. [Google Scholar] [CrossRef] [Green Version]
- Vannier, E.; Krause, P.J. Human babesiosis. N. Engl. J. Med. 2012, 366, 2397–2407. [Google Scholar] [CrossRef] [Green Version]
- Lobo, C.A.; Cursino-Santos, J.R.; Alhassan, A.; Rodrigues, M. Babesia: An emerging infectious threat in transfusion medicine. PLoS Pathog. 2013, 9, e1003387. [Google Scholar] [CrossRef] [Green Version]
- Lempereur, L.; Shiels, B.; Heyman, P.; Moreau, E.; Saegerman, C.; Losson, B.; Malandrin, L. A retrospective serological survey on human babesiosis in Belgium. Clin. Microbiol. Infect. 2015, 21, 96.e91–96.e97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arsuaga, M.; González, L.M.; Padial, E.S.; Dinkessa, A.W.; Sevilla, E.; Trigo, E.; Puente, S.; Gray, J.; Montero, E. Misdiagnosis of babesiosis as malaria, Equatorial Guinea, 2014. Emerg. Infect. Dis. 2018, 24, 1588–1589. [Google Scholar] [CrossRef]
- Rathinasamy, V.; Poole, W.A.; Bastos, R.G.; Suarez, C.E.; Cooke, B.M. Babesiosis vaccines: Lessons learned, challenges ahead, and future glimpses. Trends Parasitol. 2019, 35, 622–635. [Google Scholar] [CrossRef] [PubMed]
- Lemieux, J.E.; Tran, A.D.; Freimark, L.; Schaffner, S.F.; Goethert, H.; Andersen, K.G.; Bazner, S.; Li, A.; McGrath, G.; Sloan, L. A global map of genetic diversity in Babesia microti reveals strong population structure and identifies variants associated with clinical relapse. Nat. Microbiol. 2016, 1, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simon, M.S.; Westblade, L.F.; Dziedziech, A.; Visone, J.E.; Furman, R.R.; Jenkins, S.G.; Schuetz, A.N.; Kirkman, L.A. Clinical and molecular evidence of atovaquone and azithromycin resistance in relapsed Babesia microti infection associated with rituximab and chronic lymphocytic leukemia. Clin. Infect. Dis. 2017, 65, 1222–1225. [Google Scholar] [CrossRef] [Green Version]
- McKerrow, J.H.; Caffrey, C.; Kelly, B.; Loke, P.n.; Sajid, M. Proteases in parasitic diseases. Annu. Rev. Pathol. Mech. Dis. 2006, 1, 497–536. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Bateman, A. How to use the MEROPS database and website to help understand peptidase specificity. Protein Sci. 2021, 30, 83–92. [Google Scholar] [CrossRef]
- Barrett, A.J.; Rawlings, N.D.; Salvesen, G.; Woessner, J.F. Handbook of proteolytic enzymes introduction. In Handbook of Proteolytic Enzymes, 3rd ed.; Elsevier: Abingdon, UK, 2012; ISBN 9780123822208. [Google Scholar]
- Sojka, D.; Hartmann, D.; Bartošová-Sojková, P.; Dvořák, J. Parasite cathepsin D-like peptidases and their relevance as therapeutic targets. Trends Parasitol. 2016, 32, 708–723. [Google Scholar] [CrossRef]
- Shea, M.; Jäkle, U.; Liu, Q.; Berry, C.; Joiner, K.A.; Soldati-Favre, D. A family of aspartic proteases and a novel, dynamic and cell-cycle-dependent protease localization in the secretory pathway of Toxoplasma Gondii. Traffic 2007, 8, 1018–1034. [Google Scholar] [CrossRef]
- Coombs, G.H.; Goldberg, D.E.; Klemba, M.; Berry, C.; Kay, J.; Mottram, J.C. Aspartic proteases of Plasmodium falciparum and other parasitic protozoa as drug targets. Trends Parasitol. 2001, 17, 532–537. [Google Scholar] [CrossRef]
- Burrows, J.N.; Soldati-Favre, D. Targeting plasmepsins—an achilles’ heel of the malaria parasite. Cell Host Microbe 2020, 27, 496–498. [Google Scholar] [CrossRef] [PubMed]
- Jalovecka, M.; Urbanova, V.; Sojka, D.; Malandrin, L.; Sima, R.; Kopacek, P.; Hajdusek, O. Establishment of Babesia microti laboratory model and its experimental application. In Proceedings of the 9. Tick and Tick-borne Pathogen Conference and 1st Asia Pacific Rickettsia Conference, Cairns, Australia, 20 August 2017; p. 140. [Google Scholar]
- Pino, P.; Caldelari, R.; Mukherjee, B.; Vahokoski, J.; Klages, N.; Maco, B.; Collins, C.R.; Blackman, M.J.; Kursula, I.; Heussler, V. A multistage antimalarial targets the plasmepsins IX and X essential for invasion and egress. Science 2017, 358, 522–528. [Google Scholar] [CrossRef] [Green Version]
- Dogga, S.K.; Mukherjee, B.; Jacot, D.; Kockmann, T.; Molino, L.; Hammoudi, P.-M.; Hartkoorn, R.C.; Hehl, A.B.; Soldati-Favre, D. A druggable secretory protein maturase of Toxoplasma essential for invasion and egress. Elife 2017, 6, e27480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, F.; Bounkeua, V.; Pettersen, K.; Vinetz, J.M. Plasmodium falciparum ookinete expression of plasmepsin VII and plasmepsin X. Malar. J. 2016, 15, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ecker, A.; Bushell, E.S.; Tewari, R.; Sinden, R.E. Reverse genetics screen identifies six proteins important for malaria development in the mosquito. Mol. Microbiol. 2008, 70, 209–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jalovecka, M.; Hajdusek, O.; Sojka, D.; Kopacek, P.; Malandrin, L. The complexity of piroplasms life cycles. Front. Cell. Infect. Microbiol. 2018, 8, 248. [Google Scholar] [CrossRef] [Green Version]
- Boddey, J.A.; Carvalho, T.G.; Hodder, A.N.; Sargeant, T.J.; Sleebs, B.E.; Marapana, D.; Lopaticki, S.; Nebl, T.; Cowman, A.F. Role of plasmepsin V in export of diverse protein families from the Plasmodium falciparum exportome. Traffic 2013, 14, 532–550. [Google Scholar] [CrossRef] [PubMed]
- Russo, I.; Babbitt, S.; Muralidharan, V.; Butler, T.; Oksman, A.; Goldberg, D.E. Plasmepsin V licenses Plasmodium proteins for export into the host erythrocyte. Nature 2010, 463, 632–636. [Google Scholar] [CrossRef]
- Coffey, M.J.; Dagley, L.F.; Seizova, S.; Kapp, E.A.; Infusini, G.; Roos, D.S.; Boddey, J.A.; Webb, A.I.; Tonkin, C.J. Aspartyl protease 5 matures dense granule proteins that reside at the host-parasite interface. Toxoplasma Gondii. MBio 2018, 9, e01718–e01796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, J.C.; Cornillot, E.; McCracken, C.; Usmani-Brown, S.; Dwivedi, A.; Ifeonu, O.O.; Crabtree, J.; Gotia, H.T.; Virji, A.Z.; Reynes, C.; et al. Genome-wide diversity and gene expression profiling of Babesia microti isolates identify polymorphic genes that mediate host-pathogen interactions. Sci. Rep. 2016, 6, 35284. [Google Scholar] [CrossRef]
- Thekkiniath, J.; Kilian, N.; Lawres, L.; Gewirtz, M.A.; Graham, M.M.; Liu, X.; Ledizet, M.; Mamoun, C.B. Evidence for vesicle-mediated antigen export by the human pathogen Babesia Microti. Life Sci. Alliance 2019, 2, e201900382. [Google Scholar] [CrossRef] [Green Version]
- Rudzinska, M.A.; Trager, W.; Lewengrub, S.J.; Gubert, E. An electron microscopic study of Babesia microti invading erythrocytes. Cell Tissue Res. 1976, 169, 323–334. [Google Scholar] [CrossRef]
- Jennison, C.; Lucantoni, L.; O’Neill, M.T.; McConville, R.; Erickson, S.M.; Cowman, A.F.; Sleebs, B.E.; Avery, V.M.; Boddey, J.A. Inhibition of plasmepsin V activity blocks Plasmodium falciparum gametocytogenesis and transmission to mosquitoes. Cell Rep. 2019, 29, 3796–3806.e3794. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Gluzman, I.Y.; Drew, M.E.; Goldberg, D.E. The role of Plasmodium falciparum food vacuole plasmepsins. J. Biol. Chem. 2005, 280, 1432–1437. [Google Scholar] [CrossRef] [Green Version]
- Votýpka, J.; Modrý, D.; Oborník, M.; Šlapeta, J.; Lukeš, J. Apicomplexa. Handb. Protists 2017, 2, 567–624. [Google Scholar]
- Rinehart, M.T.; Park, H.S.; Walzer, K.A.; Chi, J.-T.A.; Wax, A. Hemoglobin consumption by P. falciparum in individual erythrocytes imaged via quantitative phase spectroscopy. Sci. Rep. 2016, 6, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Ponsuwanna, P.; Kochakarn, T.; Bunditvorapoom, D.; Kümpornsin, K.; Otto, T.D.; Ridenour, C.; Chotivanich, K.; Wilairat, P.; White, N.J.; Miotto, O. Comparative genome-wide analysis and evolutionary history of haemoglobin-processing and haem detoxification enzymes in malarial parasites. Malar. J. 2016, 15, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Nasamu, A.S.; Polino, A.J.; Istvan, E.S.; Goldberg, D.E. Malaria parasite plasmepsins: More than just plain old degradative pepsins. J. Biol. Chem. 2020, 295, 8425–8441. [Google Scholar] [CrossRef]
- Rosenthal, P.J.; Meshnick, S.R. Hemoglobin catabolism and iron utilization by malaria parasites. Mol. Biochem. Parasitol. 1996, 83, 131–139. [Google Scholar] [CrossRef]
- Lew, V.L.; Tiffert, T.; Ginsburg, H. Excess hemoglobin digestion and the osmotic stability of Plasmodium falciparum–infected red blood cells. Blood 2003, 101, 4189–4194. [Google Scholar] [CrossRef] [PubMed]
- Chugh, M.; Sundararaman, V.; Kumar, S.; Reddy, V.S.; Siddiqui, W.A.; Stuart, K.D.; Malhotra, P. Protein complex directs hemoglobin-to-hemozoin formation in Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 2013, 110, 5392–5397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, H.; Hong, S.-H.; Kim, K.; Cho, S.-H.; Lee, W.-J.; Kim, Y.; Lee, S.-E.; Park, Y. Characterizations of individual mouse red blood cells parasitized by Babesia microti using 3-D holographic microscopy. Sci. Rep. 2015, 5, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Lempereur, L.; Beck, R.; Fonseca, I.; Marques, C.; Duarte, A.; Santos, M.; Zúquete, S.; Gomes, J.; Walder, G.; Domingos, A. Guidelines for the detection of Babesia and Theileria parasites. Vector-Borne Zoonotic Dis. 2017, 17, 51–65. [Google Scholar] [CrossRef] [Green Version]
- Cursino-Santos, J.R.; Singh, M.; Senaldi, E.; Manwani, D.; Yazdanbakhsh, K.; Lobo, C.A. Altered parasite life-cycle processes characterize Babesia divergens infection in human sickle cell anemia. Haematologica 2019, 104, 2189. [Google Scholar] [CrossRef]
- Fawcett, D.W.; Conrad, P.A.; Grootenhuis, J.G.; Morzaria, S.P. Ultrastructure of the intra-erythrocytic stage of Theileria species from cattle and waterbuck. Tissue Cell 1987, 19, 643–655. [Google Scholar] [CrossRef]
- Guimarães, A.M.; Lima, J.D.; Ribeiro, M.F. Ultrastructure of Babesia equi trophozoites isolated in Minas Gerais, Brazil. Pesqui. Vet. Bras. 2003, 23, 101–104. [Google Scholar] [CrossRef] [Green Version]
- Rudzinska, M.A. Ultrastructure of intraerythrocytic Babesia microti with emphasis on the feeding mechanism. J. Protozool. 1976, 23, 224–233. [Google Scholar] [CrossRef] [PubMed]
- Conesa, J.J.; Sevilla, E.; Terrón, M.C.; González, L.M.; Gray, J.; Pérez-Berná, A.J.; Carrascosa, J.L.; Pereiro, E.; Chichón, F.J.; Luque, D.; et al. Four-dimensional characterizationof the Babesia divergens asexual life cycle, from the trophozoite to the multiparasite stage. MSphere 2020, 5, e00928-20. [Google Scholar] [CrossRef]
- Okubo, K.; Yokoyama, N.; Govind, Y.; Alhassan, A.; Igarashi, I. Babesia bovis: Effects of cysteine protease inhibitors on in vitro growth. Exp. Parasitol. 2007, 117, 214–217. [Google Scholar] [CrossRef]
- Carletti, T.; Barreto, C.; Mesplet, M.; Mira, A.; Weir, W.; Shiels, B.; Oliva, A.G.; Schnittger, L.; Florin-Christensen, M. Characterization of a papain-like cysteine protease essential for the survival of Babesia ovis merozoites. Ticks Tick-Borne Dis. 2016, 7, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Jean, L.; Long, M.; Young, J.; Péry, P.; Tomley, F. Aspartyl proteinase genes from apicomplexan parasites: Evidence for evolution of the gene structure. Trends Parasitol. 2001, 17, 491–498. [Google Scholar] [CrossRef]
- Mastan, B.S.; Narwal, S.K.; Dey, S.; Kumar, K.A.; Mishra, S. Plasmodium berghei plasmepsin VIII is essential for sporozoite gliding motility. Int. J. Parasitol. 2017, 47, 239–245. [Google Scholar] [CrossRef]
- Banerjee, R.; Liu, J.; Beatty, W.; Pelosof, L.; Klemba, M.; Goldberg, D.E. Four plasmepsins are active in the Plasmodium falciparum food vacuole, including a protease with an active-site histidine. Proc. Natl. Acad. Sci. USA 2002, 99, 990–995. [Google Scholar] [CrossRef] [Green Version]
- Favuzza, P.; de Lera Ruiz, M.; Thompson, J.K.; Triglia, T.; Ngo, A.; Steel, R.W.; Vavrek, M.; Christensen, J.; Healer, J.; Boyce, C. Dual plasmepsin-targeting antimalarial agents disrupt multiple stages of the malaria parasite life cycle. Cell Host Microbe 2020, 27, 642–658.e612. [Google Scholar] [CrossRef] [PubMed]
- Sojka, D.; Šnebergerová, P.; Robbertse, L. Protease inhibition—an established strategy to combat infectious diseases. Int. J. Mol. Sci. 2021, 22, 5762. [Google Scholar] [CrossRef]
- Meissner, M.; Ferguson, D.J.; Frischknecht, F. Invasion factors of apicomplexan parasites: Essential or redundant? Curr. Opin. Microbiol. 2013, 16, 438–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frénal, K.; Dubremetz, J.-F.; Lebrun, M.; Soldati-Favre, D. Gliding motility powers invasion and egress in Apicomplexa. Nat. Rev. Microbiol. 2017, 15, 645–660. [Google Scholar] [CrossRef] [Green Version]
- Chauvin, A.; Moreau, E.; Bonnet, S.; Plantard, O.; Malandrin, L. Babesia and its hosts: Adaptation to long-lasting interactions as a way to achieve efficient transmission. Vet. Res. 2009, 40, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Lobo, C.-A. Babesia divergens and Plasmodium falciparum use common receptors, glycophorins A and B, to invade the human red blood cell. Infect. Immun. 2005, 73, 649–651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malpede, B.M.; Tolia, N.H. Malaria adhesins: Structure and function. Cell. Microbiol. 2014, 16, 621–631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beeson, J.G.; Drew, D.R.; Boyle, M.J.; Feng, G.; Fowkes, F.J.; Richards, J.S. Merozoite surface proteins in red blood cell invasion, immunity and vaccines against malaria. FEMS Microbiol. Rev. 2016, 40, 343–372. [Google Scholar] [CrossRef] [Green Version]
- Mosqueda, J.; McElwain, T.F.; Palmer, G.H. Babesia bovis merozoite surface antigen 2 proteins are expressed on the merozoite and sporozoite surface, and specific antibodies inhibit attachment and invasion of erythrocytes. Infect. Immun. 2002, 70, 6448–6455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woods, K.; Perry, C.; Brühlmann, F.; Olias, P. Theileria’s strategies and effector mechanisms for host cell transformation: From invasion to immortalization. Front. Cell Dev. Biol. 2021, 9, 972. [Google Scholar] [CrossRef]
- Lobo, C.A.; Rodriguez, M.; Cursino-Santos, J.R. Babesia and red cell invasion. Curr. Opin. Hematol. 2012, 19, 170–175. [Google Scholar] [CrossRef]
- Besteiro, S.; Dubremetz, J.F.; Lebrun, M. The moving junction of apicomplexan parasites: A key structure for invasion. Cell. Microbiol. 2011, 13, 797–805. [Google Scholar] [CrossRef] [PubMed]
- Montero, E.; Rodriguez, M.; Oksov, Y.; Lobo, C.A. Babesia divergens apical membrane antigen 1 and its interaction with the human red blood cell. Infect. Immun. 2009, 77, 4783–4793. [Google Scholar] [CrossRef] [Green Version]
- Moitra, P.; Zheng, H.; Anantharaman, V.; Banerjee, R.; Takeda, K.; Kozakai, Y.; Lepore, T.; Krause, P.J.; Aravind, L.; Kumar, S. Expression, purification, and biological characterization of Babesia microti apical membrane antigen 1. Infect. Immun. 2015, 83, 3890–3901. [Google Scholar] [CrossRef] [Green Version]
- Gaffar, F.R.; Yatsuda, A.P.; Franssen, F.F.; de Vries, E. Erythrocyte invasion by Babesia bovis merozoites is inhibited by polyclonal antisera directed against peptides derived from a homologue of Plasmodium falciparum apical membrane antigen 1. Infect. Immun. 2004, 72, 2947–2955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González, L.M.; Estrada, K.; Grande, R.; Jiménez-Jacinto, V.; Vega-Alvarado, L.; Sevilla, E.; Barrera, J.; Cuesta, I.; Zaballos, Á.; Bautista, J.M.; et al. Comparative and functional genomics of the protozoan parasite Babesia divergens highlighting the invasion and egress processes. PLoS Negl. Trop. Dis. 2019, 13, e0007680. [Google Scholar] [CrossRef]
- Buguliskis, J.S.; Brossier, F.; Shuman, J.; Sibley, L.D. Rhomboid 4 (ROM4) affects the processing of surface adhesins and facilitates host cell invasion by Toxoplasma gondii. PLoS Pathog. 2010, 6, e1000858. [Google Scholar] [CrossRef] [Green Version]
- Shaw, M.K. Cell invasion by Theileria sporozoites. Trends Parasitol. 2003, 19, 2–6. [Google Scholar] [CrossRef]
- Shaw, M.K. Theileria development and host cell invasion. In Theileria; Springer: Boston, MA, USA, 2002; pp. 1–22. [Google Scholar]
- Thomas, J.A.; Tan, M.S.; Bisson, C.; Borg, A.; Umrekar, T.R.; Hackett, F.; Hale, V.L.; Vizcay-Barrena, G.; Fleck, R.A.; Snijders, A.P. A protease cascade regulates release of the human malaria parasite Plasmodium falciparum from host red blood cells. Nat. Microbiol. 2018, 3, 447–455. [Google Scholar] [CrossRef]
- Collins, C.R.; Hackett, F.; Atid, J.; Tan, M.S.Y.; Blackman, M.J. The Plasmodium falciparum pseudoprotease SERA5 regulates the kinetics and efficiency of malaria parasite egress from host erythrocytes. PLoS Pathog. 2017, 13, e1006453. [Google Scholar] [CrossRef] [Green Version]
- Withers-Martinez, C.; Strath, M.; Hackett, F.; Haire, L.F.; Howell, S.A.; Walker, P.A.; Christodoulou, E.; Dodson, G.G.; Blackman, M.J. The malaria parasite egress protease SUB1 is a calcium-dependent redox switch subtilisin. Nat. Commun. 2014, 5, 1–11. [Google Scholar] [CrossRef]
- Das, S.; Hertrich, N.; Perrin, A.J.; Withers-Martinez, C.; Collins, C.R.; Jones, M.L.; Watermeyer, J.M.; Fobes, E.T.; Martin, S.R.; Saibil, H.R. Processing of Plasmodium falciparum merozoite surface protein MSP1 activates a spectrin-binding function enabling parasite egress from RBCs. Cell Host Microbe 2015, 18, 433–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koussis, K.; Withers-Martinez, C.; Yeoh, S.; Child, M.; Hackett, F.; Knuepfer, E.; Juliano, L.; Woehlbier, U.; Bujard, H.; Blackman, M.J. A multifunctional serine protease primes the malaria parasite for red blood cell invasion. EMBO J. 2009, 28, 725–735. [Google Scholar] [CrossRef] [Green Version]
- Montero, E.; Gonzalez, L.M.; Rodriguez, M.; Oksov, Y.; Blackman, M.J.; Lobo, C.A. A conserved subtilisin protease identified in Babesia divergens merozoites. J. Biol. Chem. 2006, 281, 35717–35726. [Google Scholar] [CrossRef] [Green Version]
- Lempereur, L.; Larcombe, S.D.; Durrani, Z.; Karagenc, T.; Bilgic, H.B.; Bakirci, S.; Hacilarlioglu, S.; Kinnaird, J.; Thompson, J.; Weir, W.; et al. Identification of candidate transmission-blocking antigen genes in Theileria annulata and related vector-borne apicomplexan parasites. BMC Genom. 2017, 18, 438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kariu, T.; Ishino, T.; Yano, K.; Chinzei, Y.; Yuda, M. CelTOS, a novel malarial protein that mediates transmission to mosquito and vertebrate hosts. Mol. Microbiol. 2006, 59, 1369–1379. [Google Scholar] [CrossRef] [PubMed]
- Jimah, J.R.; Salinas, N.D.; Sala-Rabanal, M.; Jones, N.G.; Sibley, L.D.; Nichols, C.G.; Schlesinger, P.H.; Tolia, N.H. Malaria parasite CelTOS targets the inner leaflet of cell membranes for pore-dependent disruption. Elife 2016, 5, e20621. [Google Scholar] [CrossRef] [Green Version]
- Baldi, D.L.; Andrews, K.T.; Waller, R.F.; Roos, D.S.; Howard, R.F.; Crabb, B.S.; Cowman, A.F. RAP1 controls rhoptry targeting of RAP2 in the malaria parasite Plasmodium Falciparum. EMBO J. 2000, 19, 2435–2443. [Google Scholar] [CrossRef] [Green Version]
- Low, L.M.; Azasi, Y.; Sherling, E.S.; Garten, M.; Zimmerberg, J.; Tsuboi, T.; Brzostowski, J.; Mu, J.; Blackman, M.J.; Miller, L.H. Deletion of Plasmodium falciparum protein RON3 affects the functional translocation of exported proteins and glucose uptake. MBio 2019, 10, e01419–e01460. [Google Scholar] [CrossRef] [Green Version]
- Bargieri, D.Y.; Thiberge, S.; Tay, C.L.; Carey, A.F.; Rantz, A.; Hischen, F.; Lorthiois, A.; Straschil, U.; Singh, P.; Singh, S. Plasmodium merozoite TRAP family protein is essential for vacuole membrane disruption and gamete egress from erythrocytes. Cell Host Microbe 2016, 20, 618–630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, H.; Wang, Y.; McCarthy, D.; Wen, H.; Borchelt, D.R.; Price, D.L.; Wong, P.C. BACE1 is the major β-secretase for generation of Aβ peptides by neurons. Nat. Neurosci. 2001, 4, 233–234. [Google Scholar] [CrossRef]
- Sleebs, B.E.; Lopaticki, S.; Marapana, D.S.; O’Neill, M.T.; Rajasekaran, P.; Gazdik, M.; Günther, S.; Whitehead, L.W.; Lowes, K.N.; Barfod, L. Inhibition of Plasmepsin V activity demonstrates its essential role in protein export, PfEMP1 display, and survival of malaria parasites. PLoS Biol. 2014, 12, e1001897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Curt-Varesano, A.; Braun, L.; Ranquet, C.; Hakimi, M.A.; Bougdour, A. The aspartyl protease TgASP5 mediates the export of the Toxoplasma GRA16 and GRA24 effectors into host cells. Cell. Microbiol. 2016, 18, 151–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammoudi, P.-M.; Jacot, D.; Mueller, C.; Di Cristina, M.; Dogga, S.K.; Marq, J.-B.; Romano, J.; Tosetti, N.; Dubrot, J.; Emre, Y.; et al. Fundamental roles of the Golgi-associated Toxoplasma aspartyl protease, ASP5, at the host-parasite interface. PLoS Pathog. 2015, 11, e1005211. [Google Scholar] [CrossRef] [Green Version]
- Asada, M.; Goto, Y.; Yahata, K.; Yokoyama, N.; Kawai, S.; Inoue, N.; Kaneko, O.; Kawazu, S.-I. Gliding motility of Babesia bovis merozoites visualized by time-lapse video microscopy. PLoS ONE 2012, 7, e35227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Repnik, U.; Gangopadhyay, P.; Bietz, S.; Przyborski, J.M.; Griffiths, G.; Lingelbach, K. The apicomplexan parasite Babesia divergens internalizes band 3, glycophorin A and spectrin during invasion of human red blood cells. Cell. Microbiol. 2015, 17, 1052–1068. [Google Scholar] [CrossRef] [PubMed]
- Matz, J.M.; Beck, J.R.; Blackman, M.J. The parasitophorous vacuole of the blood-stage malaria parasite. Nat. Rev. Microbiol. 2020, 18, 379–391. [Google Scholar] [CrossRef] [PubMed]
- Egea, P.F. Crossing the vacuolar rubicon: Structural insights into effector protein trafficking in apicomplexan parasites. Microorganisms 2020, 8, 865. [Google Scholar] [CrossRef]
- Ho, C.-M.; Beck, J.R.; Lai, M.; Cui, Y.; Goldberg, D.E.; Egea, P.F.; Zhou, Z.H. Malaria parasite translocon structure and mechanism of effector export. Nature 2018, 561, 70–75. [Google Scholar] [CrossRef]
- Cygan, A.M.; Theisen, T.C.; Mendoza, A.G.; Marino, N.D.; Panas, M.W.; Boothroyd, J.C. Coimmunoprecipitation with MYR1 identifies three additional proteins within the Toxoplasma gondii parasitophorous vacuole required for translocation of dense granule effectors into host cells. Msphere 2020, 5, e00819–e00858. [Google Scholar] [CrossRef] [Green Version]
- Hakimi, H.; Templeton, T.J.; Sakaguchi, M.; Yamagishi, J.; Miyazaki, S.; Yahata, K.; Uchihashi, T.; Kawazu, S.-I.; Kaneko, O.; Asada, M. Novel Babesia bovis exported proteins that modify properties of infected red blood cells. PLoS Pathog. 2020, 16, e1008917. [Google Scholar] [CrossRef]
- Pellé, K.G.; Jiang, R.H.; Mantel, P.Y.; Xiao, Y.P.; Hjelmqvist, D.; Gallego-Lopez, G.M.; OTLau, A.; Kang, B.H.; Allred, D.R.; Marti, M. Shared elements of host-targeting pathways among apicomplexan parasites of differing lifestyles. Cell. Microbiol. 2015, 17, 1618–1639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suarez, C.E.; Alzan, H.F.; Silva, M.G.; Rathinasamy, V.; Poole, W.A.; Cooke, B.M. Unravelling the cellular and molecular pathogenesis of bovine babesiosis: Is the sky the limit? Int. J. Parasitol. 2019, 49, 183–197. [Google Scholar] [CrossRef] [PubMed]
- Jackson, A.P.; Otto, T.D.; Darby, A.; Ramaprasad, A.; Xia, D.; Echaide, I.E.; Farber, M.; Gahlot, S.; Gamble, J.; Gupta, D. The evolutionary dynamics of variant antigen genes in Babesia reveal a history of genomic innovation underlying host–parasite interaction. Nucleic Acids Res. 2014, 42, 7113–7131. [Google Scholar] [CrossRef] [Green Version]
- Cornillot, E.; Hadj-Kaddour, K.; Dassouli, A.; Noel, B.; Ranwez, V.; Vacherie, B.; Augagneur, Y.; Brès, V.; Duclos, A.; Randazzo, S. Sequencing of the smallest Apicomplexan genome from the human pathogen Babesia microti. Nucleic Acids Res. 2012, 40, 9102–9114. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.; Hu, J.; Sun, Y.; Yu, L.; He, J.; He, P.; Nie, Z.; Li, M.; Zhan, X.; Zhao, Y. A novel Babesia orientalis 135-kilodalton spherical body protein like: Identification of its secretion into cytoplasm of infected erythrocytes. Parasites Vectors 2018, 11, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Mastan, B.S.; Kumari, A.; Gupta, D.; Mishra, S.; Kumar, K.A. Gene disruption reveals a dispensable role for plasmepsin VII in the Plasmodium berghei life cycle. Mol. Biochem. Parasitol. 2014, 195, 10–13. [Google Scholar] [CrossRef] [PubMed]
- Polonais, V.; Shea, M.; Soldati-Favre, D. Toxoplasma gondii aspartic protease 1 is not essential in tachyzoites. Exp. Parasitol. 2011, 128, 454–459. [Google Scholar] [CrossRef] [Green Version]
- Francia, M.E.; Striepen, B. Cell division in apicomplexan parasites. Nat. Rev. Microbiol. 2014, 12, 125–136. [Google Scholar] [CrossRef] [PubMed]
- Jalovecka, M.; Bonsergent, C.; Hajdusek, O.; Kopacek, P.; Malandrin, L. Stimulation and quantification of Babesia divergens gametocytogenesis. Parasites Vectors 2016, 9, 439. [Google Scholar] [CrossRef] [Green Version]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Šnebergerová, P.; Bartošová-Sojková, P.; Jalovecká, M.; Sojka, D. Plasmepsin-like Aspartyl Proteases in Babesia. Pathogens 2021, 10, 1241. https://doi.org/10.3390/pathogens10101241
Šnebergerová P, Bartošová-Sojková P, Jalovecká M, Sojka D. Plasmepsin-like Aspartyl Proteases in Babesia. Pathogens. 2021; 10(10):1241. https://doi.org/10.3390/pathogens10101241
Chicago/Turabian StyleŠnebergerová, Pavla, Pavla Bartošová-Sojková, Marie Jalovecká, and Daniel Sojka. 2021. "Plasmepsin-like Aspartyl Proteases in Babesia" Pathogens 10, no. 10: 1241. https://doi.org/10.3390/pathogens10101241
APA StyleŠnebergerová, P., Bartošová-Sojková, P., Jalovecká, M., & Sojka, D. (2021). Plasmepsin-like Aspartyl Proteases in Babesia. Pathogens, 10(10), 1241. https://doi.org/10.3390/pathogens10101241





