1. Introduction
The
Anopheles (
Cellia)
subpictus sensu lato (
s.l.) Grassi 1899 species complex is the most abundant anopheline mosquito in the Indian subcontinent [
1,
2]. It acts as a vector for malaria and Japanese encephalitis in many parts of Asia [
2].
An. subpictus is the major secondary vector of malaria in Sri Lanka [
1,
3].
In India,
An. subpictus was first deemed to be a species complex based on differences in larval morphology [
4]. Further, the occurrence of two distinct types of eggs and cytological evidence has temporarily designated two forms of
An. subpictus sibling species as A and B in India [
5]. This taxon has further been categorized as a complex of four sibling species—designated as A, B, C, and D—based on stage-specific morphometric characters [
6]. The presence of four sibling species (A–D) has been confirmed through fixed inversions in the X-arm of polytene chromosomes viz. A = X+
a,+
b; species B = Xa,b; species C = Xa,+
b; species D = X + a, b [
6]. In Sri Lanka, the existence of sibling species A and B was first reported by Abhayawardana et al. [
7], based on the single inversion (X+
a/X
a) on the X chromosome. No other cytotaxonomic studies have been carried out yet to identify all the members of the
An. subpictus complex in Sri Lanka. However, all four sibling species (A–D) have been reported to occur in Sri Lanka [
8,
9] based on identification using the morphometric characteristics described by Suguna et al. [
6]. The method of Suguna et al. [
6] was confirmed by Singh et al. [
10], except for the egg morphology, and it was further concluded that a single identification character in a randomly picked individual from a population of any life stage could be used for identification of the
An. subpictus sibling species status.
Although
An. subpictus was described and designated as A–D, intraspecific variations within the taxon were first reported in India, among urban, pre-urban, and rural populations [
11]. Kirti and Kaur [
12] observed morphological differences in wings and palpi of
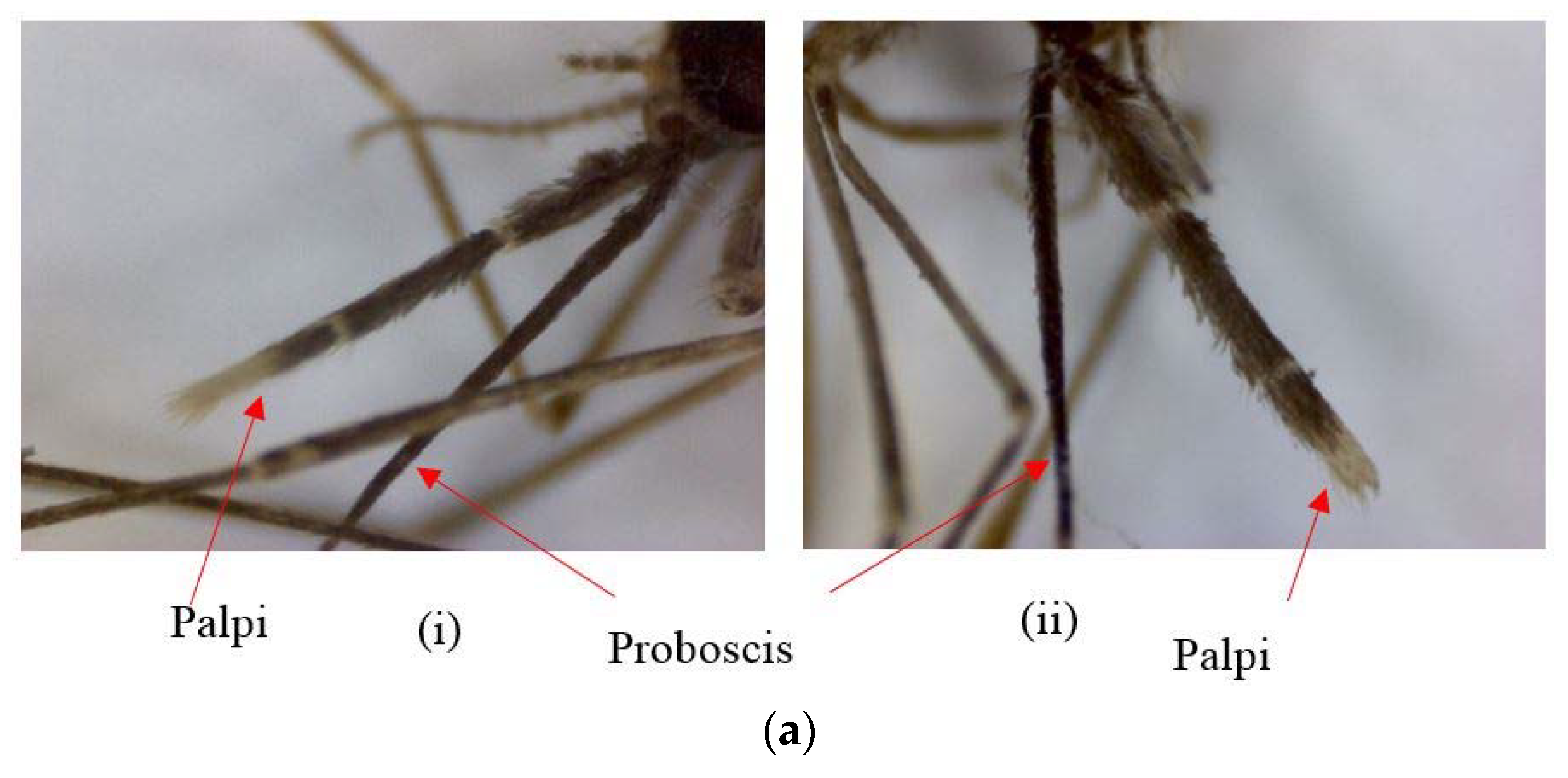
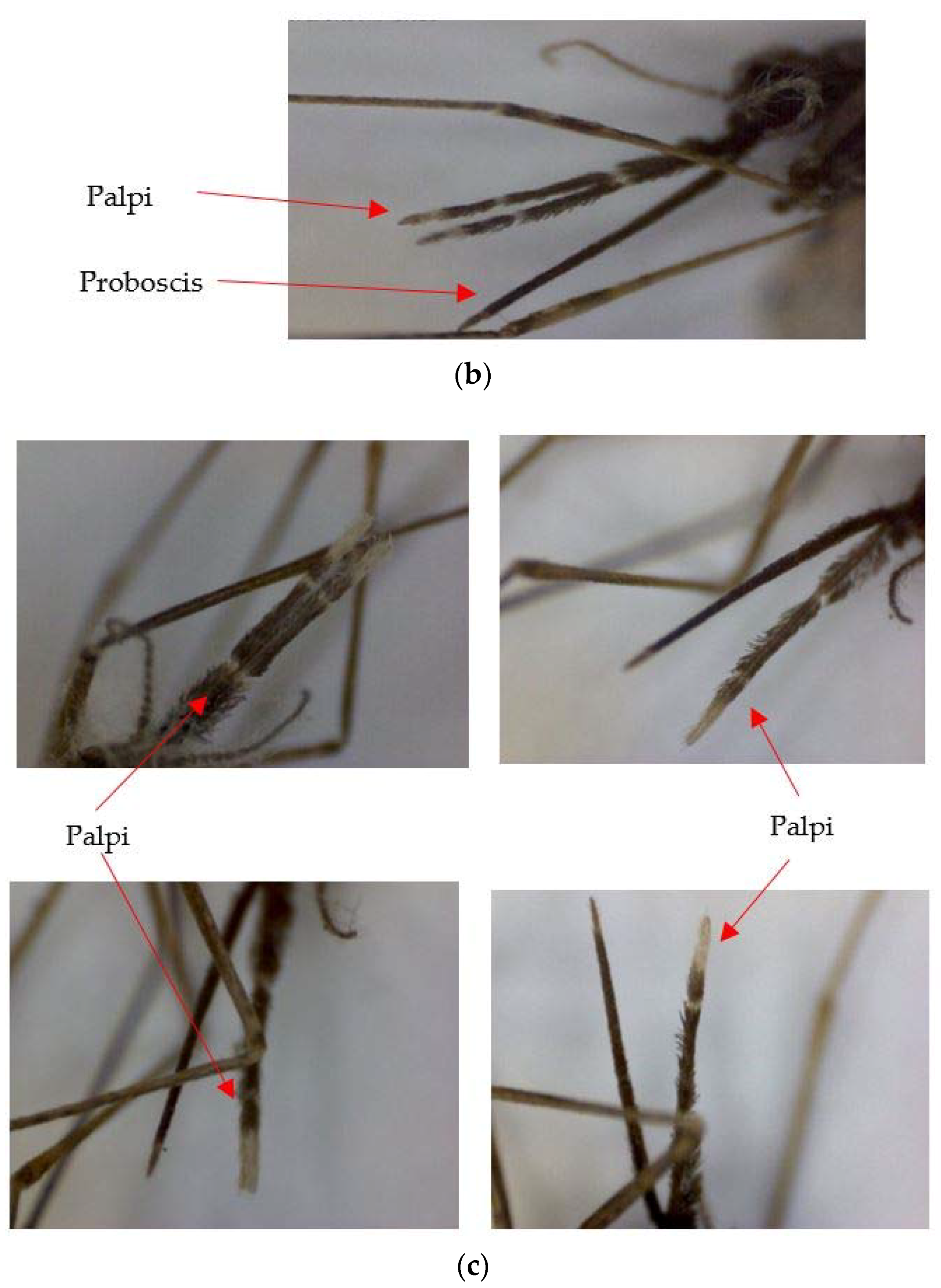
An. subpictus in India. Further, a different set of morphological variations (not reported in Suguna et al. [
6]), mainly in proboscis and palpi of
An. subpictus, was reported in Sri Lanka [
13,
14,
15]. Furthermore, the
An. subpictus complex has been designated as only A and B in Sri Lanka based on ITS2 and cytochrome
c oxidase I (
COI) sequence polymorphism [
16], showing the complexity of sibling species identification of the
An. subpictus population in Sri Lanka.
Therefore, the current study was carried out to investigate the suitability of morphological character variations and genomic variations for the identification of the An. subpictus population in Sri Lanka.
4. Discussion
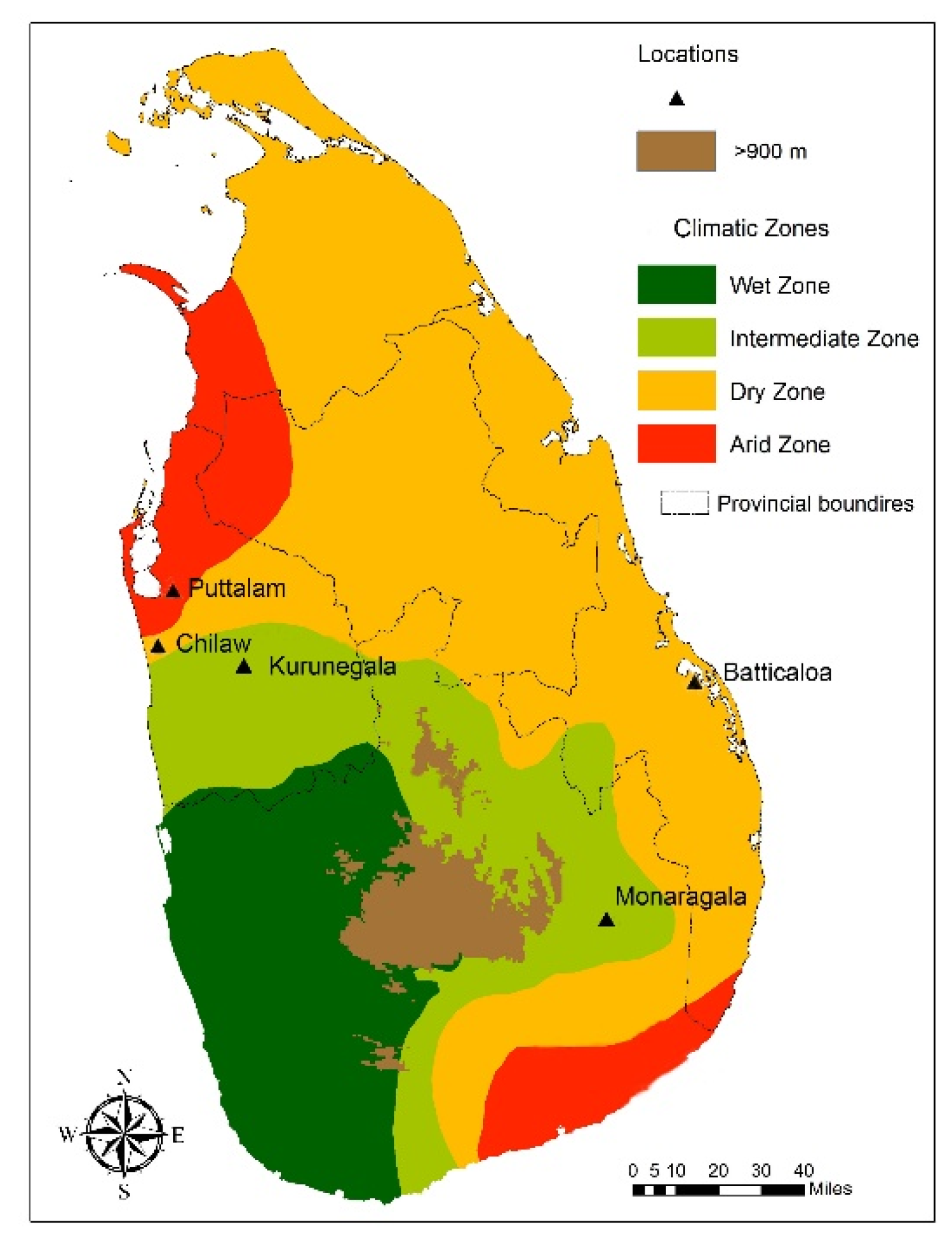
The results suggest that the
An. subpictus sibling species [
6] are distributed in five study locations, which are dry (Puttalam, Batticaloa) and intermediate (Chilaw, Kurunegala, Monaragala) climatic zones in Sri Lanka. The initial screening of wild-caught adult mosquitoes from the five selected locations for their morphological characteristics to determine sibling species status shows that the distribution of morphological variations is independent of the study locations. Morphological variations previously designated in the
An. subpictus sibling species [
13,
14,
15] and additional variations observed in the current study were commonly found from all locations. Hence, further analysis was carried out by treating the samples collected from all locations as a single pooled
An. subpictus population from Sri Lanka.
In the morphological examination of individuals, the egg morphology was excluded due to the time restriction until hatching. The eggs were hatching on the microscopic slide, prior to completing observation of the whole egg clutch. Further, it has been shown that the eggs do not serve as a good taxonomic feature due to phenotypic plasticity [
24]. Therefore, only larvae, pupal exuviae, and adult morphology were considered for further analysis.
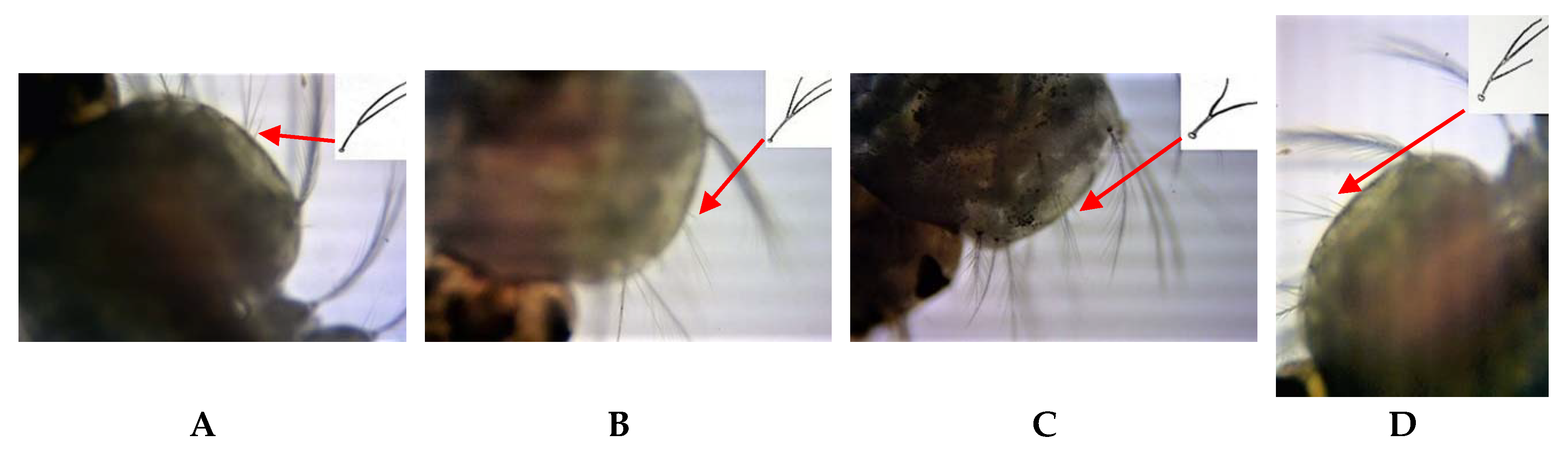
The current study shows that morphological variations of life cycle stages previously used for sibling species characterization are unreliable and polymorphic for the population found in Sri Lanka. The percentage of variation reported from the morphological identification keys of Suguna et al. [
6] was 15.87–26.10% for all life cycle stages. The extended pre-apical dark bands and the additional dark spots at the tips of palpi were first-time reports of
An. subpictus variations, which have not been reported in India or in Sri Lanka. Furthermore, deviations of the features on either the left or right side of the larvae and pupal exuviae were observed in the present study. However, no such one-sided variations were seen in the palpus of adults; instead, all variations were indicated in the pair of palpi. Parental sibling species characteristics were not consistent for the respective F
1 adults. Further, F
1 adults from a single egg clutch gave rise to a mixture of sibling species, as well as individuals with features deviating from A–D sibling species. Therefore, these findings justify the unreliability of the
An. subpictus sibling species discrimination characters [
6] for the identification of the
An. subpictus population found in Sri Lanka. Moreover, this study does not agree with the findings of Singh et al. (2010) [
10], who stated the possibility of discriminating a species by a single identification character in randomly picked individuals in a population of any life stage.
In the genomic region analysis, using nuclear and mitochondrial DNA regions, two types of sequences were reported for all analyzed DNA regions. A remarkable length variation (90 bp) was observed in the ITS2 region after annotation of the sequence using the ITS2 database. The occurrence of two types of sequences was independent from the morphological variations at any life cycle stage. This finding further confirms the unsuitability of existing morphological characterization methods [
6] to discriminate the
An. subpictus species complex found in Sri Lanka. Further, the haplotype analysis showed that the shorter and longer sequence forms of the ITS2 region are two distinct sequences which were similarly separated in all studied genomic regions.
Although India is closely related and shares a biodiversity in Western Ghats that is similar to Sri Lanka, there may have been independent and distinct evolutionary forces after the main land separation during the marine introgression in the Pleistocene period. Therefore, despite the close proximity of Sri Lanka and India, the evolutionary roots and relationships of Sri Lanka to other surrounding land masses—Madagascar, Africa, Australia, and the islands of Andaman—have to be considered. In the cases of the major malaria vector of Sri Lanka, the
An. culicifacies species complex [
25], and the major vector of leishmaniasis, the
Ph. argentipus species complex [
26,
27], the initial reports on the similarity of their composition to Indian species were later shown to have variations and deviations. Therefore, it is essential to consider the taxonomy and evolutionary studies for such species found in Sri Lanka without considering India as a basis. Unbiased analysis of species in Sri Lanka may reveal more biologically and evolutionarily important information on species. Further, the evolutionary patterns exerted on the two countries may have variations based on the different geographic and environmental influences. Since the geography and topology of the two land masses are different, it can be concluded that the two countries have different independent evolutionary processes for these mosquito species.
Finally, although the variations in vectorial capacity have been reported for the sibling species of the major vector species An. culicifacies, such variations could not be observed among the sibling species of the secondary or minor vector species of malaria. Therefore, sibling species status and the relationship of morphological variations to sibling status of An. subpictus should be reassessed to delineate the composition and the diversity of the An. subpictus species complex found in Sri Lanka.