Gene Expression Profile Analysis is Directly Affected by the Selected Reference Gene: The Case of Leaf-Cutting Atta Sexdens
Abstract
1. Introduction
2. Materials and Methods
2.1. Biological Samples
2.2. Total RNA Extraction and cDNA Synthesis
2.3. Selection of Candidate Reference Genes and Primer Design
2.4. RT-qPCR Amplification
2.5. Expression Stability Analysis of Candidate Reference Genes
2.6. Determination of the Minimum Number of Reference Genes
2.7. Validation of Reference Genes by SNF7 Expression Analysis
3. Results
3.1. Primer Specificity and Efficiency
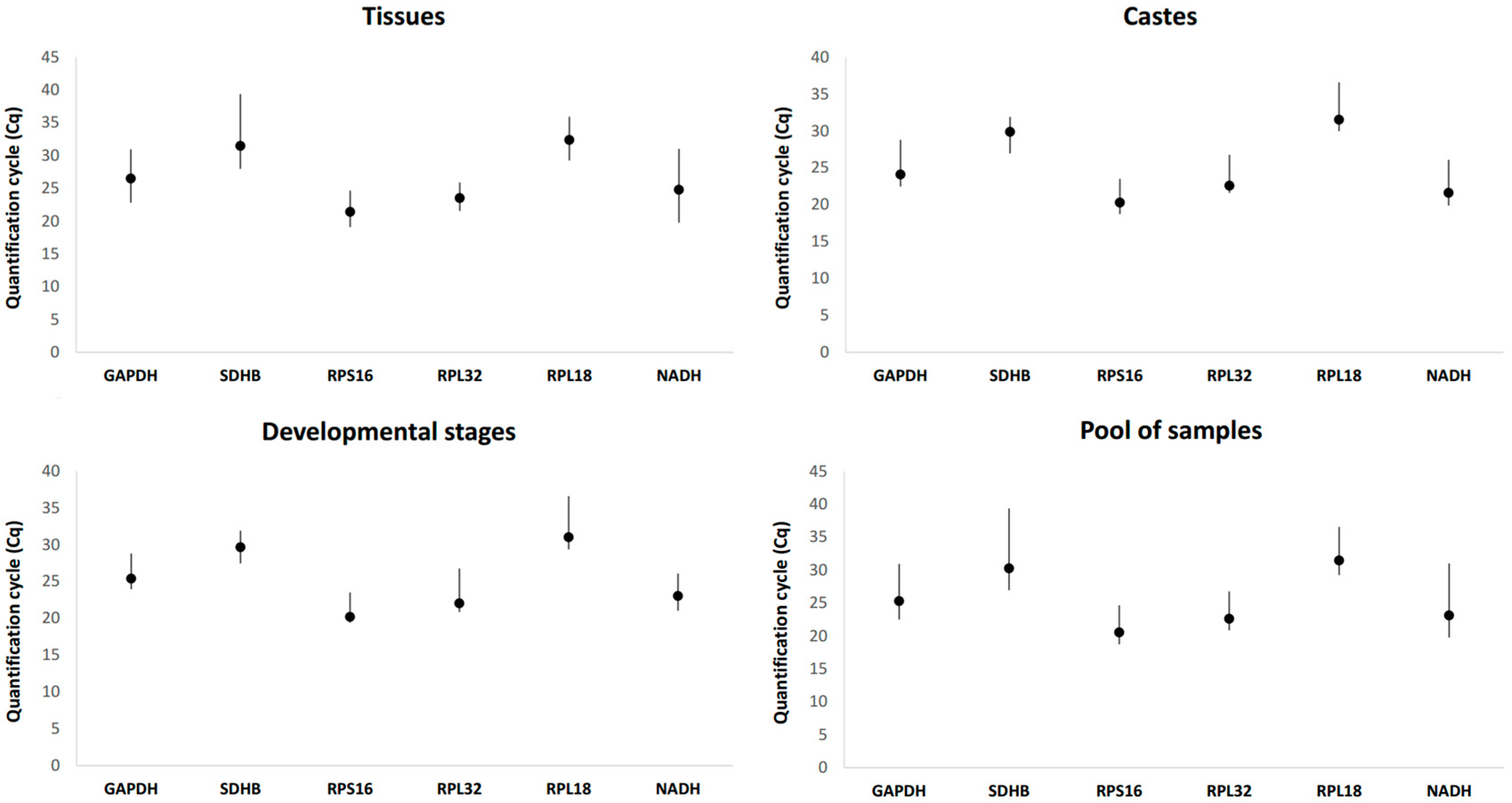
3.2. Expression Profile of Candidate Reference Genes
3.3. Expression Stabilities of Candidate Reference Genes
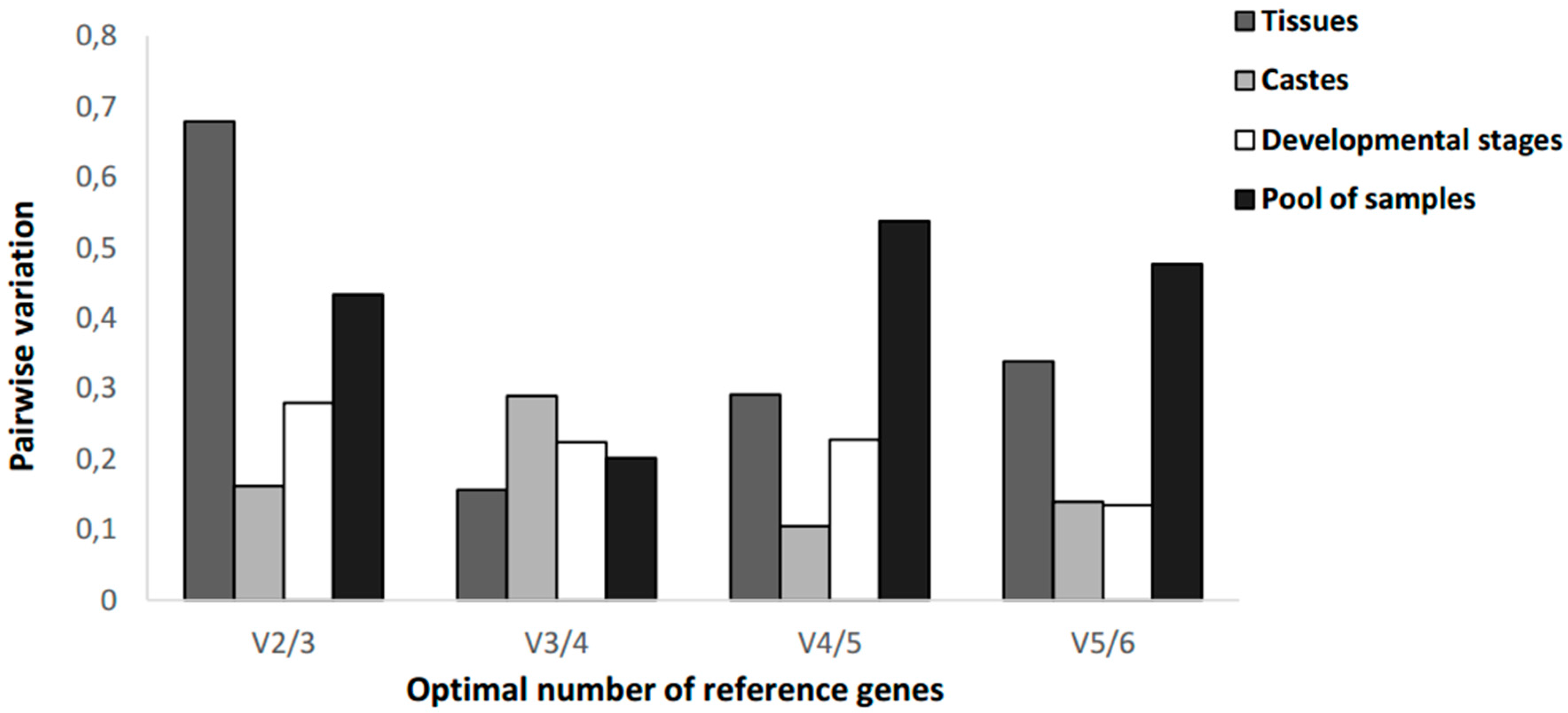
3.4. Determination of the Minimum Number of Reference Genes
3.5. Validation of the Selected Reference Genes
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Zanetti, R.; Zanuncio, J.C.; Vilela, E.F.; Leite, H.G.; Jaffé, K.; Oliveira, A.C. Level of economic damage for leaf-cutting ants (Hymenoptera: Formicidae) in eucalyptus plantations in Brazil. Sociobiology 2003, 42, 433–442. [Google Scholar]
- Matrangolo, C.A.R.; Castro, R.V.O.; Della Lucia, T.M.C.; Della Lucia, R.M.; Mendes, A.F.N.; Costa, J.M.F.N.; Leite, H.G. Crescimento de eucalipto sob efeito de desfolhamento artificial. Pesqui. Agropecu. Bras. 2010, 45, 952–957. [Google Scholar] [CrossRef]
- Asokan, R.; Sharath Chandra, G.S.; Manamohan, M.; Kumar, N.K.K.; Sita, T. Response of various target genes to diet-delivered dsRNA mediated RNA interference in the cotton bollworm, Helicoverpa armigera. J. Pest Sci. 2014, 87, 163–172. [Google Scholar] [CrossRef]
- Kolliopoulou, A.; Swevers, L. Recent progress in RNAi research in Lepidoptera: Intracellular machinery, antiviral immune response and prospects for insect pest control. Curr. Opin. Insect Sci. 2014, 6, 28–34. [Google Scholar] [CrossRef]
- Cheng, D.; Lu, Y.; Zeng, L.; Liang, G.; He, X. Si-CSP9 regulates the integument and moulting process of larvae in the red imported fire ant, Solenopsis invicta. Sci. Rep. 2015, 5, 9245. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 1–11. [Google Scholar] [CrossRef]
- Zhu, X.; Yuan, M.; Shakeel, M.; Zhang, Y.; Wang, S.; Wang, X.; Zhan, S.; Kang, T.; Li, J. Selection and Evaluation of Reference Genes for Expression Analysis Using RT-qPCR in the Beet Armyworm Spodoptera exigua (Hübner) (Lepidoptera: Noctuidae). PLoS ONE 2014, 9, e84730. [Google Scholar] [CrossRef]
- Thellin, O.; Zorzi, W.; Lakaye, B.; De Borman, B.; Coumans, B.; Hennen, G.; Grisar, T.; Igout, A.; Heinen, E. Housekeeping genes as internal standards: Use and limits. J. Biotechnol. 1999, 75, 291–295. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- RefFinder. Evaluating Reference Genes Expression. 2016. Available online: http://150.216.56.64/referencegene.php (accessed on 16 November 2017).
- Vandesompele, J.; Kubista, M.; Pfaffl, M. Reference gene validation software for improved normalization. In Real-Time Pcr: Current Technology and Applications; Logan, J., Edwards, K., Saunders, N., Eds.; Caister Academic: Norfolk, UK, 2009; pp. 47–63. ISBN 978-1-904455-39-4. [Google Scholar]
- Mamo, S.; Gal, A.B.; Bodo, S.; Dinnyes, A. Quantitative evaluation and selection of reference genes in mouse oocytes and embryos cultured in vivo and in vitro. BMC Dev. Biol. 2007, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, T.B.; Khajuria, C.; Wang, H.; Matz, N.; Cardoso, D.C.; Valicente, F.H.; Zhou, X.; Siegfried, B. Validation of reference housekeeping genes for gene expression studies in western corn rootworm (Diabrotica virgifera virgifera). PLoS ONE 2014, 9, e109825. [Google Scholar] [CrossRef] [PubMed]
- Spinsanti, G.; Panti, C.; Lazzeri, E.; Marsili, L.; Casini, S.; Frati, F.; Fossi, C.M. Selection of reference genes for quantitative RT-PCR studies in striped dolphin (Stenella coeruleoalba) skin biopsies. BMC Mol. Biol. 2006, 7, 32. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Freitas, N.C.; Barreto, H.G.; Fernandes-Brum, C.N.; Moreira, R.O.; Chalfun-Junior, A.; Paiva, L.V. Validation of reference genes for qPCR analysis of Coffea arabica L. somatic embryogenesis-related tissues. Plant Cell Tissue Organ Cult. 2017, 128, 663–678. [Google Scholar] [CrossRef]
- Cheng, D.; Zhang, Z.; He, X.; Liang, G. Validation of Reference Genes in Solenopsis invicta in Different Developmental Stages, Castes and Tissues. PLoS ONE 2013, 8, e57718. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Xue, Y.; Zhou, H.; Li, S.; Zhang, Z.; Hou, R.; Ding, Y.; Hu, K. Evaluation of reference gene suitability for quantitative expression analysis by quantitative polymerase chain reaction in the mandibular condyle of sheep. Mol. Med. Rep. 2015, 12, 5633–5640. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.A.; Beigneux, A.; Ahmad, S.T.; Young, S.G.; Gao, F.B. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr. Biol. 2007, 17, 1561–1567. [Google Scholar] [CrossRef] [PubMed]
- Chandra, G.S.; Asokan, R.; Manamohan, M.; Kumar, N.K.K.; Sita, T. Evaluation of reference genes for quantitative real-time PCR normalization in cotton bollworm Helicoverpa armigera. Mol. Biol. 2014, 48, 813–822. [Google Scholar] [CrossRef]
- Yang, C.; Pan, H.; Liu, Y.; Zhou, X. Selection of Reference Genes for Expression Analysis Using Quantitative. Real-Time PCR in the Pea Aphid, Acyrthosiphon pisum (Harris) (Hemiptera Aphidiae). PLoS ONE 2014, 9, e110454. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; An, S.; Li, Z.; Wu, F.; Yang, Q.; Liu, Y.; Cao, J.; Zhang, H.; Zhang, Q.; Liu, X. Identification and validation of reference genes for normalization of gene expression analysis using qRT-PCR in Helicoverpa armigera (Lepidoptera: Noctuidae). Gene 2015, 555, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Kosera, B.; Rapacz, M. Reference genes in real-time PCR. J. Appl. Gene. 2013, 54, 391–406. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.Y.; Seo, P.; Yang, M.S.; Xiang, F.; Park, C.M. Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. BMC Plant Biol. 2008, 8, 112. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Ma, J.; Wang, J.; Wu, X.; Li, P.; Yao, Y. Validation of suitable reference genes for gene expression analysis in the halophyte Salicornia europaea by real-time quantitative PCR. Front. Plant Sci. 2015, 5, 788. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Lu, M.X.; Tang, X.T.; Du, Y.Z. Exploring valid reference genes for quantitative real-time PCR analysis in Sesamia inferens (Lepidoptera: Noctuidae). PLoS ONE 2015, 10, e0115979. [Google Scholar] [CrossRef] [PubMed]
- Maerken, T.V.; Mestdagh, P.; De Clercq, S.; Pattyn, F.; Yigit, N.; De Paepe, A.; Marine, J.-C.; Speleman, F.; Vandesompele, J. Real-Time qPCR as a Tool for Evaluating RNAi-Mediated Gene Silencing. BioTech. Protoc. Guide 2009, 47. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.; et al. The need for transparency and good practices in the qPCR literature. Nat. Methods 2013, 10, 1063–1067. [Google Scholar] [CrossRef] [PubMed]
- Kupper, M.; Stigloher, C.; Feldhaar, H.; Gross, R. Distribution of the obligate endosymbiont Blochmannia floridanus and expression analysis of putative immune genes in ovaries of the carpenter ant Camponotus floridanus. Arthropod Struct. Dev. 2016, 45, 475–487. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Dang, L.; Xi, G.; Wang, F. Molecular Cloning, Characterization and Expression Analysis of Calreticulin Gene in the Ant Polyrhachis vicina Roger (Hymenoptera: Formicidae). Sociobiology 2013, 60, 355–361. [Google Scholar] [CrossRef]
- Lü, S.M.; Zhao, Z.; Li, K.; Zhang, Y.L.; Xi, G.S. Cloning and expression analysis of a muscarinic cholinergic receptor from the brain of ant, Polyrhachis vicina. Arch. Insect Biochem. Physiol. 2011, 78, 46–60. [Google Scholar] [CrossRef] [PubMed]
- Elsik, C.G.; Tayal, A.; Diesh, C.M.; Unni, D.R.; Emery, M.L.; Nguyen, H.N.; Hagen, D.E. Hymenoptera Genome Database: Integrating genome annotations in HymenopteraMine. Nucleic Acids Res. 2016, 44, D793–D800. [Google Scholar] [CrossRef] [PubMed]
- Nygaard, S.; Zhang, G.; Schiøtt, M.; Li, C.; Wurm, Y.; Hu, H.; Zhou, J.; Ji, L.; Qiu, F.; Rasmussen, M.; et al. The genome of the leaf-cutting ant Acromyrmex echinatior suggests key adaptations to advanced social life and fungus farming. Genome Res. 2011, 21, 1339–1348. [Google Scholar] [CrossRef] [PubMed]
- Suen, G.; Teiling, C.; Li, L.; Holt, C.; Abouheif, E. The Genome Sequence of the Leaf-Cutter Ant Atta cephalotes Reveals Insights into Its Obligate Symbiotic Lifestyle. PLoS Genet. 2011, 7, e1002007. [Google Scholar] [CrossRef] [PubMed]
- Simola, D.F.; Graham, R.J.; Brady, C.M.; Enzmann, B.L.; Desplan, C.; Ray, A.; Zwiebel, L.J.; Bonasio, R.; Reinberg, D.; Liebig, J.; et al. Epigenetic (re)programming of caste-specific behavior in the ant Camponotus floridanus. Science 2016, 351, aac6633. [Google Scholar] [CrossRef] [PubMed]
- Gotoh, A.; Shigenobu, S.; Yamaguchi, K.; Kobayashi, S.; Ito, F.; Tsuji, K. Transcriptome profiling of the spermatheca identifies genes potentially involved in the long-term sperm storage of ant queens. Sci. Rep. 2017, 7, 5972. [Google Scholar] [CrossRef] [PubMed]
- Ratzka, C.; Gross, R.; Feldhaar, H. Gene expression analysis of the endosymbiont-bearing midgut tissue during ontogeny of the carpenter ant Camponotus floridanus. J. Insect Physiol. 2013, 59, 611–623. [Google Scholar] [CrossRef] [PubMed]
- Moreira, A.C.; Santos, A.M.; Carneiro, R.L.; Bueno, O.C.; Souza, D.H.F. Validation of reference genes in leaf-cutting ant Atta sexdens rubropilosa in different developmental stages and tissues. Int. J. Environ. Agric. Biotechnol. 2017, 2, 743–755. [Google Scholar] [CrossRef]
- Ratzka, C.; Gross, R.; Feldhaar, H. Systemic gene knockdown in Camponotus floridanus workers by feeding of dsRNA. Insectes Sociaux 2013, 60, 475–484. [Google Scholar] [CrossRef]
- Gao, F.B.; Brenman, J.E.; Jan, L.Y.; Jan, Y.N. Genes regulating dendritic outgrowth, branching, and routing in Drosophila. Genes Dev. 1999, 13, 2549–2561. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.W.; Sung, H.; Shin, D.; Shen, H.; Ahnn, J.; Lee, S.K.; Lee, S. Differential physiological roles of ESCRT complexes in Caenorhabditis elegans. Mol. Cells 2011, 31, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Winter, V.; Hauser, M.T. Exploring the ESCRTing machinery in eukaryotes. Trends Plant Sci. 2006, 11, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Urquhart, W.; Mueller, G.M.; Carleton, S.; Song, Z.; Perez, T.; Uffman, J.P.; Jensen, P.D.; Levine, S.L.; Ward, J. A novel method of demonstrating the molecular and functional equivalence between in vitro and plant-produced double-stranded RNA. Regul. Toxicol. Pharmacol. 2015, 73, 607–612. [Google Scholar] [CrossRef] [PubMed]
- Vaccari, T.; Rusten, T.E.; Menut, L.; Nezis, I.P.; Brech, A.; Stenmark, H.; Bilder, D. Comparative analysis of ESCRT-I, ESCRT-II and ESCRT-III function in Drosophila by efficient isolation of ESCRT mutants. J. Cell Sci. 2009, 122, 2413–2423. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, N.T.; Brenman, J.E.; Jan, Y.N.; Gao, F.B. The coiled-coil protein shrub controls neuronal morphogenesis in Drosophila. Curr. Biol. 2006, 16, 1006–1011. [Google Scholar] [CrossRef] [PubMed]
- Bolognesi, R.; Ramaseshadri, P.; Anderson, J.; Bachman, P.; Clinton, W.; Flannagan, R.; Ilagan, O.; Lawrence, C.; Levine, S.; Moar, W. Characterizing the Mechanism of Action of Double-Stranded RNA Activity against Western Corn Rootworm (Diabrotica virgifera virgifera LeConte). PLoS ONE 2012, 7, e47534. [Google Scholar] [CrossRef] [PubMed]
- Ramaseshadri, P.; Segers, G.; Flannagan, R.; Wiggins, E.; Clinton, W.; Ilagan, O.; McNulty, B.; Clark, T.; Bolognesi, R. Physiological and Cellular Responses Caused by RNAi-Mediated Suppression of Snf7 Orthologue in Western Corn Rootworm (Diabrotica virgifera virgifera) Larvae. PLoS ONE 2013, 8, e54270. [Google Scholar] [CrossRef] [PubMed]
- Christiaens, O.; Swevers, L.; Smagghe, G. DsRNA degradation in the pea aphid (Acyrthosiphon pisum) associated with lack of response in RNAi feeding and injection assay. Peptides 2014, 53, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Prentice, K.; Christiaens, O.; Pertry, I.; Bailey, A.; Niblett, C.; Ghislain, M.; Gheysen, G.; Smagghe, G. RNAi-based gene silencing through dsRNA injection or ingestion against the African sweetpotato weevil Cylas puncticollis (Coleoptera: Brentidae). Pest Manag. Sci. 2016, 73, 44–52. [Google Scholar] [CrossRef] [PubMed]

| Gene Abbreviation | Accession Number | Primer Sequence (Forward/Reverse 5′–3′) | Concentration (uM) | Tm (°C) | Amplicon (pb) | E (%) | R2 |
|---|---|---|---|---|---|---|---|
| RPL18 | EH413666 | CTCTGTCGTTTCCGCTGTCTCACCTTCCGATCATGCTTATG | 0.4 | 60.59 60.48 | 108 | 102.07 | 0.973 |
| RPL32 | JQ744274.1 | TTCTGCCTTTCTGTTTTTCGTTTGGGTCGATAAACTGGTC | 1.0 | 58.15 57.52 | 91 | 99.627 | 0.997 |
| RPS16 | EH413178.1 | GAAACAAAAAGAGCCGATCCTCCACGTCCACGTTTACAAT | 0.4 | 58.77 58.90 | 88 | 99.223 | 0.988 |
| GAPDH | EH413647 | CGTGGTATGACAACGAGTACGGGAGTTAGGAGGACGCAGATGAA | 0.4 | 62.62 60.76 | 120 | 99.239 | 0.986 |
| NADH | NM_001162323 | GGAAAAATCGCACTAGGAGGATGTGTAGTTGCTGCTTCCATAA | 0.4 | 60.57 58.52 | 137 | 93.529 | 0.99 |
| SDHB | NM_001162436 | GCTAATGTGAGCCAAAAGCCGATGCTGCGTTGTGTCATCT | 0.4 | 59.85 59.87 | 139 | 99.346 | 0.984 |
| SNF-7 a | XM_012199288.1 | GAGCCAACTGCTCCTTCAACTTCGACGCATTTTTCTTCG | 0.4 | 60.31 59.12 | 134 | 91.605 | 0.996 |
| Ranking | geNorm | NormFinder | BestKeeper | Delta-Ct | RefFinder | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Stability M-Value | Gene | Stability SV-Value | Gene | Stability SD-Value | Gene | Stability ΔCt-Value | Gene | Overall Stability Value | Gene | |
| 1 | 0.132 | GAPDH/NADH | 0.056 | RPS16 | 0.168 | SDHB | 0.204 | RPS16 | 1.565 | RPS16 |
| 2 | - | - | 0.176 | NADH | 0.189 | RPS16 | 0.246 | NADH | 2.060 | NADH |
| 3 | 0.192 | RPS16 | 0.183 | SDHB | 0.212 | NADH | 0.249 | SDHB | 2.213 | SDHB |
| 4 | 0.207 | SDHB | 0.188 | RPL18 | 0.218 | RPL18 | 0.253 | RPL18 | 3.344 | GAPDH |
| 5 | 0.234 | RPL18 | 0.207 | GAPDH | 0.241 | GAPDH | 0.260 | GAPDH | 4.229 | RPL18 |
| 6 | 0.248 | RPL32 | 0.228 | RPL32 | 0.247 | RPL32 | 0.275 | RPL32 | 6.000 | RPL32 |
| Ranking | geNorm | NormFinder | BestKeeper | Delta-Ct | RefFinder | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Stability M-Value | Gene | Stability SV-Value | Gene | Stability SD-Value | Gene | Stability ΔCt-Value | Gene | Overall Stability Value | Gene | |
| 1 | 0.079 | RPL18/NADH | 0.073 | RPS16 | 0.204 | RPL18 | 0.243 | RPS16 | 1.414 | RPL18 |
| 2 | - | - | 0.081 | RPL18 | 0.208 | NADH | 0.244 | RPL18 | 1.968 | RPS16 |
| 3 | 0.121 | GAPDH | 0.111 | GAPDH | 0.226 | RPL32 | 0.251 | GAPDH | 2.060 | NADH |
| 4 | 0.144 | RPS16 | 0.113 | NADH | 0.248 | GAPDH | 0.270 | NADH | 4.000 | GAPDH |
| 5 | 0.176 | RPL32 | 0.217 | RPL32 | 0.269 | RPS16 | 0.309 | RPL32 | 5.045 | RPL32 |
| 6 | 0.322 | SDHB | 0.599 | SDHB | 0.300 | SDHB | 0.613 | SDHB | 5.233 | SDHB |
| Ranking | geNorm | NormFinder | BestKeeper | Delta-Ct | RefFinder | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Stability M-Value | Gene | Stability SV-Value | Gene | Stability SD-Value | Gene | Stability ΔCt-Value | Gene | Overall Stability Value | Gene | |
| 1 | 0.122 | GAPDH/NADH | 0.106 | RPS16 | 0.182 | RPS16 | 0.208 | RPS16 | 1.316 | RPS16 |
| 2 | - | - | 0.135 | NADH | 0.187 | SDHB | 0.218 | NADH | 1.861 | NADH |
| 3 | 0.189 | RPS16 | 0.154 | GAPDH | 0.231 | NADH | 0.227 | GAPDH | 2.590 | GAPDH |
| 4 | 0.201 | SDHB | 0.181 | RPL32 | 0.237 | RPL32 | 0.245 | RPL32 | 3.761 | SDHB |
| 5 | 0.222 | RPL32 | 0.209 | SDHB | 0.241 | GAPDH | 0.257 | SDHB | 4.229 | RPL32 |
| 6 | 0.237 | RPL18 | 0.218 | RPL18 | 0.251 | RPL18 | 0.267 | RPL18 | 6.000 | RPL18 |
| Ranking | geNorm | NormFinder | BestKeeper | Delta-Ct | RefFinder | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Stability M-Value | Gene | Stability SV-Value | Gene | Stability SD-Value | Gene | Stability ΔCt-Value | Gene | Overall Stability Value | Gene | |
| 1 | 0.163 | RPL32/RPL18 | 0.114 | RPS16 | 0.174 | RPS16 | 0.261 | RPS16 | 1.316 | RPS16 |
| 2 | - | - | 0.208 | RPL32 | 0.208 | GAPDH | 0.293 | RPL32 | 2.115 | RPL32 |
| 3 | 0.196 | RPS16 | 0.213 | RPL18 | 0.211 | SDHB | 0.296 | RPL18 | 2.449 | RPL18 |
| 4 | 0.253 | SDHB | 0.230 | GAPDH | 0.229 | RPL18 | 0.316 | GAPDH | 3.557 | GAPDH |
| 5 | 0.285 | GAPDH | 0.247 | SDHB | 0.243 | RPL32 | 0.329 | SDHB | 4.162 | SDHB |
| 6 | 0.308 | NADH | 0.293 | NADH | 0.274 | NADH | 0.353 | NADH | 6.000 | NADH |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Livramento, K.G.d.; Freitas, N.C.; Máximo, W.P.F.; Zanetti, R.; Paiva, L.V. Gene Expression Profile Analysis is Directly Affected by the Selected Reference Gene: The Case of Leaf-Cutting Atta Sexdens. Insects 2018, 9, 18. https://doi.org/10.3390/insects9010018
Livramento KGd, Freitas NC, Máximo WPF, Zanetti R, Paiva LV. Gene Expression Profile Analysis is Directly Affected by the Selected Reference Gene: The Case of Leaf-Cutting Atta Sexdens. Insects. 2018; 9(1):18. https://doi.org/10.3390/insects9010018
Chicago/Turabian StyleLivramento, Kalynka G. do, Natália C. Freitas, Wesley P. F. Máximo, Ronald Zanetti, and Luciano V. Paiva. 2018. "Gene Expression Profile Analysis is Directly Affected by the Selected Reference Gene: The Case of Leaf-Cutting Atta Sexdens" Insects 9, no. 1: 18. https://doi.org/10.3390/insects9010018
APA StyleLivramento, K. G. d., Freitas, N. C., Máximo, W. P. F., Zanetti, R., & Paiva, L. V. (2018). Gene Expression Profile Analysis is Directly Affected by the Selected Reference Gene: The Case of Leaf-Cutting Atta Sexdens. Insects, 9(1), 18. https://doi.org/10.3390/insects9010018