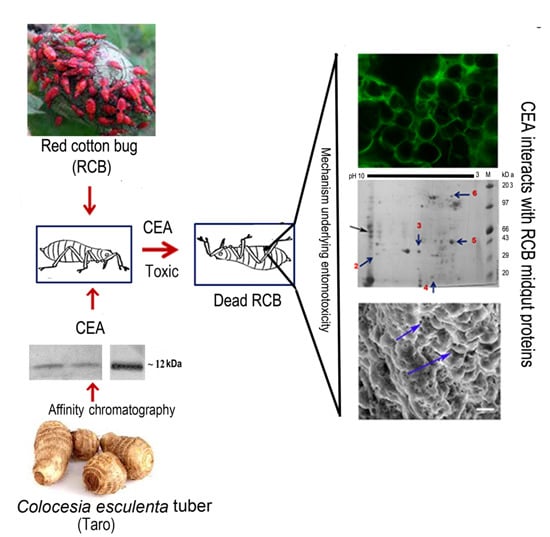
Molecular Mechanism Underlying the Entomotoxic Effect of Colocasia esculenta Tuber Agglutinin against Dysdercus cingulatus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Insects Used
2.2. Extraction and Isolation of the Lectin from Colocasia sp
2.3. SDS-PAGE and Western Analysis
2.4. Characterization of CEA through Agglutination Assays
2.5. Insect Bioassay on Artificial Diet
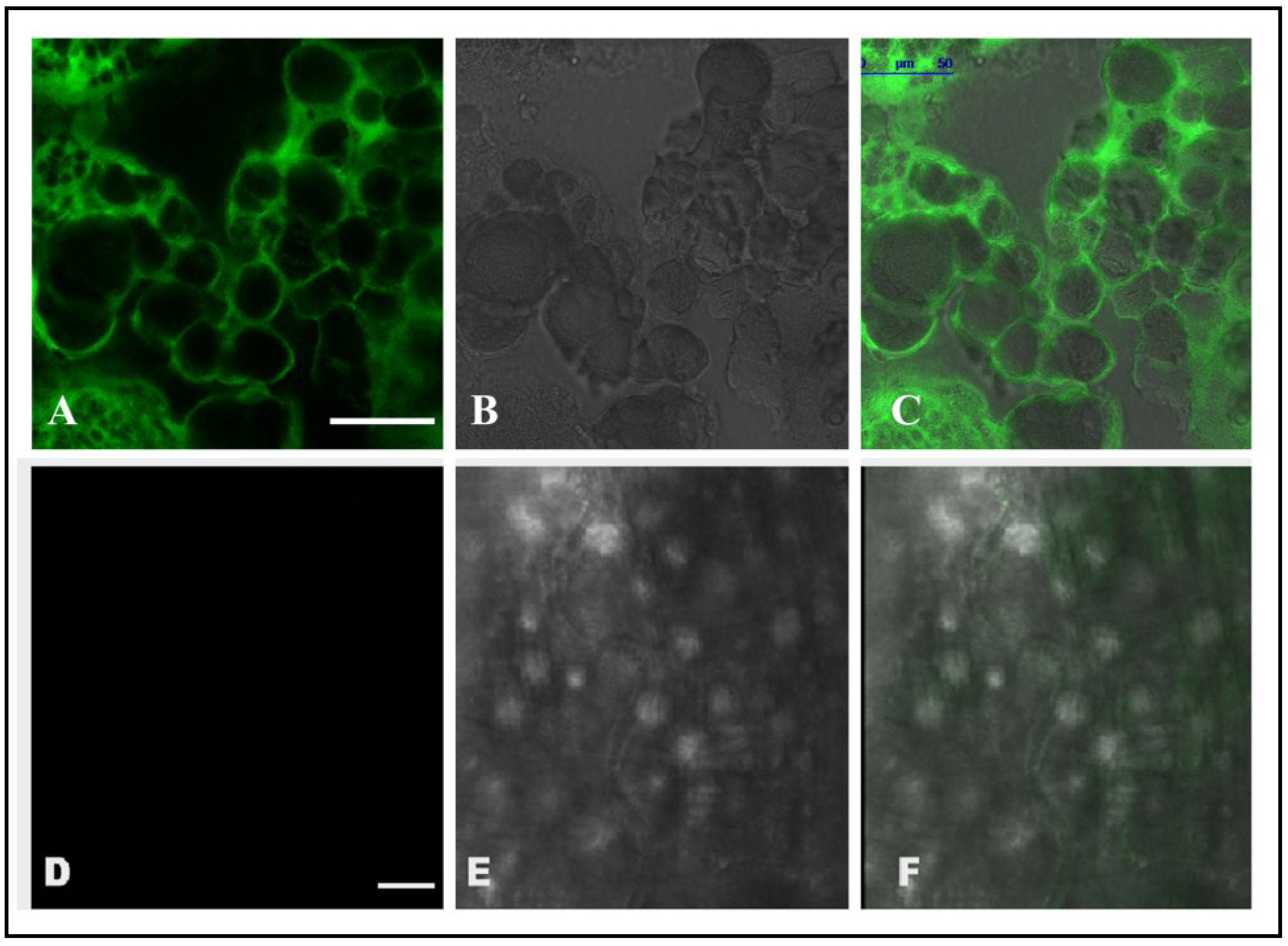
2.6. Localization of CEA in RCB Midgut through Confocal Microscopy
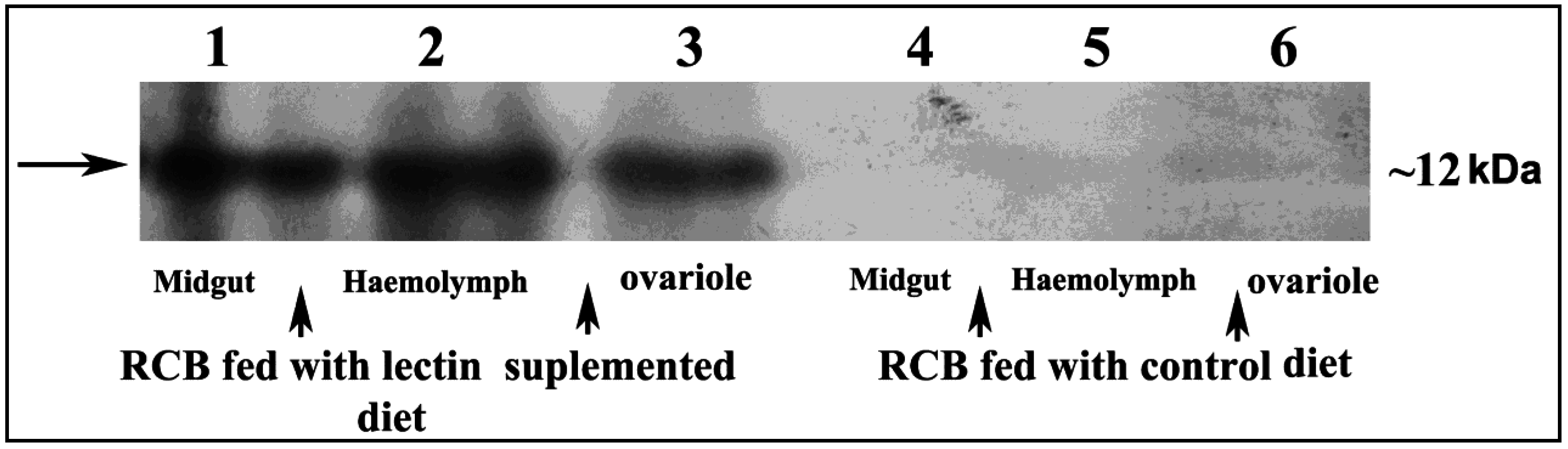
2.7. Detection of CEA in Midgut, Haemolymph and Ovary Extract of RCB
2.8. Scanning Electron Microscopic (SEM) Analysis with the Midgut of RCB
2.9. Ligand Blot Assay
2.10. Characterization of CEA Binding Proteins
2.10.1. Carbohydrate Specific Staining
2.10.2. Deglycosylation and Mannose Inhibition Assay
2.11. Two-Dimensional Gel Analyses of Total Midgut Protein Followed by Ligand Blot Assay
2.12. MALDI TOF/TOF Analyses
2.13. Immunoprecipitation of Binding Partners and Subsequent Ligand Blot Analyses
3. Results
3.1. Purification, Characterization, and Monitoring of Insecticidal Efficacy of CEA on RCB
3.2. Localization and Internalization of CEA in RCB Midgut
3.3. Scanning Electron Microscopy Reveals Alteration in Midgut Morphology

3.4. Ligand Blot Analysis Highlighted the Midgut Binding Partners
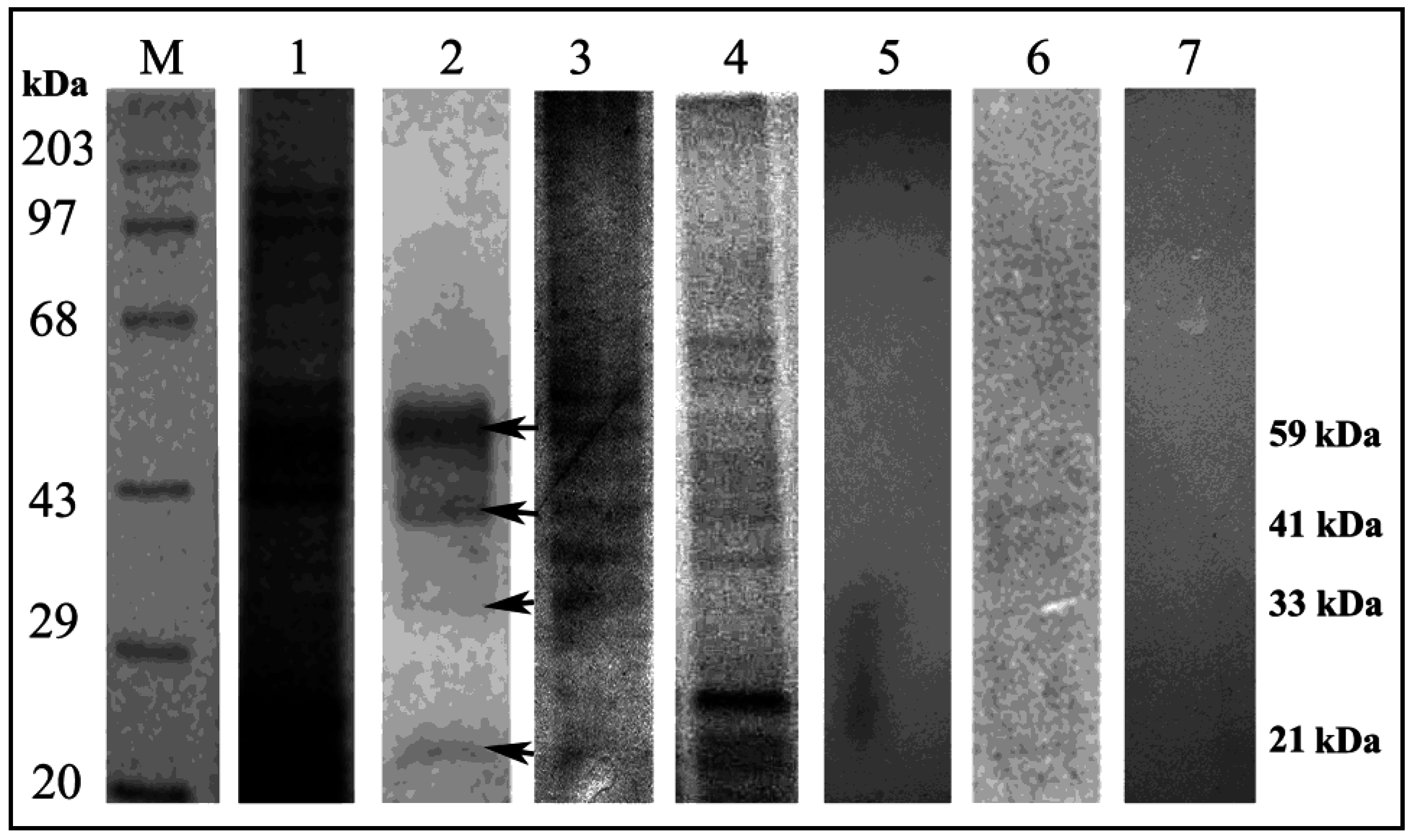
3.5. Characterization of Binding Partners
3.6. Mass Spectrometric Identification of Binding Partners from RCB

| Spot No | Method of Identification | Accession (NCBI nr/MSDB Database) | Protein Name/Organism | Predicted kDa/pI | Observed kDa/pI | MOWSE Score: PMF/MS-MS | No of Peptide Match/Coverage (%) |
|---|---|---|---|---|---|---|---|
| 1 * | PMF and MS/MS | Q5XUA1_9HEMI | ATP synthase α subunit/Brown citrus aphid | 59/9.14 | 58/8.9 | 169/232 | 14/25% |
| 2 | PMF | Q1HPZ3_BOMMO | Inorganic phosphate carrier/Silk moth | 35/9.3 | 33/8.7 | 87 | 6/20% |
| 3 * | PMF and MS/MS | Q16E72_AEDAE | Cytochrome P450/Yellow fever mosquito | 48/6.4 | 44/6.3 | 118/182 | 10/23% |
| 4 | PMF | Q17LH0_AEDAE | RNA binding protein/Yellow fever mosquito | 15/5.7 | 19/5.3 | 120 | 8/33% |
| 5 * | PMF and MS/MS | Q16VS2_AEDAE | Actin/Yellow fever mosquito | 41/5.3 | 43/5.1 | 99/312 | 9/25% |
| 6 | PMF | GI:170040984 | 26s proteasome non ATPase regulatory subunit 1/Culex mosquito | 112/5.5 | 110/5.0 | 106 | 10/14% |
3.7. Authentication of Mass Spectrometric Identification of the Binding Partners
4. Discussion
4.1. CEA Binding to RCB Gut Membrane and its Internalization: the Crucial Factor for Toxicity
4.2. CEA Alters Midgut Perimicrovillar Membrane Morphology
4.3. Sugar Mediated Interaction with Midgut Proteins
4.4. Mechanism of CEA Entomotoxicity
4.4.1. Actin Mediated Toxicity
4.4.2. ATP Synthase Mediated Toxicity
4.4.3. Cytochrome P450 Mediated Toxicity
5. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| ASAL | Allium sativum leaf agglutinin; |
| BBMV | Brush border membrane vesicle; |
| Bt | Bacillus thuringiensis; |
| CEA | Colocasia esculenta tuber agglutinin; |
| GNA | Galanthus nivalis agglutinin; |
| IPM | Insect pest management; |
| MALDI | Matrix-assisted laser desorption ionization; |
| MMBL | Monocot mannose binding lectin; |
| PMF | Peptide mass fingerprinting; |
| PMM | Perimicrovillar membrane; |
| RCB | Red cotton bug; |
| TOF | Time of flight |
References
- Staniscuaski, F.; Ferreira-DaSilva, C.T.; Mulinari, F.; Pires-Alves, M.; Carlini, C.R. Insecticidal effects of canatoxin on the cotton stainer bug (Hemiptera, Pyrrhocoridae). Toxicon 2005, 45, 753–760. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Banerjee, S.; Majumder, P.; Das, S. Evaluation of the efficiency of different Mannose-binding plant lectins in controlling homopteran insect red cotton bug. J. Agric. Food Chem. 2002, 50, 6775–6779. [Google Scholar] [CrossRef] [PubMed]
- Barton, K.A.; Whiteley, H.R.; Yang, N.S. Bacillus thuringiensis delta-endotoxin expressed in transgenic Nicotiana tabacum provides resistance to lepidopteran insects. Plant Physiol. 1987, 85, 1103–1109. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, S.K.; Chandrashekar, K. Interaction of salivary and midgut proteins of Helicoverpa armigera with soybean trypsin inhibitor. Protein J. 2012, 31, 259–264. [Google Scholar] [CrossRef] [PubMed]
- Chougule, N.P.; Bonning, B.C. Toxins for transgenic resistance to hemipteran pests. Toxins 2012, 4, 405–429. [Google Scholar] [CrossRef] [PubMed]
- Vandenborre, G.; Smagghe, G.; van Damme, E.J.M. Plant lectins as defense proteins against phytophagous insects. Phytochemistry 2011, 72, 1538–1550. [Google Scholar] [CrossRef] [PubMed]
- Powell, K.S.; Gatehouse, A.M.R.; Hilder, V.A.; Gatehouse, A.J. Antifeedant effects of plant lectins and an enzyme on the adult stage of the rice brown planthopper, Nilaparvata lugens. Entomol. Exp. Appl. 1995, 75, 51–59. [Google Scholar] [CrossRef]
- Dutta, I.; Saha, P.; Majumder, P.; Sarkar, A.; Chakraborti, D.; Banerjee, S.; Das, S. The efficacy of a novel insecticidal protein, Allium sativum leaf lectin (ASAL) against homopteran insect monitored in transgenic tobacco. Plant Biotechnol. J. 2005, 3, 601–611. [Google Scholar] [CrossRef] [PubMed]
- Bala, A.; Roy, A.; Behura, N.; Hess, D.; Das, S. Insight to the mode of action of Allium sativum leaf agglutinin (ASAL) expressing in T3 rice lines on brown planthopper. Am. J. Plant Sci. 2013, 4, 400–407. [Google Scholar] [CrossRef]
- Lu, Y.; Wu, K.; Jiang, Y.; Guo, Y.; Desneux, N. Wide spread adoption of Bt cotton and insecticide decrease promotes biocontrol services. Nature 2012, 487, 362–365. [Google Scholar] [CrossRef] [PubMed]
- Oscarsson, K.V.; Savage, G.P. Composition and availability of soluble and insoluble oxalates in raw and cooked taro (Colocasia esculenta var. Schott) leaves. Food Chem. 2007, 101, 559–562. [Google Scholar] [CrossRef]
- Van Damme, E.J.M.; Allen, A.K.; Peumans, W.J. Isolation and characterization of a lectin with exclusive specificity towards mannose from snowdrop (Galanthus nivalis) bulbs. FEBS Lett. 1987, 215, 140–144. [Google Scholar] [CrossRef]
- Pereira, P.R.; Winter, H.C.; Verícimo, M.A.; Meagher, J.L.; Stuckey, J.A.; Goldstein, I.J.; Pascho-alin, V.M.; Silva, J.T. Structural analysis and binding properties of isoforms of tarin, the GNA-related lectin from Colocasia esculenta. Biochim. Biophys. Acta 2015, 185, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Thakur, K.; Kaur, M.; Kaur, S.; Kaur, A.; Kamboj, S.S.; Singh, J. Purification of Colocasia esculenta lectin and determination of its anti-insect potential towards Bactrocera cucurbitae. J. Environ. Biol. 2013, 34, 31–36. [Google Scholar] [PubMed]
- Arannilewa, S.T.; Odeyemi, O.O. Insecticidal evaluation of some plant materials as grain protect ants against the maize weevil, Sitophilus zeamais (Mots) (Coleoptera: Curculionidae). Agric. J. 2007, 2, 155–159. [Google Scholar]
- Das, A.; Roy, A.; Hess, D.; Das, S. Characterization of a highly potent insecticidal lectin from Colocasia esculenta tuber and cloning of its coding sequence. Am. J. Plant Sci. 2013, 4, 408–416. [Google Scholar] [CrossRef]
- Van Damme, E.J.M.; Goossens, K.; Smeets, K.; Leuven, F.V.; Verheart, P.; Peumans, W.J. The major tuber storage protein of Araceae species is a lectin. Plant Physiol. 1995, 107, 1147–1158. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Gupta, S.; Hess, D.; Das, K.P.; Das, S. Binding of insecticidal lectin Colocasia esculenta tuber agglutinin (CEA) to midgut receptors of Bemisia tabaci and Lipaphis erysimi provides cues to its insecticidal potential. Proteomics 2014, 14, 1646–1659. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–688. [Google Scholar] [CrossRef] [PubMed]
- Dadd, R.; Mittler, T. Permanent culture of an aphid on a totally synthetic diet. Experientia 1966, 22, 832–833. [Google Scholar] [CrossRef] [PubMed]
- Chi, H. Computer Program for Probit Analysis; National Chung Hsing University: Taichung, Taiwan, 1997. [Google Scholar]
- Cilia, M.; Fish, T.; Yang, X.; Mclaughlin, M.; Thannhauser, T.W.; Gray, S. A comparison of protein extraction methods suitable for gel-based proteomic studies of aphid proteins. J. Biomol. Tech. 2009, 20, 201–215. [Google Scholar] [PubMed]
- Feder, D.; Salles, J.M.; Garcia, E.S.; Azambuja, P. Haemolymph and fat body metallo-protease associated with Enterobacter cloacae infection in the bloodsucking insect, Rhodnius prolixus. Mem. Inst. Oswaldo. Cruz 1998, 93, 823–826. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Damasceno-Sa, J.C.; Carneiro, C.N.B.; Damatta, R.A.; Samuels, R.I.; Terra, W.R.; Silva, C.P. Biphasic perimicrovillar membrane production following feeding by previously starved Dysdercus peruvianus (hemiptera, pyrrhocoridae). J. Insect Physiol. 2007, 53, 592–600. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, S.; Roy, A.; Das, S. Binding of garlic (Allium sativum) leaf lectin to the gut receptors of homopteran pests is correlated to its insecticidal activity. Plant Sci. 2001, 161, 1025–1033. [Google Scholar] [CrossRef]
- Bayyareddy, K.; Andacht, T.M.; Abdullah, M.A.; Adang, M.J. Proteomic identification of Bacillus thuringiensis subsp. israelensis toxin Cry4Ba binding proteins in midgut membranes from Aedes (Stegomyia) aegypti Linnaeus (Diptera.; Culicidae) larvae. Insect Biochem. Mol. Biol. 2009, 39, 279–286. [Google Scholar] [PubMed]
- Moller, H.J.; Poulsen, J.H. Staining of Glycoproteins/Proteoglycans in SDS-Gels, in Protein Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 1996; pp. 627–632. [Google Scholar]
- Banerjee, S.; Hess, D.; Majumder, P.; Roy, D.; Das, S. The interaction of Allium sativum leaf agglutinin with a chaperonin group of unique receptor protein isolated from a bacterial endosymbiont of the mustard aphid. J. Biol. Chem. 2004, 279, 23782–23789. [Google Scholar] [CrossRef] [PubMed]
- Mondal, H.A.; Roy, A.; Gupta, S.; Das, S. Exploring the insecticidal potentiality of Amorphophallus paeonifolius tuber agglutinin in hemipteran pest management. Am. J. Plant Sci. 2012, 3, 780–790. [Google Scholar] [CrossRef]
- Shevchenko, A.; Tomas, H.; Havlis, J.; Olsen, J.V.; Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2007, 1, 2856–2860. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, M.; Gupta, S.; Bhar, A.; Das, S. Optimization of an efficient protein extraction protocol compatible with two-dimensional electrophoresis and mass spectrometry from recalcitrant phenolic rich roots of chickpea (Cicer arietinum L.). Int. J. Proteom. 2012. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, S.K.; Mishra, M.; Singh, H.; Ranjan, A.; Chandrashekar, K.; Verma, P.C.; Singh, P.K.; Tuli, R. Interaction of Allium sativum leaf agglutinin with midgut brush border membrane vesicles proteins and its stability in Helicoverpa armigera. Proteomics 2010, 10, 4431–4440. [Google Scholar] [CrossRef] [PubMed]
- Cristofoletti, P.T.; de Sousa, F.A.M.; Rahbé, Y.; Terra, W.R. Characterization of a membrane-bound aminopeptidase purified from Acyrthosiphon pisum midgut cells. FEBS J. 2006, 273, 5574–5588. [Google Scholar] [CrossRef] [PubMed]
- Araujo-Filho, J.H.; Vasconcelos, I.M.; Martins-Miranda, A.S.; Gondim, D.M.F.; Oliveira, J.T.A. A ConA-like lectin from Dioclea guianensis benth has antifungal activity against Colletotrichum gloeosporioides, unlike its homologues, ConM and ConA. J. Agric. Food Chem. 2010, 58, 4090–4096. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.; Ng, T.B. Preparation and biological properties of a melibiose binding lectin from Bauhinia variegata seeds. J. Agric. Food Chem. 2008, 56, 10481–10486. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, S.K.; Saurabh, S.; Singh, R.; Rai, P.; Dubey, N.K.; Chandrashekar, K.; Negi, K.S.; Tuli, R.; Singh, P.K. Purification and characterization of a lectin with high hemagglutination property isolated from Allium altaicum. Protein J. 2011, 30, 374–383. [Google Scholar] [CrossRef] [PubMed]
- Majumder, P.; Mondal, H.A.; Das, S. Insecticidal activity of Arum maculatum tuber lectin and its binding to the glycosylated insect gut receptors. J. Agric. Food Chem. 2005, 53, 6725–6729. [Google Scholar] [CrossRef] [PubMed]
- Ohizumi, Y.; Gaidamashvili, M.; Ohwada, S.; Matsuda, K.; Kominami, J.; Nakamura-Tsuruta, S.; Hirabayashi, J.; Naganuma, T.; Ogawa, T.; Muramoto, A. Mannose-binding lectin from yam (dioscorea batatas) tubers with insecticidal properties against Helicoverpa armigera (Lepidoptera, Noctuidae). J. Agric. Food Chem. 2009, 57, 2896–2902. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, S.K.; Singh, P.K. Receptors of Garlic (Allium sativum) lectins and their role in insecticidal action. Protein J. 2012, 31, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Dangl, J.L.; McDowell, J.M. Two modes of pathogen recognition by plants. Proc. Natl. Acad. Sci. USA 2006, 103, 8575–8576. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.S. Inducible direct plant defence against insect herbivores: A review. Insect Sci. 2008, 15, 101–114. [Google Scholar] [CrossRef]
- Lagarda-Diaz, I.; Guzman-Partida, A.M.; Urbano-Hernandez, G.; Ortega-Nieblas, M.M.; Robles-BurguenO, M.R.; Winzerling, J.; Vazqez-Moreno, A. Insecticidal action of PF2 lectin from Olneya tesota (palo fierro) against Zabrotes subfasciatus larvae and midgut glycoconjugate binding. J. Agric. Food Chem. 2009, 57, 689–694. [Google Scholar] [CrossRef] [PubMed]
- Powell, K.S.; Spence, J.; Bharathi, M.; Gatehouse, J.A.; Gatehouse, A.M.R. Immunohisto-chemical and developmental studies to elucidate the mechanism of action of the snow drop lectin on the rice brown planthopper, Nilaparvata lugens (Stal). J. Insect Physiol. 1998, 44, 529–539. [Google Scholar] [CrossRef]
- Caccia, S.; van Damme, E.J.M.; de Vos, W.H.; Smagghe, G. Mechanism of entomotoxicity of the plant lectin from Hippeastrum hybrid (Amaryllis) in Spodoptera littoralis larvae. J. Insect Physiol. 2012, 58, 1177–1183. [Google Scholar] [CrossRef] [PubMed]
- Fitches, E.; Woodhouse, S.D.; Edwards, J.P.; Gatehouse, J.A. In vitro and in vivo binding of snowdrop (Galanthus nivalis agglutinin; GNA) and jackbean (Canavalia ensiformis; Con A) lectins within tomato moth (Lacanobia oleracea) larvae: Mechanisms of insecticidal action. J. Insect Physiol. 2001, 47, 777–787. [Google Scholar] [CrossRef]
- Down, R.E.; Gatehouse, A.M.R.; Hamilton, W.D.O.; Gatehouse, J.A. Snowdrop lectin inhibits development and decreases fecundity of the glasshouse potato aphid (Aulacorthum solani) when administered in vitro and via transgenic plants both in laboratory and glasshouse trials. J. Insect physiol. 1996, 42, 1035–l045. [Google Scholar] [CrossRef]
- Walski, T.; van Damme, E.J.M.; Smagghe, G. Penetration through the peritrophic matrix is a key to lectin toxicity against Tribolium castaneum. J. Insect Phsiol. 2014, 70, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Guerrieri, E.; Digilio, M.C. Aphid-plant interactions, a review. J. Plant Interact. 2008, 3, 223–232. [Google Scholar] [CrossRef]
- Murdock, L.L.; Shade, R.E. Lectins and protease inhibitors as plant defenses against insects. J. Agric. Food Chem. 2002, 50, 6605–6611. [Google Scholar] [CrossRef] [PubMed]
- McNall, R.J.; Adang, M.J. Identification of novel Bacillus thuringiensis Cry1Ac binding proteins in Manduca sexta midgut through proteomic analysis. Insect Biochem. Mol. Biol. 2003, 33, 999–1010. [Google Scholar] [CrossRef]
- Krishnamoorthy, M.; Jurat-Fuentes, J.L.; McNall, R.J.; Andacht, T.; Adang, M.J. Identification of novel Cry1Ac binding proteins in midgut membranes from Heliothis virescens using proteomic analyses. Insect Biochem. Mol. Biol. 2007, 37, 189–201. [Google Scholar] [CrossRef] [PubMed]
- Bement, W.M.; Mooseker, M.S. The cytoskeleton of the intestinal epithelium: Components, assembly, and dynamic rearrangements. In The Cytoskeleton: A Multi-Volume Treatise; Hesketh, J.E., Pryme, J.F., Eds.; JAI Press: Greenwich, UK, 1996; Volume 3, pp. 359–404. [Google Scholar]
- Hopmann, R.; Cooper, J.A.; Miller, K.G. Actin organization, bristle morphology and viability are affected by actin capping protein mutations in Drosophila. J. Cell Biol. 1996, 133, 1293–1305. [Google Scholar] [CrossRef] [PubMed]
- Younis, H.M.; Abo-El-Saad, M.M.; Abdel-Razik, R.K.; Abo-Seda, S.A. Resolving the DDT target protein in insects as a subunit of the ATP synthase. Biotechnol. Appl. Biochem. 2002, 35, 9–17. [Google Scholar] [PubMed]
- Sohail, S.K.; Rup, P.J.; Sehgal, R.; Kaur, S. Activity of eleven enzymes in nymphs of Lipaphis erysimi as affected by 2,4-D (herbicide) treatment. Bull. Insectol. 2008, 61, 239–244. [Google Scholar]
- Terra, W.R.; Ferreira, C. Insect digestive enzymes, properties, compartmentalization and function. Comp. Biochem. Phys. B 1994, 109, 1–62. [Google Scholar] [CrossRef]
- Scott, J.G. Cytochromes P450 and insecticide resistance. Insect Biochem. Mol. Biol. 1999, 29, 757–777. [Google Scholar] [CrossRef]
- Enayati, A.; Ranson, H.; Hemingway, J. Insect glutathione transferases and insecticide resistance. Insect Mol. Biol. 2005, 14, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Tao, X.Y.; Xue, X.Y.; Huang, Y.P.; Chen, X.Y.; Mao, Y.B. Gossypol-enhanced P450 gene pool contributes to cotton bollworm tolerance to a pyrethroid insecticide. Mol. Ecol. 2012, 21, 4371–4385. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
#, A.R.; Das, S. Molecular Mechanism Underlying the Entomotoxic Effect of Colocasia esculenta Tuber Agglutinin against Dysdercus cingulatus. Insects 2015, 6, 827-846. https://doi.org/10.3390/insects6040827
# AR, Das S. Molecular Mechanism Underlying the Entomotoxic Effect of Colocasia esculenta Tuber Agglutinin against Dysdercus cingulatus. Insects. 2015; 6(4):827-846. https://doi.org/10.3390/insects6040827
Chicago/Turabian Style#, Amit Roy, and Sampa Das. 2015. "Molecular Mechanism Underlying the Entomotoxic Effect of Colocasia esculenta Tuber Agglutinin against Dysdercus cingulatus" Insects 6, no. 4: 827-846. https://doi.org/10.3390/insects6040827
APA Style#, A. R., & Das, S. (2015). Molecular Mechanism Underlying the Entomotoxic Effect of Colocasia esculenta Tuber Agglutinin against Dysdercus cingulatus. Insects, 6(4), 827-846. https://doi.org/10.3390/insects6040827