Exploring Phylogeographic Congruence in a Continental Island System
Abstract
: A prediction in phylogeographic studies is that patterns of lineage diversity and timing will be similar within the same landscape under the assumption that these lineages have responded to past environmental changes in comparable ways. Eight invertebrate taxa from four different orders were included in this study of mainland New Zealand and Chatham Islands lineages to explore outcomes of island colonization. These comprised two orthopteran genera, one an endemic forest-dwelling genus of cave weta (Rhaphidophoridae, Talitropsis) and the other a grasshopper (Acrididae, Phaulacridum) that inhabits open grassland; four genera of Coleoptera including carabid beetles (Mecodema), stag beetles (Geodorcus), weevils (Hadramphus) and clickbeetles (Amychus); the widespread earwig genus Anisolabis (Dermaptera) that is common on beaches in New Zealand and the Chatham Islands, and an endemic and widespread cockroach genus Celatoblatta (Blattodea). Mitochondrial DNA data were used to reconstruct phylogeographic hypotheses to compare among these taxa. Strikingly, despite a maximum age of the Chathams of ∼4 million years there is no concordance among these taxa, in the extent of genetic divergence and partitioning between Chatham and Mainland populations. Some Chatham lineages are represented by insular endemics and others by haplotypes shared with mainland populations. These diverse patterns suggest that combinations of intrinsic (taxon ecology) and extrinsic (extinction and dispersal) factors can result in apparently very different biogeographic outcomes.1. Introduction
A null hypothesis in biogeography is that different taxon groups will show similar patterns of distribution and phylogeny if their evolution has responded to the same historic processes. Some biogeographers have used such a proposal as the basis of a putative test of the role of vicariance in biogeography, under the assumption that patterns associated with dispersal would not be coincident [1–3]. However, early in the application of phylogeographic approaches it was expected that dispersal could also be expected to yield congruent biogeographic patterns, where the taxa involved were responding to a common cause [4] and in many contexts the perceived distinction between vicariance and dispersal processes in biogeography is illusional [5]. In continental systems the past location of species' refugia has been identified using phylogeographic data and found to coincide for many taxa, as does the general trend of expansion from refugia [6,7]. On oceanic islands a general trend in the phylogeographic history of taxa colonizing successive islands as they emerged, has been revealed (e.g., [8,9]). However, discordance is also found and whilst this might be attributed to an overwhelming influence of stochastic events, it might also reflect, at least in part, differences associated with mobility, population size and reproductive strategy as these might influence establishment success [10,11]. Distinguishing between random effects and those linked to species traits is very difficult, especially because traits associated with dispersal might be selected against after colonization [10,12,13].
The study of New Zealand biogeography has in the past focused on the role of plate tectonic vicariance in the origins of New Zealand lineages, but more recently molecular data have shown that many lineages have arrived relatively recently and that diversification is young in many taxon groups [14,15]. Furthermore, it is increasingly evident that there is no uniform pattern in the phylogeographic structuring of the biota within New Zealand [16], with so far, no consistent linkage between lineage formation, landscape history or distribution of taxa being evident. Although New Zealand was influenced by Pleistocene climate cycling, this does not by itself explain the phylogeographic structure of taxa we see today. Not every lineage or population in New Zealand is younger than the last glacial maximum (LGM) [14,16], even though every species and population must have been affected by climate cycling. Although some similarity can be found in lineage ages for different taxa, the pattern of spatial structuring is not consistent among lineages [17]. This was a plausible expectation of species that have similar ecological requirements (e.g., habitat, microclimate, diet) and have experienced range retraction and expansion in a similar time frame (e.g., since the LGM). Instead, what we observe among the extant biota in New Zealand is that some endemic taxa have retained high genetic and taxonomic diversity more consistent with pre-Pleistocene events. On the other hand there are endemic taxa that are widespread with low genetic diversity and in many cases comprising a single mainland New Zealand species (e.g., [18,19]; and see [20]) or a combination of both patterns with widespread species showing low genetic diversity and localized populations/species that retained high genetic diversity within small ranges (e.g., Phaulacridium grasshopper [21]; weta, [15,22]). Understanding why different taxon groups have responded so differently requires comparison of taxa across a variety of diverse ecological and evolutionary backgrounds. Their genetic, taxonomic and spatial structuring will help development of more robust hypotheses about the relative role of intrinsic, extrinsic and stochastic processes in the present day biota of New Zealand. Maximising geophysical heterogeneity in such studies allows contrasts to be made between those intrinsic and extrinsic influences and this can be achieved in New Zealand by examining taxa distributed across the mainland and the largest offshore islands, the Chathams.
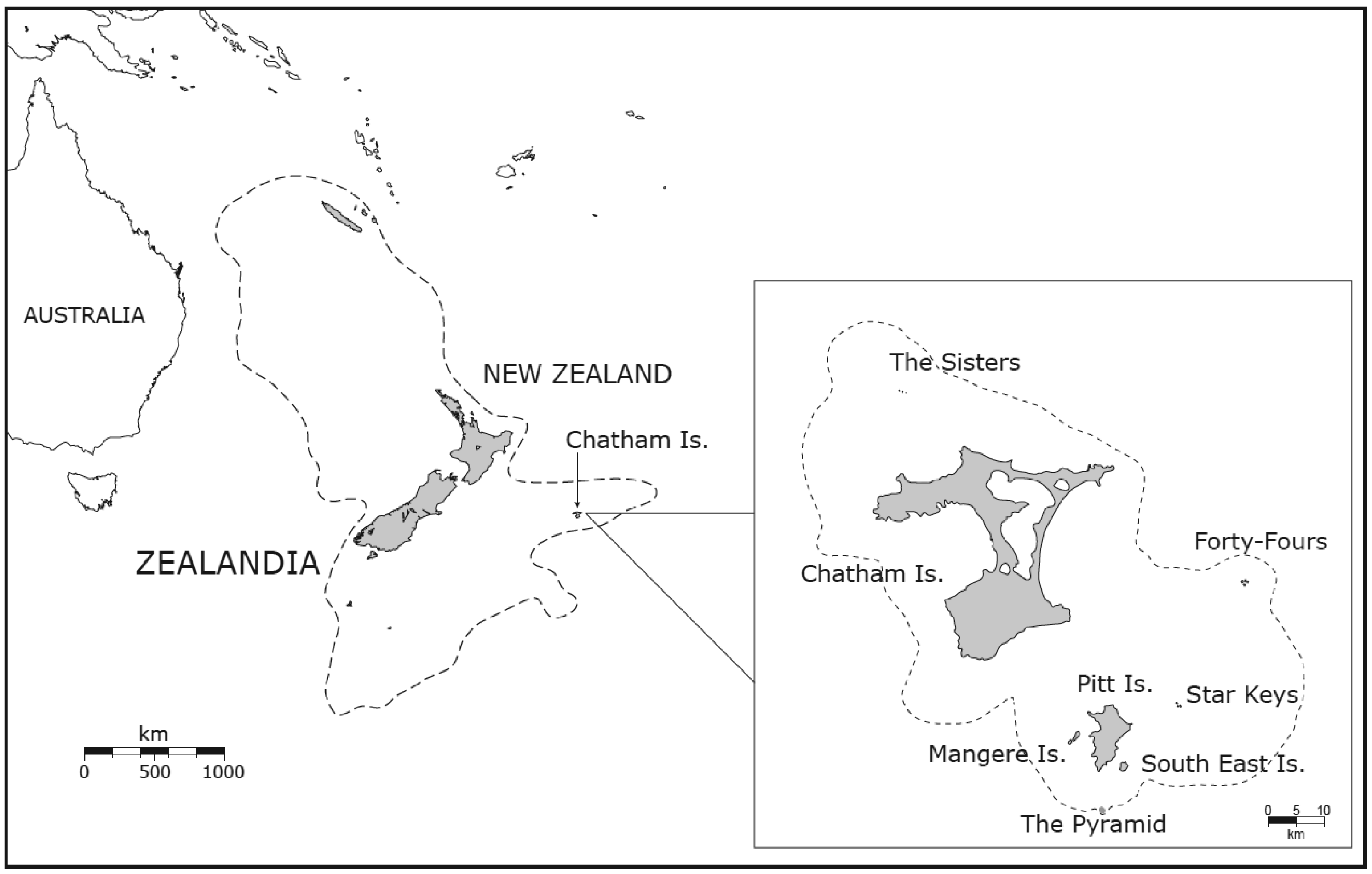
The Chatham Islands are located on the Pacific Plate (S 44°03′47.16″ and W 175°57′35.73″) approximately 850 km east of mainland New Zealand (Figure 1) at the eastern end of the Chatham Rise, a submerged ridge-structure extending from mid-South Island. The ridge, together with the Chathams, is part of the same continental crust on which New Zealand is located, and only 10% of the Chathams landmass is above sea level nowadays [23]. The present land above sea level consists of two inhabited islands (Main or Chatham and Pitt Island) and several smaller islets and rock-stacks (Figure 1). Paleomagnetic studies provide evidence that the position of the islands on the eastern tip of the Chatham Rise has been more or less fixed since the break up of Gondwanaland [24]. The geology of the Chathams is unlike most of the other small Pacific island groups because their basement consists of old metamorphic rocks (Chatham schist), similar to the old schists known from the foot of the Southern Alps of Otago and Canterbury, New Zealand. Today the Chatham schist is only exposed in the northern part of the Chathams and on the Forty Fours and its metamorphosis is dated to 160 million years (Myr). On top of this old layer are several younger deposits from the Cretaceous and Cenozoic, which contain the oldest fossils on the Chathams. These fossils indicate a sustained isolation of the Chatham area for the last 65 Myr [24]. In contrast to its settled behavior today, the Chatham area has a long history of volcanic activity, mainly between 60–40 million years ago (Ma) (Eocene to Oligocene) and 5–1.6 Ma (Pliocene). In the latter period most of the smaller islands were formed.
Throughout this history, the Chatham Islands have been submerged and emerged several times in response to regional tectonism affecting the entire eastern end of the Chatham Rise, culminating in the last emergence event no more than 4 million years ago [25,26]. Since then, phases of climate cooling in the Pleistocene (<2 Ma) have influenced the connectivity and land area of the islands [24,27] (Figure 1). Connection of islands during lowered sea level may have enabled population extension and gene flow within the Chathams and mainland New Zealand separately, but could not have influenced New Zealand–Chatham connectivity.
The Chathams therefore provide a convenient context for studying New Zealand biogeography in terms of both spatial (two land areas separated by ∼800 km of ocean) and temporal (emergence ∼4 Ma) scale [28]. Mainland New Zealand is about 270-times larger than the Chathams, and the geographic distance from the mainland to Chathams is about half the length of the main islands (∼1600 km), and 4 Myr is an appropriate time period for exploring species evolution [29]. Furthermore, this Plio/Pleistocene period was one of significant geophysical activity in New Zealand [30]. Thus, there is a nice contrast of highly disjunct versus near continuous habitat. Mitochondrial data from taxa in several insect orders present on the Chatham Islands and New Zealand were examined. These data allow assessment of the degree of congruence among genetic diversity, taxonomic status and spatial distribution of mitochondrial lineages within this ecologically diverse set of invertebrate taxa.
2. Materials and Methods
This study reports on the mtDNA phylogeography of eight New Zealand invertebrate taxa (Figure 2). As such, each component does not assess variation that might exist among genes. While these data represent just one heritable unit within each taxon group, our focus has been on large-scale differences among ecologically diverse taxa. Taxonomic diversity, distribution and abundance of each taxon differs considerably and the scale of analyses varies accordingly, but our sampling reflects approximately their abundance and the strategy was to sample each genus as intensely as possible to include possible sister lineages to the Chatham taxa. The taxa compared share the characteristics of relative large size and flightlessness (with one partial exception), but they differ in their foraging habits, abundance, and landscapes that they occupy. Some of them are subjects of ongoing research, incorporating non-New Zealand outgroup taxa and additional genetic markers, the results of which are in peer-review at the moment and will be published separately elsewhere. Here, we first report data and phylogenetic structure within each genus, we then summarize and compare phylogeographic structure across New Zealand/Chathams.
The invertebrate specimens for this study were collected mainly by hand and preserved in 95% ethanol. They are housed and catalogued in the collection of the Phoenix Lab at Massey University, Palmerston North, with unique voucher numbers. In some cases data were supplemented by specimens from other collections. DNA extraction used leg tissue and a standard salting-out method [31], with whole genomic DNA extractions stored at −20 °C.
The polymerase chain reaction (PCR) was used to target the mitochondrial cytochrome oxidase I (COI) gene region, using in most cases the published oligonucleotide primers C1-J-1718, C1-J2195 and L2-N-3014 [32]. PCR amplifications were performed in a total volume of 10 μL using Red Hot Taq (ABgene). After an initial denaturation at 94 °C for 3 min. DNA was amplified during 35 cycles of 30 s at 94 °C, 45 s at 50 °C and 30 s at 72 °C, followed by a final extension step at 72 °C for 4 min. The PCR cycle conditions varied from above as follows: 35 cycles of 1 min. at 94 °C, 1 min. at 42 °C and 1.5 min. at 72 °C, followed by a final extension step at 72 °C for 5 min. The amplified products were checked on 1% Agarose gels and purified using SAP/EXO1 (USB Corporation) enzyme digest following the manufacturer's instructions. Purified DNA fragments were used for cycle sequencing with Big Dye terminators under standard conditions and read on an ABI 377 sequencer (ABI).
2.1. Genetic Analysis
Sequences were edited using Sequencher 4.9 (Gene Codes Corporation) and aligned using SeAl [33]. Mr. Bayes 3.1.2 [34]) was used under a six parameter model selected by jMODELTEST 3.5 [35,36] to reconstruct phylogenetic relationships within each taxon group. MrBayes implemented models incorporating gamma-distributed rate variation across sites and a proportion of invariable sites as appropriate. Four independent MCMC runs for ten million generations with a burn in of 10% were employed for each analysis (results not shown). Unrooted Neigbour-Joining (NJ) networks were also generated for each ingroup dataset of each taxon using HKY distances as implemented in Geneiuos 5.4.3 (Biomatters Ltd.). Uncorrected p-distances for all taxa were calculated using PAUP*4 [37]. Where appropriate, DnaSP v5.0 [38] was used to calculate nucleotide diversity (π, [39]), haplotype diversity (h) and average number of nucleotide differences (k).
2.2. Taxa
2.2.1. Talitropsis (Orthoptera: Raphidophoridae)-Cave Weta
The family Raphidophoridae (called cave weta in New Zealand) comprises approx. 300 species worldwide [40] with ∼18 genera endemic to New Zealand. Talitropsis is a New Zealand endemic consisting of three recognized species, two of which are allopatric endemic species on the Chatham Islands, T. crassicruris (Hutton, 1897) and T. megatibia Trewick, 1999, and one is widespread throughout mainland New Zealand, T. sedilloti Bolivar, 1882. Talitropsis is found in forested areas where it hides during the day in holes and cavities and is active at night. On the Chathams, and especially on the smaller islands that lack trees, Talitropsis burrows into peaty soil under rocks.
2.2.2. Phaulacridium (Orthoptera: Acrididae)-Grasshopper
Phaulacridium Brunner v. Wattenwyl, 1893 (Orthoptera: Acrididae) comprises five closely related species, two in New Zealand [P. marginale (Walker, 1870) and P. otagoense Westerman and Ritchie, 1984], two in Australia [P. vittatum (Sjöstedt, 1920) and P. crassum Key,1992] and one on Lord Howe Island (P. howeanum Key, 1992) [41]. Phaulacridium are lowland grasshoppers that inhabit native and mixed exotic herb-/grasslands [41,42]. Most have non-functional reduced wings, but micropterous individuals of P. vittatum, P. crassum and P. marginale do occur [41,42]. Phaulacridium marginale is widespread in open grasslands in mainland New Zealand and the Chatham Islands, whereas Phaulacridium otagoense is restricted to small semi-arid environments in central Otago and central Canterbury in South Island, New Zealand.
2.2.3. Anisolabis (Dermaptera: Labiduridae)-Earwig
Anisolabis Fieber 1853 is an earwig genus distributed around the Pacific. However, Anisolabis littorea (White, 1846) is an endemic New Zealand species that lives and breeds under rocks and logs on beaches around mainland New Zealand and islands including the Chathams [43]. There are two other species known from New Zealand. A. kaspar (Hudson, 1973) is endemic to the Three Kings Islands, northern New Zealand and A. occidentalis that has been introduced from Australia, but is restricted to the Hawkes Bay in eastern North Island.
2.2.4. Celatoblatta (Blattodea: Blattidae)-Cockroach
The genus Celatoblatta Johns 1966 is endemic to the New Zealand region and comprises 13 described species. These nocturnal cockroaches are flightless and species occupy habitats from the subalpine zone to coastal forests. They hide during the day in rotting logs, leaf litter, under bark and rocks. Most of the diversity in this genus is found in South Island, New Zealand and one endemic species (C. brunni) is known from the Chatham Islands [44].
2.2.5. Mecodema (Coleoptera: Carabidae)-carabid beetle
Mecodema (Blanchard, 1843) is a diverse endemic genus of large, flightless carabid beetles (tribe Broscini). Mecodema is one of six endemic genera of the tribe Broscini recognized in New Zealand and comprises about 66 species [45,46] and taxa are distributed throughout mainland New Zealand from alpine to coastal habitats. The adults of these beetles are nocturnal, flightless (with fused elytra) and their larvae are predatory [47]. They avoid light and forage within soil and decaying logs for worms and other invertebrate prey. The genus is represented on the Chatham Islands by a population of the species, M. alternans.
2.2.6. Geodorcus (Coleoptera: Lucanidae)-Stag Beetle
The members of the genus Geodorcus are flightless and relatively uncommon. There are 10 described species in Geodorcus [48], two of which are endemic to the Chatham Islands, G. capito Deyrolle, 1873 and G. sororum Holloway, 2007. Geodorcus larvae forage on and in decaying logs, but adults sometimes emerge at night onto trees and leaflitter. Most species are now rare and restricted and subject to protection.
2.2.7. Hadramphus (Coleoptera: Curculionidae)-Weevil
The endemic weevil genus Hadramphus comprises five species with a sparse distribution, including one species restricted to the Chatham Islands (H. spinipennis (Broun, 1911)). Taxonomically they belong to the tribe Molytini with the New Zealand genus Lyperobius [49]. Hadramphus are relatively large flightless weevils (11–23 mm), and both adults and larvae feed on the plants of the families Apiaceae, Araliaceae and one on Pittosporaceae and are therefore restricted to the distribution of these plants. All of the species are endangered and mainly confined to offshore islands or remote areas in Fiordland, South Island. One species, Hadramphus tuberculatus (Pascoe, 1877) was rediscovered in Canterbury region during 2004 having last been recorded in 1922 and assumed extinct. Hadramphus stilbocarpae (Kuschel, 1971) occurs in Fiordland and Southland, New Zealand, and on the Chatham Islands the endemic H. spinipennis feeds and lives on the endemic speargrass Aciphylla dieffenbachii.
2.2.8. Amychus (Coleoptera: Elateridae)-Click Beetle
Amychus are large, flightless click beetles [50]. All species are endangered and are restricted to small predator-free offshore islands. The genus is endemic to New Zealand with three extant species known, including one endemic to the Chatham Islands (Amychus candezei Pascoe, 1876). The two other species are restricted to Three Kings Island, (Amychus manawatawhi Marris and Johnson, 2010) and islands in Cook Strait (Amychus granulatus Broun, 1883). The current distribution suggests that they were probably more abundant and widespread prior to the introduction of mammalian predators, and the populations on Cook Strait islands were connected to one another and to mainland New Zealand as little as 15,000 years ago. Amychus generally conceal themselves during the day within and amongst leaf litter and decaying logs or under rocks on peaty soils.
Details of sampling locations and voucher numbers for all taxa employed in this study can be found in the Supplementary Material (Table S1). Details of number of included species per taxon, habitat specification, etc., are listed in Table 1.
3. Results
3.1. Talitropsis (Orthoptera: Raphidophoridae)-Cave Weta
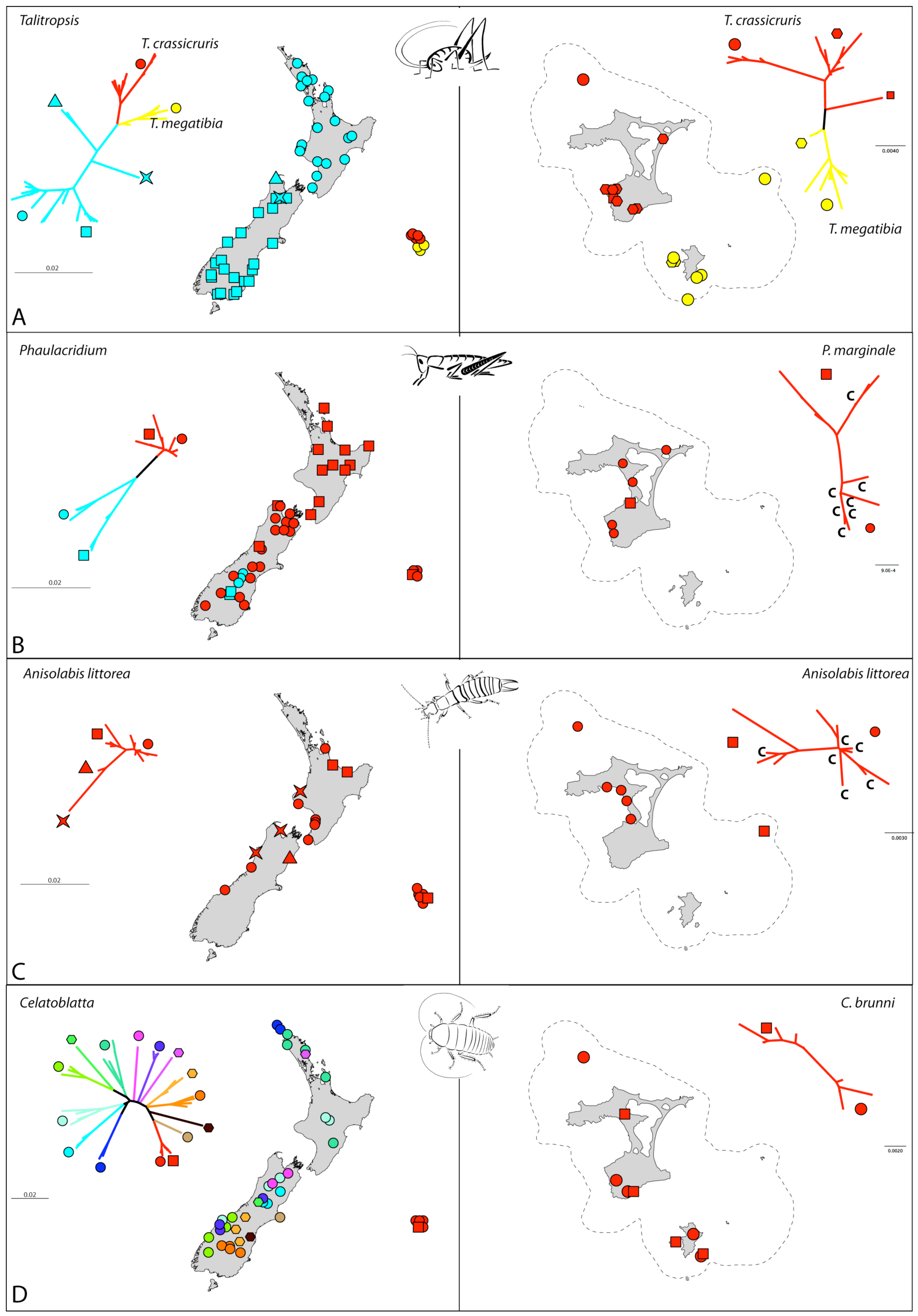
The sampling of Talitropsis cave weta included all three recognized species of the genus. The two species on the Chathams are currently isolated from one another with T. crassicruris inhabiting Main Island and The Sisters and T. megatibia the southern islands of the archipelago including the 44s (Figure 1). The widespread T. sedilotti on the other hand occurs throughout the length of mainland New Zealand and several small offshore islands (Figure 2A). A set of 90 specimens comprising all three species of Talitropsis were sequenced for a partial fragment (857 bp) of COI (Table S1). Phylogenetic reconstruction of the genus with a suituable outgroup shows distinct monophyletic clades for the species [14]. In T. sedilotti there are two additional lineages represented by few individuals from NW South Island, and aside from these the species exhibits some broad regional structuring with samples from North Island and South Islands grouping together but with shallow divergence (Figure 2A and Figure 3A). Within the Chatham species the network splits into two clusters, corresponding to the two endemic species. T. crassicruris shows division between the Sisters and Main Island populations, but there is no similar structure apparent among individuals of T. megatibia in the southern part of the archipelago. Population genetic statistics calculated for the different species of Talitropsis show that nucleotide diversity (π) is similar in the New Zealand species T. sedilotti and the Chatham Talitropsis together but is lower within each Chatham species (Table 2). Haplotype diversity (h) within and between species, on the other hand, is more or less similar. Genetic pairwise distances within T. sedilotti were up to 3.12% and almost as high as the distance between the two endemic Chatham species (3.4%). The genetic distance within these two species was up to 2.38% in T. crassicruris and 0.98% in T. megatibia. Notably there was more genetic diversity between the two taxa in the small Chatham area than within the entire extent of T. sedilotti on the mainland. Between the Chathams and New Zealand taxa the genetic distances reached 4.67%. Sequences are deposited in Genbank (JN409905 - JN409993).
3.2. Phaulacridium (Orthoptera: Acrididae)-grasshopper
In total 76 individuals, including outgroup taxa (Table S1), were sequenced for COI (763 bp). The two New Zealand species form separate clades with outgroup analysis confirming that P. marginale samples form a monophyletic cluster [21]. Haplotypes of P. marginale from the Chatham Islands are varied and fall throughout this clade. In contrast to the homogenous and shallow pattern in P. marginale throughout New Zealand mainland and the Chatham Islands, there is a deep split within P. otagoense, dividing the sampling from two adjacent areas in central South Island (Figure 2B and Figure 3B). Population genetic statistics calculated for the different species of Phaulacridium showed that nucleotide diversity (π) was much lower in P. marginale including the Chatham samples than within P. oatgoense (Table 2). The same is apparent for the haplotype diversity (h). Genetic pairwise distances within P. marginale were up to 1.8% including Chatham specimens. The genetic distance within P. otagoense was similar to this, and 3.5% between the two populations in South Island. Sequences are deposited in Genbank (JN409741 - JN409816).
3.3. Anisolabis (Dermaptera: Labiduridae)-Earwig
In total 27 specimens of the widespread species Anisolabis littorea and its New Zealand sister taxa were sequenced for a fragment (828 bp) of COI. For A. kaspar, unique primer pairs targeting short overlapping sections of COI were designed for the scarce museum material (Table S2) and phylogenetic reconstruction including this species placed it as sister to A. littorea [21]. Chatham Island A. littorea haplotypes did not form a monophyletic cluster (Figure 2C and Figure 3C), and several haplotypes present in the Chathams had their closest relatives scattered through mainland New Zealand. Genetic distances within the Chatham Island samples were up to 1.2%, and up to 2.9% among all mainland New Zealand and Chathams specimens. Although restricted to a narrow coastal environment this species appears to have had extensive gene flow around New Zealand including the Chathams. This might be due to ongoing exchange or a recent wave of migration involving numerous individuals. Sequences are deposited in Genbank (JN409619 - JN409644).
3.4. Celatoblatta (Blattodea: Blattidae)-Cockroach
For Celatoblatta, mitochondrial COI (611 bp) was sequenced for 52 specimens comprising 11 species. The Chatham C. brunni is sister to C. peninsularis, which occupies habitat on Banks Peninsula, the nearest point on the New Zealand mainland to the Chatham Islands [51] (Figure 2D and Figure 3D). Genetic distances between these two species reach a maximum of 3.7%. Some Celatoblatta species, in particular in South Island, show strong allopatry to distinct mountain ranges, but other species occupying forest habitats have wider, sometimes overlapping ranges. Sequences are deposited in Genbank (JN409645 - JN409696).
3.5. Mecodema (Coleoptera: Carabidae)-Carabid Beetle
Sampling comprised 89 Mecodema specimens, representing 37 described and 4 undescribed species (I. Townsend, pers. comm.). COI (788 bp) sequences were obtained, where necessary using specific PCR primers designed to target short overlapping fragments [21]. Bayesian analysis showed strong structure among species groups within Mecodema confirming that Chatham M. alternans are sister to mainland M. alternans in Dunedin [21] (Figure 2E and Figure 3E). In New Zealand, neither species nor species groups are entirely allopatric, suggesting extensive range shifting following speciation. In the case of M. alternans, range expansion has included dispersal to the Chatham Islands, although the distribution of the species there is limited. Genetic distances between New Zealand and Chatham Island specimens of M. alternans reached 2.9%. Sequences are deposited in Genbank (JN409817 - JN409904).
3.6. Geodorcus (Coleoptera: Lucanidae)-Stag Beetle
Mitochondrial COI (777 bp) was sequenced for 20 individuals representing five species, two of which are endemic to the Chatham Islands. Several other species in this genus from mainland New Zealand are exceptionally rare and localized and were not available for analysis. Bayesian analysis with representatives of the closest sister of Geodorcus in New Zealand, Paralissotes, confirmed the Chatham samples form a monophyletic group apparently sister to the North Island species in the analysis G. novaezealandiae [21]. Among the Chatham samples, Geodorcus capito from Main Is., South East Is. and Mangere Is., form two clades, so that this species appears to be paraphyletic with respect to the species from The Sisters (G. sororum) (Figure 2F and Figure 3F). The genetic distances were up to 0.96% among G. sororum haplotypes and 6.9% among G. capito haplotypes with as much as 6.75% between these two endemic species. Distances between Chatham and New Zealand species were up to 14.1%. Sequences are deposited in Genbank (JN409697 - JN409716).
3.7. Hadramphus (Coleoptera: Curculionidae)-Weevil
The genus Hadramphus comprises four species in New Zealand. Three were available for this study, with a forth restricted to an offshore island in northern New Zealand. Hadramphus spinipennis is endemic to the Chatham Islands but only occurs on Mangere and South East Islands, were its host plant (Aciphylla dieffenbachii) is established. Partial (816 bp) COI was sequenced for 24 individuals of the three species of Hadramphus plus additional six species of its closest sister taxon in New Zealand (Lyperobius). Bayesian analysis confirmed distinct monophyletic groups for the genera and species, with comparatively shallow diversity in Hadramphus [21]. The Chatham Island H. spinipennis is, in our sample, sister to Canterbury, New Zealand H. tuberculatus (Figure 2G and Figure 3G). However, within H. spinipennis there is a very shallow split between the two populations. Genetic distance within the Chatham species was up to 0.5%, with up to 4% between the New Zealand and Chatham species. Sequences are deposited in Genbank (JN409717 - JN409740).
3.8. Amychus (Coleoptera: Elateridae)-Click Beetle
Partial (784 bp) COI was sequenced for 17 individuals of three species of the genus Amychus, mostly from the Chatham population. These beetles are extremely rare, especially those associated with mainland New Zealand so only one specimen of the Three Kings Is. species, A. manawatawhi and two Cook Strait A. granulatus were available for analysis. The Chatham endemic species Amychus candezei was collected on four islands within the Chatham archipelago (Figure 2H and Figure 3H). The genetic diversity within the Chatham species was up to 1.2%, with up to 4.7% and 9.6% between Amychus candezei and A. granulatus, and A. manawatawhi respectively. Sequences are deposited in Genbank (JN409602 - JN409618).
4. Discussion
One prediction widely made by biogeographers is that distribution patterns and phylogeographic structure of different taxon groups are likely to be congruent where they are the product of the same geophysical history. In the present case, emergence of the Chatham Island archipelago from the southern ocean about 4 Ma might be expected to have yielded a set of Chatham Island endemic taxa that were similarly divergent in terms of genetics and morphology, from neighboring New Zealand populations from which they were derived. Although the signature of Chatham diversity is consistently youthful, in keeping with the young age of the islands [28], we found that similarity of mainland New Zealand and Chatham sister lineages ranged from sharing identical haplotypes (e.g., Phaulacridium grasshoppers) to having distinct haplotypes differing by as much as 14.1% (Geodorcus beetles). Such results are in keeping with previous studies of taxa that incorporate New Zealand and Chatham Island sampling, where divergence estimates range from populations with identical cpDNA haplotypes in the fern Asplenium hookerianum [52], to 2% mtDNA sequence divergence in a skink Oligosoma nigriplantare nigriplantare [53], and plants with up to 6.4% sequence divergence [54]. Among these examples are spiders [55], insects (cicada [56]; cockroach [51]; damselfly [57]; stick insect [58]), amphipods [59], isopods [60], and birds (rails [61]; robin [62]; parakeet [63,64]; pigeon [18]). Given the wide taxonomic and ecological diversity represented in these studies, perhaps this wide range in divergence estimates reflects differences in biology rather than history, or a combination of the two.
In reality, there are numerous plausible reasons for the variation observed among studies of different taxa, including different rates of lineage extinction; different times of arrival in the Chatham Islands; differences in effective population size; and different rates of molecular evolution. As far as genetic distance is concerned the sampling effect of lineage extinction will always tend to lengthen the inferred distance to a common ancestor from surviving taxa, and the same can result from accidental failure to include close relatives in analyses [5,14,20]. In Amychus beetles, for example, biogeographic inference is likely to be strongly influenced by extinction as their present narrow range appears to be a remnant of former diversity [50]. Similarly, high observed divergence in Geodorcus was likely exacerbated by gaps in sampling. Some traction is gained in distinguishing these effects in our study when taxonomy (as a proxy for morphological variation) is taken into account, providing an opportunity for comparison that is not available in the majority of published studies that deal with single taxa. Three parameters together contribute information, taxonomy, genealogy and geography, and we note there is no readily discernable concordance of these traits among the taxon groups we studied. Unlike single taxon studies or those with few Chatham samples, here we are able to demonstrate how different the biogeographic patterns expressed by separate taxon groups are (Figure 3). While spatial population structuring within the Chathams might not be expected for taxa that are not also structured between New Zealand and Chathams (e.g., Phaulacridium and Anisolabis), those that are extensively partitioned into species at the source (NZ) might be expected to show equivalent spatial structure at a finer scale (e.g., Celatoblatta and Mecodema). The Chatham Islands do contain endemic species and sometimes endemic sister species, but these are not consistently associated with genera that are speciose in New Zealand (e.g., Geodorcus, Talitropsis). Additionally, there is no consistent geographic relationship between New Zealand and Chatham taxa in each group; New Zealand lineages sister to those in the Chathams are in South Island (Mecodema, Celatoblatta, Hadramphus, Amychus) or the North Island (Geodorcus)—although this is sensitive to taxon sampling, or in the case of the widespread species (Anisolabis, Phaulacridium) in both of the main islands of New Zealand.
Given the young geological age for the Chatham Islands (Pliocene) and much younger connection among them (Pleistocene low sea level), the incidence of multiple endemics such as Talitropsis is intriguing, especially where this contrasts strongly with the situation of their sibling taxa in New Zealand. This is borne out by the respective distribution of genetic diversity, which in the case of Talitropsis is indicative of recent southwards range expansion throughout New Zealand (i.e., less genetic diversity in southern parts of the range), whereas in the Chathams, genetic diversity and morphological traits are tightly partitioned over a narrow spatial scale across 50 km of small islands. This is despite the fact that New Zealand and Chathams would each have existed as continuous landscapes during the LGM. Evidently, spatial and temporal effects are not consistent even among sister lineages, and this suggests a strong influence of stochastic processes.
The observation that land area and age might not be accurate predictors of genetic or taxonomic diversification is significant because these are the very traits that are routinely used in biogeographic analyses [5]. Similarly, the demonstration that not only are taxa with “good” dispersal characteristics (e.g., flying rails [61]; pigeons [18]) and windblown fern spores [52] able to reach the Chathams, but so are taxa that lack obvious dispersal attributes. In our study, the presence of flightless insects in the Chathams that are sister to flightless species in New Zealand shows that such animals are not prevented from oversea dispersal. This suggests that passive mechanisms are influential. It might be relevant, for instance, that all the taxa studied spend a substantial amount of their lives in logs, and logs are known to be transported down rivers and to the sea, and to arrive on beaches having drifted in the ocean sometimes carrying animals and plants with them [65]. Records of logs and other flotsam arriving in the Chatham Islands from New Zealand indicate this passage is frequent and rapid. New Zealand Anisolabis earwigs are most readily found in and around drift wood on beaches, stag beetle larvae and pupa live and feed in logs, and Talitropsis cave weta of all ages occupy holes in wood. Interestingly, in the Chathams the opportunities for log dwelling are fewer on some islands (Forty Fours and The Sisters) where trees are absent. Here, Talitropsis occupy cavities in the peaty soil formed between rocks and Geodorcus and Amychus must complete their life cycles in the humus-rich soil. If passive dispersal is as influential as it appears, we predict that biotic assembly may be mostly contingent upon opportunities for establishment instead.
From examination of eight insect taxon groups comprising many species we conclude that lack of congruence is likely the result of observing evolution at different stages. Anisolabis littorea appears to be in or just completing a wave of expansion around the New Zealand coastline including Chathams. There is no indication from the distribution of genetic diversity, that colonizing the Chathams was any more problematic than expansion along the mainland coast; several separate matrilines have made the 800 km trip successfully. The situation in Phaulacridium marginale is similar with many genealogical lineages shared across New Zealand and across 800 km of ocean. However, Phaulacridium history in New Zealand is intriguing with an endemic species in South Island that contains, in a small geographic area, more genetic diversity than the widespread P. marginale. In Talitropsis, morphogenesis matches mtDNA partitioning, but genetic distance is not correlated with geographic distance. Talitropsis sedilotti appears to have expanded through New Zealand since arrival of the genus on the Chathams, perhaps following local extinction of prior diversity as indicated by distinct localized New Zealand lineages in our data (Figure 3A).
In contrast to this, we see that Celatoblatta has maintained taxonomic diversity in New Zealand that may date from before colonization of the Chathams. Whatever process removed most Talitropsis diversity (possibly Pleistocene climate cycling) appears to have left Celatoblatta relatively intact. We note that in New Zealand, many cockroach species are allopatric but some, especially those associated with forest, have come to occupy relatively wide overlapping ranges. Perhaps even further along in this process, Mecodema reveals a well developed New Zealand radiation, apparently retained over protracted time (since Miocene- [21]). Extensive range shifting resulting in sympatry suggests well developed ecological partitioning of the species concerned. In this case arrival in the Chatham Islands appears to have been relatively late in the diversification of the genus. Thus, colonization and speciation appear to have occurred throughout the short geological lifetime of the Chatham Islands, and extinction in New Zealand has influenced the resulting biogeographic patterns.
5. Conclusions
Finding biogeographic congruence among taxa, at small or large geographic distances, with or without obvious habitat distinctions and in a time frame that encompasses substantial geogphysical changes is difficult [5]. For the Chatham Islands, the frailty of biogeographic analysis is starkly evident in the former assertion that biological evidence of taxon distribution in the New Zealand/Chatham system was consistent with a process of ancient vicariance [67], whereas it is now clear that neither geology nor biology support this view or are necessary [28,52,54,68]. Two important conclusions arise from this. The first is that if hypotheses about the influence of the size, age and distribution of land on the partitioning of biodiversity cannot be upheld when data about land history are available, what confidence can there be in inferences made about the biogeography of taxa where such data are missing? Secondly, if such landscape traits are not the most important predictors of phylogeographic and phylogenetic partitioning as appears to be the case, then considerable work is needed to describe ecological and behavioral traits of plants and animals so that tests for their biogeographic influence can be made [10,69]. Furthermore, these data are required to refute the hypothesis that stochasticism is the primary force in biogeography.
Future, more intensive sampling of populations and loci may allow assessment of population size and even rates of molecular evolution operating in this system. Natural history studies will contribute to knowledge of population size, behavior and important traits such as longevity and fecundity; all of which influence underlying population genetics. Quantifying these ecological traits will be an important step in elucidating the extent of stochastic processes including extinction and colonization, which are likely to exert strong influence on biogeographic outcomes.


| Order | Common name | Taxon | a | b | c | NZ habitat | Ch.Is. habitat | Genetic dist. (%) |
|---|---|---|---|---|---|---|---|---|
| Orthoptera | cave cricket | Talitropsis | (3/2) | 2 | 3 | forest | not specific | 4.7 |
| Orthoptera | grasshopper | Phaulacridium | (2/1) | 0 | 2 | open grassland | open grassland | 1.8 |
| Dermaptera | earwig | Anisolabis | (3/1) | 0 | 3 | coastal | coastal | 2.9 |
| Blattodea | cockroach | Celatoblatta | (13/1) | 1 | 12 | not specific | not specific | 3.7 |
| Coleoptera | carabid beetle | Mecodema | (66/1) | 0 | 35 | not specific | not specific | 2.9 |
| Coleoptera | stag beetle | Geodorcus | (7/2) | 2 | 5 | forest | not specific | 14.1 |
| Coleoptera | weevil | Hadramphus | (4/1) | 1 | 3 | host plant | host plant | 4.0 |
| Coleoptera | click beetle | Amychus | (3/1) | 1 | 3 | forest | not specific | 9.6 |
| area | n | Nhaps | k | π (10−3) | h |
|---|---|---|---|---|---|
| T. sedilotti (NZ) | 56 | 33 | 8.486 | 1.28 | 0.948 |
| T. crassicruris (ChIs) | 22 | 17 | 5.381 | 0.81 | 0.974 |
| T. megatibia (ChIs) | 13 | 10 | 4.295 | 0.65 | 0.962 |
| total ChIs pop. | 35 | 27 | 8.356 | 1.26 | 0.985 |
| Ph. marg. (total pop.) | 65 | 5 | 0.274 | 0.62 | 0.180 |
| Ph. ota. (NZ Alex) | 7 | 3 | 2.095 | 3.50 | 0.905 |
| Ph. ota. (NZ Mack) | 4 | 5 | 4.667 | 7.79 | 0.833 |
| Ph. ota. (NZ total pop.) | 11 | 8 | 9.800 | 16.36 | 0.945 |
| (a) Talitropsis. | ||
|---|---|---|
| Sample ID | Species | Location |
| CW 48.1 | T. sedilloti | N.I.,Hawkes Bay,Mohi Bush |
| CW 47.1 | T. sedilloti | N.I.,Hawkes Bay,Hastings,Mohi Bush |
| Tsed4 | T. sedilloti | S.I., Dunedin, Frasers Bush |
| Tsed7 | T. sedilloti | N.I.,Northland,Whatitiri Scenic Res. |
| Tsed9 | T. sedilloti | S.I.,Nelson,Nelson Lakes, Mt. Roberts, Carpark |
| Tsed24 | T. sedilloti | S.I.,Te Anau,Rainbow Reach |
| Tsed26 | T. sedilloti | S.I.,Catlins,Matai Falls |
| CW 209.1 | T. sedilloti | N.I.,Te Urewera,Lake Waikaremoana, Hinerau Walk |
| CW 211 | T. sedilloti | N.I.,Waikato,Waitomo Caves,Short Bush Walk |
| CW 210 | T. sedilloti | N.I.,BOP,nr. Mangatoi,Otanewainuku Forest,Rimu Tr. |
| CW 207 | T. sedilloti | N.I.,Mt Taranaki,East-Taranaki,Patea Track |
| CW 482.1 | T. sedilloti | S.I., Southland, Takitimu Ra., Pinchester Bush |
| CW 482.2 | T. sedilloti | S.I., Southland, Takitimu Ra., Pinchester Bush |
| CW 482.3 | T. sedilloti | S.I., Southland, Takitimu Ra., Pinchester Bush |
| CW 21 | T. sedilloti | N.I., Auckland, Waitakere, Opanuku Rd |
| CW 23 | T. sedilloti | N.I., Northland,Prescot Rd nr Ruakaka |
| CW 29 | T. sedilloti | N.I.,Auckland,Waitakere,Arataki |
| CW 275 | T. sedilloti | S.I., Otago, Leith Valley, Dunedin |
| CW 20 | T. sedilloti | N.I.,Northland, Hukatere |
| CW 331 | T. sedilloti | N.I., Coromandel, Cuvier Island |
| CW 350 | T. sedilloti | S.I., Nelson Lakes NP, Lake Rotoroa, loop Track |
| CW 351 | T. sedilloti | S.I., Lewis Pass NP, Lake Daniells Track |
| CW 352 | T. sedilloti | S.I., Fiordland NP, Te Anau, Kepler Track |
| CW 353 | T. sedilloti | S.I., Fiordland NP, Te Anau, Kepler Track |
| CW 354 | T. sedilloti | S.I., Otago, Queenstown, Kinloch |
| CW 355 | T. sedilloti | S.I., Otago, Catlins Coast, Papatowai |
| CW 45.1 | T. sedilloti | N.I.,Te Urewera, L. Waikaremoana, Black Beech Track |
| CW 276 | T. sedilloti | S.I., Otago, Hampden, Kurinui |
| CW 371 | T. sedilloti | N.I., Wellington, Eastbourne |
| CW 377 | T. sedilloti | N.I., Waikato, Te Awamutu, Maungatauturi |
| CW 423 | T. sedilloti | N.I., Wanganui |
| CW 457 | T. sedilloti | N.I., Lady Alice Island |
| Tsed21 | T. sedilloti | S.I.,Invercargill, Otarara Scenic Res. |
| Tsed22 | T. sedilloti | S.I.,Central Otago, nr. Beaumont |
| Tsed23 | T. sedilloti | S.I.,Westland, Haast River |
| CW 128 | T. sedilloti | N.I., Northland |
| CW 428 | T. sedilloti | N.I., Wanganui |
| CW 469 | T. sedilloti | N.I., Taranaki |
| CW 499.2 | T. sedilloti | S.I., Invercargill, Bluff Scenic Res. |
| CW 499.1 | T. sedilloti | S.I., Invercargill, Bluff Scenic Res. |
| CW 356.1 | T. sedilloti | S.I.,Kahurangi NP, Golden Bay,Start of Heaphy Track |
| CW 356.3 | T. sedilloti | S.I.,Kahurangi NP,Golden Bay,Start of Heaphy Track |
| CW 356.2 | T. sedilloti | S.I.,Kahurangi NP,Golden Bay,Start of Heaphy Track |
| CW 209.2 | T. sedilloti | N.I.,Te Urewera,Lake Waikaremoana, Hinerau Walk |
| CW 209.3 | T. sedilloti | N.I.,Te Urewera,Lake Waikaremoana, Hinerau Walk |
| CW 274 | T. sedilloti | S.I., Otago, Leith Valley, Dunedin |
| CW 469.2 | T. sedilloti | N.I., Taranaki |
| CW 358.1 | T. sedilloti | N.I., Levin, Tararua Ra., Makahika Lodge |
| CW 160 | T. sedilloti | N.I., Manawatu, Levin, Papaitonga Reserve |
| CW 5 | T. sedilloti | S.I.,West Coast, Lake Matheson |
| CW 191 | T. sedilloti | S.I., Lake Wakatipu, Te Kere Haka Reserve |
| CW 469.1 | T. sedilloti | N.I., Taranaki |
| CW 208 | T. sedilloti | N.I.,Manawatu, Pohangina Valley, Camp Rangi Woods |
| CW 481 | T. sedilloti | N.I., Coromandel, Cuvier Island |
| CW 481z | T. sedilloti | N.I., Coromandel, Cuvier Island |
| CW 83 | T.crassicruris | Ch.Is., Main Is., Tuku Reserve, Taiko Camp |
| CW 102.1a | T.crassicruris | Ch. Is., Main Is., Awatotora |
| CW 102 | T.crassicruris | Ch. Is., Main Is., Awatotora |
| CW 102.2 | T.crassicruris | Ch. Is., Main Is., Awatotora |
| CW 102.3 | T.crassicruris | Ch. Is., Main Is., Awatotora |
| CW 101 | T.crassicruris | Ch. Is., Main Is., Awatotora |
| CW 101.1 | T.crassicruris | Ch. Is., Main Is., Awatotora |
| CW 101.2 | T.crassicruris | Ch. Is., Main Is., Awatotora |
| CW 104 | T.crassicruris | Ch. Is., Te Whanga Lagoon, Te Mataroe Bush |
| CW 216 | T.crassicruris | Ch. Is.,The Sisters,Middle Sister |
| CW 216.1 | T.crassicruris | Ch. Is.,The Sisters,Middle Sister |
| CW 216.2 | T.crassicruris | Ch. Is.,The Sisters,Middle Sister |
| CW 212 | T.crassicruris | Ch. Is.,Main Is.,Hapupu Reserve |
| CW 214 | T.crassicruris | Ch. Is.,Main Is.,Southern Tablelands |
| CW 214.1 | T.crassicruris | Ch. Is.,Main Is.,Southern Tablelands |
| CW 205 | T. megatibia | Ch. Is.,South East Is. |
| CW 204 | T. megatibia | Ch. Is.,South East Is. |
| CW 357 | T. megatibia | Ch. Is., The Pyramid, Camp Flat |
| CW 203.1 | T. megatibia | Ch. Is.,Mangere Is.,Robin Bush |
| CW 215 | T.crassicruris | Ch. Is.,The Sisters,Middle Sister |
| CW 218 | T.crassicruris | Ch. Is.,Main Is.,Tuku Reserve |
| CW 02 | T.crassicruris | Ch. Is.,Main Is.,Whakamarino |
| CW 213 | T. megatibia | Ch. Is.,The 44s |
| CW 219.1 | T. megatibia | Ch. Is.,South East Is. |
| CW 219.2 | T. megatibia | Ch. Is.,South East Is. |
| CW 219.3 | T. megatibia | Ch. Is.,South East Is. |
| CW 206.1 | T. megatibia | Ch. Is,Mangere Is.,Robin Bush |
| CW 206.2 | T. megatibia | Ch. Is,Mangere Is.,Robin Bush |
| CW 217.1 | T.crassicruris | Ch. Is.,The Sisters,Middle Sister |
| CW 217.3 | T.crassicruris | Ch. Is.,The Sisters,Middle Sister |
| CW 217.2 | T.crassicruris | Ch. Is.,The Sisters,Middle Sister |
| CW 8 | T. megatibia | Ch. Is.,South East Is. |
| CW 7 | T.crassicruris | Ch. Is.,Main Is.,Whakamarino |
| CW 13 | T. megatibia | Ch. Is.,Little Mangere Is. |
| CW 14 | T. megatibia | Ch. Is.,Little Mangere Is. |
| (b) Phaulacridium. | ||
|---|---|---|
| Sample ID | Species | Location |
| PH02 | P. marginale | N.I., Taupo, Rangipo Desert |
| PH03 | P. marginale | N.I., Wellington, Makara |
| PH05.1 | P. marginale | S.I., Marlborough, Pelorus Sound |
| PH06 | P. marginale | Ch.I., Henga Scenic Res. |
| PH06.1 | P. marginale | Ch.I., Henga Scenic Res. |
| PH04 | P. marginale | N.I., Tongariro NP, Rangipo Desert |
| PH07 | P. marginale | N.I., Wellington, Mt. Kaukau |
| PH08 | P. marginale | Ch.I., Fakey's Quarry |
| PH09 | P. marginale | S.I., Able Tasman NP, Awaroa |
| PH10 | P. marginale | Ch.I., Tuku Res. |
| PH11 | P. marginale | Ch.I., Matarakau |
| PH12.1 | P. marginale | N.I., Te Urewera, Lake Waikaremoana |
| PH13 | P. marginale | Ch.I., Awatotora |
| PH31.1 | P. marginale | S.I., Fiordland, Borland Lodge |
| PH14.1 | P. marginale | S.I., Otago, Dunedin, Kurinui Hampden |
| PH14..2 | P. marginale | S.I., Otago, Dunedin, Kurinui Hampden |
| PH17.1 | P. marginale | Ch.I., Maipito Road |
| PH18.1 | P. marginale | Ch.I., Tuku Res., Trapline |
| PH19.1 | P. marginale | Ch.I., Tuku Res., Abyssinia Track |
| PH19.2 | P. marginale | Ch.I., Tuku Res., Abyssinia Track |
| PH22.1 | P. marginale | S.I., Able Tasman NP, Awaroa, Dacha |
| PH22.2 | P. marginale | S.I., Able Tasman NP, Awaroa, Dacha |
| PH22.3 | P. marginale | S.I., Able Tasman NP, Awaroa, Dacha |
| PH23.1 | P. marginale | S.I., Lewis Pass NP, Marble Hill Picnic Area |
| PH24 | P. marginale | S.I., Canterbury, Hunters Hills, Mackenzie Pass |
| PH26 | P. marginale | S.I., Mt Cook NP, Mt Cook Village |
| PH27.1 | P. marginale | S.I., Mt. Cook NP, Mt. Cook Village |
| PH27.2 | P. marginale | S.I., Mt. Cook NP, Mt. Cook Village |
| PH27.3 | P. marginale | S.I., Mt. Cook NP, Mt. Cook Village |
| PH28.1 | P. marginale | S.I., Canterbury, Hunters Hills, Myer's Pass |
| PH29.1 | P. marginale | S.I., Canterbury, Hunters Hills, Myer's Pass |
| PH29.2 | P. marginale | S.I., Canterbury, Hunters Hills, Myer's Pass |
| PH32.1 | P. marginale | S.I., Otago, Queenstown, Coronet Peak Skifield |
| PH35.1 | P. marginale | S.I., Canterbury, Cave Stream Scenic Res. |
| PH36 | P. marginale | S.I., Canterbury, Arthurs Pass |
| PH37 | P. marginale | S.I., Seaward Kaikoura Range, Mt. Fyffe |
| PH38 | P. marginale | S.I., Marlborough, Clarence River |
| PH39.1 | P. marginale | S.I., Marlborough, Kekerengu, Dee Stream |
| PH49 | P. marginale | N.I., BOP, Te Puke |
| PH50.1 | P. marginale | N.I., Waikato, Maungatatua |
| PH52 | P. marginale | N.I., Manawatu, Levin |
| PH54.1 | P. marginale | N.I., Hawkes Bay, SH5 |
| PH55.2 | P. marginale | N.I., Coromandel, Whitianga |
| PH55.3 | P. marginale | N.I., Coromandel, Whitianga |
| PH57.2 | P. marginale | S.I., Canterbury, Lake Tekapo, Burkes Pass |
| PH71.1 | P. marginale | N.I., Whirinaki Forest, Rata Road |
| PH71.2 | P. marginale | N.I., Whirinaki Forest, Rata Road |
| PH71.3 | P. marginale | N.I., Whirinaki Forest |
| PHM1 | P. marginale | S.I., Marlborough, Mt. Patriarch |
| PHM2 | P. marginale | S.I., Nelson, St. Arnaud, Mt. Roberts |
| PHM3 | P. marginale | S.I., Seaward Kaikoura Range, Mt. Fyffe |
| PHM4 | P. marginale | S.I., Otago, Awakino |
| PHM5 | P. marginale | S.I., Marlborough, Mt Lyford |
| PHM6 | P. marginale | S.I., Otago, Awakino |
| PHM7 | P. marginale | S.I., Otago, Awakino |
| PHM10 | P. marginale | S.I., Otago, St. Bathans |
| PHM13 | P. marginale | S.I., Canterbury, Old Man Range |
| PHM16 | P. marginale | N.I., East Cape, East Island |
| PHM26 | P. marginale | N.I., Great Barrier Is, Copper Mine |
| PHMwell | P. marginale | N.I., Wellington, Newlands |
| PHMco2 | P. marginale | N.I., Coromandel |
| PHMco3 | P. marginale | N.I., Coromandel |
| PHBurk4 | P. marginale | S.I., Canterbury, Lake Tekapo, Burks Pass |
| PHoM11 | P. otagoense | S.I., Otago, Alexandra, Graveyard Gully |
| PHoM12 | P. otagoense | S.I., Otago, Alexandra, Graveyard Gully |
| PHoM14 | P. otagoense | S.I., Otago, Alexandra, Manor Burn |
| PHoM8 | P. otagoense | S.I., Otago, Alexandra, Conroys Dam |
| PHoM15 | P. otagoense | S.I., Otago, Alexandra, Bridge Hill |
| PHoM15.1 | P. otagoense | S.I., Otago, Alexandra, Bridge Hill |
| PHo51 | P. otagoense | S.I., Mackenzie, Lake Tekapo |
| PHo59 | P. otagoense | S.I., Mackenzie, Benmore Range, Twizel |
| PHo60 | P. otagoense | S.I., Mackenzie, Simons Pass Hill, Tekapo River |
| PHoCON1 | P. otagoense | S.I., Otago, Alexandra, Conroys Dam |
| PHoMtJohn | P. otagoense | S.I., Mackenzie, Lake Tekapo, Mt John |
| (c) Anisolabis | ||
|---|---|---|
| Sample ID | Species | Location |
| EW 01 | A. littorea | N.I.,Wellington, The Sirens Rocks |
| EW 02 | A. littorea | Ch. Is.,Main, Long Beach South end |
| EW 03 | A. littorea | Ch. Is.,Main, The Crossing |
| EW 04 | A. littorea | N.I., Manawatu. Tangimoana |
| EW 05.1 | A. littorea | N.I.,Bay of Plenty, Papamoa, Tauranga |
| EW 09.1 | A. littorea | S.I., Punakaiki, Pahautane Bay |
| EW 15 | A. littorea | N.I.,BOP, Ohope Beach,West End |
| EW 16 | A. littorea | N.I.,Coromandel,Whitianga,Buffalo Beach |
| EW 17 | A. littorea | N.I.,Taranaki,Ohawe Beach |
| EW 19.1 | A. littorea | Ch. Is.,Mangere Is.,Robin Bush |
| EW 21 | A. littorea | Ch. Is.,Main,Waitangi,Maipito Rd. |
| EW 23.1 | A. littorea | Ch. Is.,Main,Hapupu Reserve |
| EW 25.1 | A. littorea | Ch. Is.,Main,Ohira Bay, Basalt Columns |
| EW 27.1 | A. littorea | Ch. Is.,The 44's |
| EW 27.2 | A. littorea | Ch. Is.,The 44's |
| EW 28.1 | A. littorea | Ch. Is.,The Sisters,Middle Sister |
| EW 28.2 | A. littorea | Ch. Is.,The Sisters,Middle Sister |
| EW 30.2 | A. littorea | S.I., Westland, Haast, Shipwreck Beach |
| EW 34 | A. littorea | N.I., Manawatu, Foxton Beach |
| EW 35 | A. littorea | S.I., Westland, Hokitika Beach |
| EW 38 | A. littorea | N.I., Taranaki, New Plymouth |
| EW 40 | A. littorea | N.I., Manawatu, Levin |
| EW 41.1 | A. littorea | S.I., Seaward Kaikoura, Marfell Beach |
| EW 42 | A. littorea | S.I., Able Tasman, Awaroa |
| EW 39 | A. kaspar | N.I., Three Kings Islands |
| EW 06.1 | A. occidentalis | N.I.,Hawkes Bay, Ngaruroro River mouth |
| EW 29.1 | A. occidentalis | N.I., Hawkes Bay, Ocean Beach |
| (d) Celatoblatta. | ||
|---|---|---|
| Sample ID | Species | Location |
| CK 01 | C. brunni | ChIs, South East Is., Woolshed Bush |
| CK 02 | C. quinquemaculata | S.I., Otago, Old Man Range |
| CK 03 | C. quinquemaculata | S.I., Otago, Rock&Pillar Range |
| CK04 | C. subcorticaria | S.I., West Coast, North of Haast |
| CK 05 | C. vulgaris | S.I., West Coast, Okuru River |
| CK 06 | C. sedilloti | N.I., Northland, Cape Reinga |
| CK 07 | C. vulgaris | S.I., Nelson Lakes, St. Arnaud |
| CK 08 | C. notialis | S.I., Southlan, Te Anau |
| CK 09 | C. montana | S. I. Canterbury, Arthurs Pass, Fog Peak |
| CK 10 | C. notialis | S.I., Riverton, Mores Scenic Res. |
| CK 11 | C. hesperia | N.I., Northlan, Kaitai, Herekino Forest |
| CK 12 | C. vulgaris | N.I., Lake Taupo, Miller's Rd. |
| CK 13 | C. notialis | S.I., West Coast, Haast River |
| CK 14 | C. quinquemaculata | S.I., Otago, Rock &Pillar Range |
| CK 15 | C. notialis | S.I., West Coast, Lake Matheson |
| CK 16 | C. undulivitta | N.I.,Whangarei, Mt. Manaia |
| CK 17 | C. sedilloti | N.I., Far North, Lake Wahakari |
| CK 18 | C. sedilloti | N.I., Whangarei, Waipapa Stream |
| CK 19 | C. brunni | ChIs, South East Is., Woolshed Bush |
| CK 20 | C. quinquemaculata | S.I., Harris Saddle, Conical Hill |
| CK 21 | C. anisoptera | S.I., Otagao, St. Bathans |
| CK 21b | C. quinquemaculata | S.I., Otago, Cromwell, Nevis Rd. |
| CK 22 | C. montana | S.I., Canterbury, Craigeburn, Mt. Ida |
| CK 23 | C. anisoptera | S.I., Otago, Ohau, Mt. Sutton |
| CK 26 | C. montana | S.I., Marlborough, Mt. Lyford, Ammi Range |
| CK 27 | C. sp. | S.I., Neslon, Mt. Patriarch, Rochmond Range |
| CK 28 | C. vulgaris | S.I., Nelson, Springs Junction |
| CK 29 | C. sp. | S.I., Nelson, Springs Junction |
| CK 69 | C. sp. | N.I., Northland, Matai Bay |
| CK 70 | C. undulivitta | N.I., Northland, Karikari Pensinsula, Lake Ohia |
| CK 92.1 | C. brunni | ChIs, Mangere Is., Robin Bush |
| CK 93 | C. brunni | ChIs., South East Is., Woolshed Bush |
| CK 99.1 | C. brunni | ChIs., The Sisters, Middle Sister |
| CK 100.1 | C. brunni | ChIs., Main Is., Nikau Reserve |
| CK 100.2 | C. brunni | ChIs., Main Is., Nikau Reserve |
| CK 103 | C. brunni | ChIs., Main Is., Tuku Reserve |
| CK 114.1 | C. brunni | ChIs., Main Is., Southern Tablelands |
| CK 115.1 | C. brunni | ChIs., Main Is., Southern Tablelands |
| CK 116 | C. anisoptera | S.I., Mt. Cook Village |
| CK 129.1 | C. anisoptera | S.I., Mt. Cook, Kea Point Track |
| CK 130 | C. sp. | S.I., Canterbury, Hunters Hills, Meyer's Pass |
| CK 131 | C. vulgaris | N.I., Lake Taupo, Kinloch, Kawakawa Track |
| CK 132 | C. notialis | N.I., Fiordland, Te Anau, Kepler Track |
| CK 133 | C. notialis | S.I., Fiordland, Milfors Rd., Key Summit |
| CK 135 | C. subcorticaria | S.I., Canterbury, Arthurs Pass, Cockayne Nature Walk |
| CK 138 | C. subcorticaria | S.I., Otagao, Mt. Aspiring, Makarora |
| CK 141.1 | C. brunni | ChIs., Pitt Is., North Head |
| CK 147 | C. hesperia | S.I., Canterbury, Arthurs Pass, Cockayne Nature Walk |
| CK 148.1 | C. undulivitta | N.I., Wairarapa, Norsewood, ANZAC Reserve |
| CK 155 | C. undulivitta | N.I., Coromandel, Kauri Grove, Waiau |
| CK 158.1 | C. pensinsularis | S.I., Canterbury, Banks Pensinsula |
| CK 158.2 | C. pensinsularis | S.I., Canterbury, Banks Pensinsula |
| (e) Mecodema. | ||
|---|---|---|
| Sample ID | Species | Location |
| MB 01 | M. alternans | Chatham Is., South East Is. |
| MB 02 | M. alternans | Chatham Is., South East Is. |
| MB 70 | M. alternans | Chatham Is., Mangere Is. |
| MB 71 | M. alternans | Chatham Is., South East Is. |
| MB 86 | M. alternans | Chatham Is., Mangere Is. |
| MB 87 | M. alternans | Chatham Is., South East Is. |
| MB 14 | M. alternans | S.I., Dunedin, Taieri Mouth |
| MB 16 | M. alternans | S.I., Dunedin, Sandfly Bay |
| MB 79 | M. crenicolle | S.I., Marlborough Sounds, Pelorus Bridge |
| MB 103 | M. crenicolle | S.I., Nelson Lakes, Wairau River |
| MB 66 | M. curvidens | N.I., BOP, Rotorua |
| MB 110 | M. fulgidum | S.I., Clarence Valley, Mt. Percival |
| MB 91 | M. cf fulgidum | S.I., Seaward Kaikoura Ra., Mt. Lyford |
| MB 98 | M. howittii | S.I., Canterbury, Banks Peninsula |
| MB 99 | M. howittii | S.I., Canterbury, Banks Peninsula |
| MB 63 | M. longicolle | N.I., Ruahine Ra., Pohangina Valley |
| MB 19 | M. lucidum | S.I., Otago, Carrick Range |
| MB 11 | M. nsp. | S.I., Central Otago, Old Man Range |
| MB 37 | M. nsp. | S.I., Arthurs Pass NP, Dome |
| MB 51.1 | M. nsp. | N.I., Hawkes Bay, Havelock North |
| MB 68 | M. occiputale | N.I., Mangatoi, Otanewainuku Forest |
| MB 23 | M. cf oconnori | N.I., Wellington, Levin, Ohou |
| MB 90 | M. oregoides | S.I., Christchurch, Akuriri Scenic Res. |
| MB 03 | M. rugiceps | S.I., Fiordland, Lake Harris |
| MB 45 | M. sculpturatum | S.I., Dunedin, Ross Reserve |
| MB 108 | M. cf huttense | S.I., Canterbury, Peel Forest |
| MB 25 | M. simplex | N.I., Manawatu, Pahiatua Track |
| MB 64 | M. simplex | N.I., Manawatu, Palmerston North |
| MB 50 | M. spinifer | N.I., Hawkes Bay, Mohi Bush |
| MB 18 | M. spinifer | N.I., Auckland, Waitakeres, Arataki |
| MB 96 | M. strictum | S.I., Nelson, Takaka Hill, Canaan |
| MB 95 | M. sulcatum | S.I., Kaikoura, North of Ohau Point |
| MB 69 | M. validum | N.I., Tongariro NP, Whakapapanui Track |
| MB 86.1 | M. alternans | Chatham Is., Mangere Is |
| MB 88 | M. alternans | Chatham Is., South East Is. |
| MB 88.1 | M. alternans | Chatham Is., South East Is. |
| MB 190 | M. alternans | S.I. Otago, Takahopa River Mouth |
| MB185 | M. alternans hudsoni | The Snares [LUNZ 00002804] |
| MB 176 | M. rectolineatum | S.I., Remarkables Range, Wye Creek |
| MB 196 | M. politanum | S.I., The Remarkables, Rastus Burn |
| MB 128 | M. impressum | S.I., Queenstown, Kinloch |
| MB 125 | M. sculpturatum | S.I., Catlins Forest Park, River Walk |
| MB 111 | M. lucidum | S.I., Pisa Range |
| MB 123 | M. fulgidum | S.I., Hamner Springs, Forest Park, |
| MB 134 | M. constrictum | S.I., Craigeburn Forest P., Education Ct. |
| MB 100 | M. costellum lewisi | S.I., Road nr. Mt. White Station |
| MB 101 | M. costellum obesum | S.I., Nelson, Takaka Hill, Canaan |
| MB 124 | M. nsp. | S.I., Lewis Pass, Lewis Tops |
| MB 195 | M. allani | S.I., Nelson Lakes, Matakitaki V. |
| MB 197 | M. laterale | S. I., Fiordland, Routeburn Track |
| MB 178 | M. minax | S.I., Catlins, Mt. Pye Summit |
| MB 180 | M. minax | S.I., Catlins, Mt. Pye Summit |
| MB 160 | M. elongatum | S.I., Otago, Kinloch |
| MB 143 | M. metallicum | S.I., Buller, Fox River |
| MB 117 | M. crenicolle | S.I., Lake Rotoroa, Braeburn Walk |
| MB 163 | M. crenaticolle | N.I., Taranaki, Kaiteke Ra., Lucy's Gully |
| MB 186 | M. crenaticolle | N.I., Waikato, Skyline Cave |
| MB 121 | M. ducale | S.I., Lewis Pass, Lake Daniels Walk |
| MB 147.1 | M. oregoides | S.I., Canterbury, Ahuriri Scenic Res. |
| MB 187 | M. undet. | S.I., Otago, nr Cromwell |
| MB 30 | M. undet. | S.I., Able Tasman, Awaroa |
| MB 38 | M. undet. | S.I., Takaka, The Grove |
| MB 49 | M. undet. | S.I., Takaka, Rameka Track |
| MB 65 | M. occiputale | N.I., BOP,Ohope Scenic Res. |
| MB 67 | M. occiputale | N.I., Mangatoi,Otanewainuku Forest, |
| MB 61 | M. crenaticolle | N.I., Taranaki, Lake Rotokare |
| MB 62 | M. crenaticolle | N.I., Taranaki, Lake Rotokare |
| MB 72 | M. crenaticolle | N.I., Tongariro, Ohakune |
| MB 12 | M. crenicolle | S.I., Nelson, Shenandoah |
| MB 44 | M. crenicolle | S.I., Able Tasman, Awaroa |
| MB 82 | M. crenicolle | S.I., Able Tasman, Awaroa |
| MB 54 | M. morio | S.I., Catlins, Purakaunui Falls |
| MB 105 | M. infimate | S.I., Codfish Island |
| MB 76 | M. simplex | N.I., Tararua Forest Park, Putara |
| MB 77 | M. simplex | N.I., Tararua Ra., Mt Holdsworth |
| MB 35 | M. constrictum | S.I., Fog Peak, Porter's Pass |
| MB 50.1 | M. spinifer | N.I., Hawkes Bay, Mohi Bush |
| MB 81 | M. fulgidum | S.I., Cobb Valley, Lake Sylvester Tr. |
| MB 27 | M. constrictum | S.I., Canterbury, Craigeburn Rec. area |
| MB 07 | M. nsp. | S.I., Seaward Kaikoura Ra., Mt. Fyffe |
| MB 20 | M. cf oconnori | N.I., Levin, 30B The Avenue |
| MB 73 | M. cf oconnori | N.I., Te Urewera, Ngamoko Trig Tr. |
| MB75 | M. cf oconnori | N.I., Dannevirke, Norsewood Res. |
| MB 10 | M. spinifer | N.I., Auckland, Waitakares Ra. |
| MB 17 | M. punctatum | S.I., Rock&Pillar Range |
| MB 04 | M. sculpturatum | S.I., Dunedin, Leith Saddle |
| MB 06 | M. sculpturatum | S.I., Dunedin, Mosgeil, Silver St. |
| MB 09 | M. huttense | S.I., Canterbury, Peel Forest |
| MB 46 | M. cf huttemse | S.I., Canterbury, Peel Forest |
| (f) Geodorcus | ||
|---|---|---|
| Sample ID | Species | Location |
| SB 03 | G. capito | Ch. Is., South East Is. |
| SB 06 | G. capito | Ch. Is., Taiko Camp,Mt. Albert |
| SB 25 | G. capito | Ch. Is., South East Is., Woolshed |
| SB 41 | G. capito | Ch. Is., Mangere Is., Robin Bush |
| SB 42 | G. capito | Ch. Is., South East Is., Woolshed |
| SB 47 | G. capito | Ch. Is.,Main Is.,Tuku Reserve |
| SB 02 | G. sororum | Ch. Is., The Sisters |
| SB 24 | G. sororum | Ch. Is., The Sisters |
| SB 46 | G. sororum | Ch. Is.,The Sisters,Middle Sister |
| SB 46.1 | G. sororum | Ch. Is.,The Sisters,Middle Sister |
| SB 46.2 | G. sororum | Ch. Is.,The Sisters,Middle Sister |
| SB 54 | G. sororum | Ch. Is.,The Sisters,Middle Sister |
| SB 29.1 | G. philpotti | S.I., Southland, Borland Saddle |
| SB 27 | G. novaezealandiae | N.I., Wellington, Catchpole |
| SB 72.1 | G. novaezealandiae | N.I., Wellington, Eastbourne, Butterfly Creek |
| SB 72.2 | G. novaezealandiae | N.I., Wellington, Eastbourne, Butterfly Creek |
| SB 01 | G. helmsi | S.I., West Coast, Copeland Track |
| SB 04 | G. helmsi | S.I., Salmon Farm,North of L. Paringa |
| SB 21 | G. helmsi | S.I., Southland, Riverton, Mores Scenic Res. |
| SB 69 | G. helmsi | S.I., Otago, Catlins Coast, Papatowai |
| (g) Hadramphus. | ||
|---|---|---|
| Sample ID | Species | Location |
| Wv 02 | H. stilbocarpae | S.I.,Fiordland, Breaksea Is. headland |
| Wv 03 | H. stilbocarpae | S.I.,Fiordland, Breaksea Is. headland |
| Wv 04 | H. stilbocarpae | S.I.,Fiordland, South Breaksea Islet |
| Wv 05 | H. stilbocarpae | S.I.,Fiordland, South Breaksea Islet |
| Wv 06 | H. spinipennis | Ch. Is.,South East Island II |
| Wv 07 | H. spinipennis | Ch. Is.,South East Island II |
| Wv 08 | H. spinipennis | Ch. Is., South East Island |
| Wv 09 | H. spinipennis | Ch. Is.,Mangere Island patch |
| Wv 10 | H. spinipennis | Ch. Is.,Mangere Island patch |
| Wv 11 | H. spinipennis | Ch. Is.,South East Island I |
| Wv 12 | H. spinipennis | Ch. Is.,South East Island I |
| Wv 13 | H. spinipennis | Ch. Is.,Mangere Island patch |
| Wv 14 | H. spinipennis | Ch. Is., 28 Keefo van patch 3 |
| Wv 15 | H. spinipennis | Ch. Is.,Mangere Island patch |
| Wv 16 | H. spinipennis | Ch. Is.,Mangere Island patch |
| Wv 17 | H. spinipennis | Ch. Is.,South East Island |
| Wv 23 | H. stilbocarpae | S.I., Fiordland, Secretary Is. |
| Wv 24 | H. stilbocarpae | S.I., Fiordland, Hawea Is. |
| Wv 25 | H. stilbocarpae | S.I., Fiordland, Puysegur Point |
| Wv 26 | H. stilbocarpae | S.I., Fiordland, Chalky Is. |
| Wv 27 | H. stilbocarpae | S.I., Fiordland, Wairaki Is. |
| Wv 28 | H. stilbocarpae | S.I.,Fiordland, Breaksea Is. |
| Wv 18 | H. tuberculatus | S.I., Canterbury |
| Wv 31 | H. tuberculatus | S.I., Canterbury |
| (h) Amychus | ||
|---|---|---|
| Sample ID | Species | Location |
| CB 04 | A. manawatawhi | N.I., Three Kings Is., Great Island, Tasman Va. |
| CB 14.1 | A. candezei | Ch. Is., South East Is.,Woolshed Bush |
| CB 14.a/2 | A. candezei | Ch. Is., South East Is.,Woolshed Bush |
| CB 14.3 | A. candezei | Ch. Is., South East Is.,Woolshed Bush |
| CB 15 | A. candezei | Ch. Is.,Main, Hapupu Reserve |
| CB 16.1 | A. candezei | Ch. Is.,The 44s |
| CB 16.1/2 | A. candezei | Ch. Is.,The 44s |
| CB 16.a/2 | A. candezei | Ch. Is.,The 44s |
| CB 16.3 | A. candezei | Ch. Is.,The 44s |
| CB 16.4 | A. candezei | Ch. Is.,The 44s |
| CB 17.1 | A. candezei | Ch. Is.,The Sisters,Middle Sister |
| CB 17.2 | A. candezei | Ch. Is.,The Sisters,Middle Sister |
| CB 17.2a | A. candezei | Ch. Is.,The Sisters,Middle Sister |
| CB 17a | A. candezei | Ch. Is.,The Sisters,Middle Sister |
| CB 17a/2 | A. candezei | Ch. Is.,The Sisters,Middle Sister |
| CB 22 | A. granulatus | S.I.,Cook Strait,Maud Is.,DoC house |
| CB 23 | A. granulatus | S.I.,Cook Strait,Maud Is.,DoC house |
ChIs. = Chatham Islands; N.I. = North Island, New Zealand; S.I. = South Island, New Zealand.
| Primer name | Sequence | Gene |
|---|---|---|
| EW_114R | 3′ GTAGGTACAGCAATAATT | COI |
| EW_115F | 5′ ATTATTGCTGTACCTACMG | COI |
| EW_322R | 3′ AKACTGCTCCTATAGAAAGAAC | COI |
| EW_293F | 5′ CTTATTATGTTGTWGCTCAC | COI |
| EW_514R | 3′ CAACAWATATAAGCATCAGG | COI |
| EW_489F | 5′ GATACCTCGWCGATAYTCAG | COI |
| EW_679R | 3′ CTATGRTCTGMTGGTGGA | COI |
Acknowledgments
We are indebted to many people who supplied specimens for this study and/or helped during collection trips over the years, especially R. Emberson, I. Townsend, M. Westerman, P. Johns, S.M. Pawson, S. Morris, P. Sirvid, J. Recsei, R. Harris, I. Millar, A. Paterson, J. Marris, P. Lillywhite, M. Morgan-Richards, F. Wieland and everybody involved in the ChEAr (Chatham Emergent Ark) research project, in particular the geologists H. Campbell, J. Begg, C. Wallace and K. Holt. This research was supported by the Marsden Research Fund (GNS-003). We are grateful to the Chatham Islands Conservation Board and the Department of Conservation area offices for their support and provision of necessary collection permits for the Chathams and mainland New Zealand. We would like to thank the people of the Chatham Islands for their hospitality, support, and enthusiasm and for granting us access to their properties. Many thanks to Frank Wieland for providing the insect pictograms in Figure 3. We appreciate the friendly collegiality of the Phoenix evolutionary ecology and genetics group [70].
References
- Rosen, D.E. Vicariant patterns and historical explanation in biogeography. Syst. Zool. 1978, 27, 159–188. [Google Scholar]
- Zink, R.M.; Blackwell-Rago, R.C.; Ronquist, F. The shifting roles of dispersal and vicariance in biogeography. Proc. R. Soc. B. 2000, 267, 497–503. [Google Scholar]
- Ebach, M.; Humphries, C.J.; Williams, D.M. Phylogenetic biogeography deconstructed. J. Biogeogr. 2003, 30, 1285–1296. [Google Scholar]
- Taberlet, P.; Fumagalli, L.; Wust-Saucy, A.-G.; Cosson, J.-F. Comparative phylogeography and postglacial colonization routes in Europe. Mol. Ecol. 1998, 7, 453–464. [Google Scholar]
- Crisp, M.; Cook, L.G.; Trewick, S.A. Hypothesis testing in biogeography. Trends Ecol. Evol. 2011, 26, 66–72. [Google Scholar]
- Avise, J.C. Phylogeography: The History and Formation of Species; Harvard University Press: Cambridge, MA, USA, 2000. [Google Scholar]
- Hewitt, G.M. Genetic consequences of climatic oscillations in the Quaternary. Philos. Trans. R. Soc. B 2004, 359, 183–195. [Google Scholar]
- Fleischer, R.C.; McIntosh, C.E.; Tarr, C.L. Evolution on a volcanic conveyor belt: Using phylogeographic reconstructions and K–Ar-based ages of the Hawaiian Islands to estimate molecular evolutionary rates. Mol. Ecol. 1998, 7, 533–545. [Google Scholar]
- Jordan, S.; Simon, C.; Foote, D; Polhemus, D. Molecular systematics and adaptive radiation in Hawaii's endemic damselfly genus Megalagrion (Odonata: Coenagrionidae). Syst. Biol. 2003, 52, 89–109. [Google Scholar]
- Carlquist, S. Island Biology; Columbia University Press: New York, NY, USA, 1974. [Google Scholar]
- Moritz, C.; Patton, J.L.; Schneider, C.J.; Smith, T.B. Diversification of rainforest faunas: An integrated molecular approach. Annu. Rev. Ecol. Syst. 2000, 31, 533–563. [Google Scholar]
- Vittoz, P.; Engler, R. Seed dispersal distances: A typology based on dispersal modes and plant traits. Bot. Helv. 2007, 17, 109–124. [Google Scholar]
- Toon, A.; Hughes, J.M.; Joseph, J. Multilocus analysis of Honeyeaters (Aves: Meliphagidae) highlights spatio-temporal heterogeneity in the influence of biogeographic barriers in the Australian monsoonal zone. Mol. Ecol. 2010, 19, 2980–2994. [Google Scholar]
- Goldberg, J.; Trewick, S.A.; Paterson, A.M. Evolution of New Zealand's terrestrial fauna: A review of molecular evidence. Philos. Trans. R. Soc. B 2008, 363, 3319–3334. [Google Scholar]
- Trewick, S.A.; Morgan-Richards, M. New Zealand biology. In Encyclopedia of Islands; Gillespie, R.G., Clague, D.A., Eds.; University of California Press: Berkeley, CA, USA, 2009; pp. 665–673. [Google Scholar]
- Wallis, G.P.; Trewick, S.A. New Zealand phylogeography: Evolution on a small continent. Mol. Ecol. 2009, 18, 3548–3580. [Google Scholar]
- Trewick, S.A.; Wallis, G.P.; Morgan-Richards, M. The invertebrate life of New Zealand: A phylogeographic approach. Insects 2011, 2, 297–325. [Google Scholar]
- Goldberg, J.; Trewick, S.A.; Powlesland, R.G. Population structure and biogeography of Hemiphaga pigeons (Aves: Columbidae) on islands in the New Zealand region. J. Biogeogr. 2011, 38, 285–298. [Google Scholar]
- Morgan-Richards, M.; Trewick, S.A.; Stringer, I.A. Geographic parthenogenesis and the common tea-tree stick insect of New Zealand. Mol. Ecol. 2010, 19, 1227–1238. [Google Scholar]
- Trewick, S.A.; Gibb, G.C. Vicars, tramps and assembly of the New Zealand avifauna: A review of molecular phylogenetic evidence. Ibis 2010, 152, 226–253. [Google Scholar]
- Goldberg, J. Speciation and Phylogeography in the New Zealand Archipelago. Ph.D. Thesis, Massey University, Palmerston North, New Zealand, 2010. [Google Scholar]
- Trewick, S.A.; Morgan-Richards, M. After the deluge: Mitochondrial DNA indicates Miocene radiation and Pliocene adaptation of tree and giant weta (Orthoptera: Anostostomatidae). J. Biogeogr. 2005, 32, 295–309. [Google Scholar]
- Stevens, G.R. New Zealand Adrift: The Theory of Continetal Drift in a New Zealand Setting; A H. & A.W. Reed LTD: London, UK, 1980. [Google Scholar]
- Campbell, H.J.; Andrews, P.B.; Beu, A.G.; Maxwell, P.A.; Edwards, A.R.; Laird, M.G.; de Hornibrook, N.B.; Mildenhall, D.C.; Watters, W.A.; Buckeridge, J.S.; et al. Cretaous-Cenozoic Geology and Biostratigraphy of the Chatham Islands, New Zealand; Institute of Geology & Nuclear Science Monograph; INGS: Lower Hutt, New Zealand, 1993; Volume 2. [Google Scholar]
- Campbell, H.J. Fauna and flora of the Chatham Islands: Less than 4 MY old? Geol. Soc. N. Z. Misc. Publ. 1998, 97, 15–16. [Google Scholar]
- Campbell, H.J.; Begg, J.G.; Beu, A.G.; Carter, R.M.; Davies, G.; Holt, K.; Landis, C.; Wallace, C. On the turn of a scallop. Geol. Soc. N. Z. Misc. Publ. 2006, 121, 9. [Google Scholar]
- Holt, K.A. The Quaternary History of Chatham Islands, New Zealand. Ph.D. Thesis, Massey University, Palmerston North, New Zealand, 2008. [Google Scholar]
- Trewick, S.A. Molecular evidence for dispersal rather than vicariance as the origin of flightless insect species on the Chatham Islands, New Zealand. J. Biogeogr. 2000, 27, 1189–1200. [Google Scholar]
- Hewitt, G.M. Speciation, hybrid zones and phylogeography—or seeing genes in space and time. Mol. Ecol. 2001, 10, 537–549. [Google Scholar]
- Trewick, S.A.; Bland, K.J. Fire and slice: Paleogeography for biogeography at New Zealand's North Island/South Island juncture. J. R. Soc. N. Z. 2011. in press. [Google Scholar]
- Sunnucks, P.; Hales, D.F. Numerous transposed sequences of mitochondrial Cytochrome Oxidase I-II in aphids of the genus Sitobion (Hemiptera: Aphididae). Mol. Biol. Evol. 1996, 13, 510–524. [Google Scholar]
- Simon, C.; Frati, F.; Beckenbach, A.; Crespi, B.; Liu, H.; Flook, P. Evolution, weighting and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann. Entomol. Soc. Am. 1994, 87, 651–701. [Google Scholar]
- Rambaut, A. Se-Al: Sequence Alignment Editor, 1996. Available online: http://tree.bio.ed.ac.uk/software/seal/ (accessed on 2008).
- Ronquist, F.; Huelsenbeck, J.P. MrBayes3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar]
- Guindon, S.; Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar]
- Posada, D. jModelTest: Phylogenetic model averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar]
- Swofford, D.L. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods) Version 4; Sinauer Associates: Sunderland, MA, USA, 1998. [Google Scholar]
- Rozas, J.; Sánchez-DelBarrio, J.C.; Messequer, X.; Rozas, R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 2003, 19, 2496–2497. [Google Scholar]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987. [Google Scholar]
- Groll, E.K.; Günther, K.K. Ordnung Saltatoria (Orthoptera), Heuschrecken, Springschrecken. In Lehrbuch der Speziellen Zoologie, Band 1: Wirbellose Tiere, 5.Teil: Insecta. 2.Aufl.; Dathe, H.H., Ed.; Spektrum Verlag Heidelberg: Berlin, Germany, 2003; pp. 261–290. [Google Scholar]
- Key, K.H.L. Taxonomy of the genus Phaulacridium and a related new genus (Orthoptera: Acrididae). Invertebr. Syst. 1992, 6, 197–243. [Google Scholar]
- Westerman, M.; Ritchie, J.M. The taxonomy, distribution and origins of two species of Phaulacridium (Orthoptera: Acrididae) in the South Island of New Zealand. Biol. J. Linn. Soc. 1984, 21, 283–298. [Google Scholar]
- Hudson, L. A systematic revision of the New Zealand Dermaptera. J. R. Soc. N. Z. 1973, 3, 219–254. [Google Scholar]
- Johns, P.M. The cockroaches of New Zealand. Rec. Canterbury Mus. 1966, 8, 93–136. [Google Scholar]
- Britton, E.B. The Carabidae (Coleoptera) of New Zealand, Part III–A revision of the tribe Broscini. Trans. R. Soc. N. Z. 1949, 77, 533–581. [Google Scholar]
- Larochelle, A.; Larivière, M.-C. Carabidae (Insecta: Coleoptera): Catalogue; Fauna of New Zealand; Manaaki Whenua Press: Lincoln, Canterbury, New Zealand, 2001; Volume 43, pp. 44–61. [Google Scholar]
- Hutchison, M.A.S. Habitat Use, Seasonality and Ecology of Carabid Beetles (Coleoptera: Carabidae) in Native Forest Remnants, North Island, New Zealand. Master Thesis, Massey University, Palmerston North, New Zealand, 2001. [Google Scholar]
- Holloway, B.A. A Systematic Revision of the New Zealand Lucanidae (Insecta: Coleoptera); Dominion Museum: Wellington, New Zealand, 1961. [Google Scholar]
- Craw, R.C. Molytini (Insecta: Coleoptera: Curculionidae: Molytinae); Fauna of New Zealand; Manaaki Whenua Press: Lincoln, Canterbury, New Zealand, 1999; Volume 39. [Google Scholar]
- Marris, J.W.M.; Johnson, P.J. A revision of the New Zealand click beetle genus Amychus Pascoe 1876 (Coleoptera: Elateridae: Denticollinae): With a description of a new species from the Three Kings Islands. Zootaxa 2010, 2331, 35–56. [Google Scholar]
- Chinn, W.G.; Gemmell, N.J. Adaptive radiation within New Zealand endemic species of the cockroach genus Celatoblatta Johns (Blattidae): A response to Plio-Pleistocene mountain building and climate change. Mol. Ecol. 2004, 13, 1507–1518. [Google Scholar]
- Shepherd, L.D.; de Lange, P.J.; Perrie, L.R. Multiple colonizations of a remote oceanic archipelago by one species: How common is long-distance dispersal? J. Biogeogr. 2009, 36, 1972–1977. [Google Scholar]
- Liggins, L.; Chapple, D.G.; Daugherty, C.H.; Ritchie, P.A. Origin and post-colonization evolution of the Chatham Islands skink (Oligosoma nigriplantare nigriplantare). Mol. Ecol. 2008, 17, 3290–3305. [Google Scholar]
- Heenan', P.B.; Mitchell, A.D.; de Lange, P.J.; Keeling, J.; Paterson, A.M. Late-Cenozoic origin and diversification of Chatham Islands endemic plant species revealed by analyses of DNA sequence data. N. Z. J. Bot. 2010, 48, 83–136. [Google Scholar]
- Vink, C.J.; Paterson, A.M. Combined molecular and morphological phylogenetic analyses of the New Zealand wolf spider genus Anoteropsis (Araneae: Lycosidae). Mol. Phylogenet. Evol. 2003, 28, 576–587. [Google Scholar]
- Arensburger, P.; Simon, C.; Holsinger, K. Evolution and phylogeny of the New Zealand cicada genus Kikihia Dugdale (Homoptera: Auchenorrhyncha: Cicadidae) with special reference to the origin of the Kermadec and Norfolk Islands' species. J. Biogeogr. 2004, 31, 1769–1783. [Google Scholar]
- Nolan, L.; Hogg, I.A.; Sutherland, D.L.; Stevens, M.I.; Schnabel, K.E. Allozyme and mitochondrial DNA variability within the New Zealand damselfly genera Xanthocnemis, Austrolestes, and Ischnura (Odonata). N. Z. J. Zool. 2007, 34, 371–380. [Google Scholar]
- Trewick, S.A.; Goldberg, J.; Morgan-Richards, M. Fewer species of Argosarchus and Clitarchus stick insects (Phasmida, Phasmatinae): Evidence from nuclear and mitochondrial DNA sequence data. Zool. Scr. 2005, 34, 438–491. [Google Scholar]
- Stevens, M.I.; Hogg, I.D. Population genetic structure of New Zealand's endemic corophiid amphipods: evidence for allopatric speciation. Biol. J. Linn. Soc. 2004, 81, 119–133. [Google Scholar]
- McGaughran, A.; Hogg, I.D.; Stevens, M.I.; Chadderton, W.L.; Winterbourn, M.J. Genetic divergence of three freshwater isopod species from southern New Zealand. J. Biogeogr. 2006, 33, 23–30. [Google Scholar]
- Trewick, S.A. Sympatric flightless rails Gallirallus dieffenbachii and G. modestus on the Chatham Islands, New Zealand; morphometrics and alternative evolutionary scenarios. J. R. Soc. N. Z. 1997, 27, 451–464. [Google Scholar]
- Miller, H.C.; Lambert, D.M. A molecular phylogeny of New Zealand's Petroica (Aves: Petroicidae) species based on mitochondrial DNA sequences. Mol. Phylogenet. Evol. 2006, 40, 844–855. [Google Scholar]
- Boon, W.M.; Kearvell, J.C.; Daugherty, C.H.; Chambers, G.K. Molecular Systematics and Conservation of Kakariki (Cyanoramphus spp.); Department of Conservation: Wellington, New Zealand, 2001; Volume 176, pp. 1–46. [Google Scholar]
- Boon, W.M.; Robinet, O.; Rawlence, N.; Bretagnolle, V.; Norman, J.A.; Christidis, L.; Chambers, G.K. Morphological, behavioural and genetic differentiation within the Horned Parakeet (Eunymphicus cornutus) and its affinities to Cyanoramphus and Prosopeia. Emu 2008, 108, 251–260. [Google Scholar]
- Johansen, S.; Hytteborn, H. A contribution to the discussion of biota dispersal with drift ice and driftwood in the North Atlantic. J. Biogeogr. 2001, 28, 105–115. [Google Scholar]
- Trewick, S.A.; Paterson, A.M.; Campbell, H.J. Hello New Zealand. J. Biogeogr. 2007, 34, 1–6. [Google Scholar]
- Craw, R.C. Continuing the synthesis between panbiogeography, phylogenetic systematics and geology as illustrated by empirical studies on the biogeography of New Zealand and the Chatham Islands. Syst. Zool. 1988, 37, 291–310. [Google Scholar]
- Campbell, H.J. Geology Chatham Islands: Heritage and Conservation, 2nd ed.; Miskelly, C., Ed.; Department of Conservation: Wellington, New Zealand, 2008; pp. 35–52. [Google Scholar]
- Skarpaas, O.; Silverman, E.J.; Jongejans, E.; Shea, K. Are the best dispersers the best colonizers? Seed mass, dispersal and establishment in Carduus thistles. Evol. Ecol. 2011, 25, 155–169. [Google Scholar]
- Homepage of Phoenix evolutionary ecology and genetics group. Available online: http://www.massey.ac.nz/∼strewick/ (accessed on 4 July 2011).
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Goldberg, J.; Trewick, S.A. Exploring Phylogeographic Congruence in a Continental Island System. Insects 2011, 2, 369-399. https://doi.org/10.3390/insects2030369
Goldberg J, Trewick SA. Exploring Phylogeographic Congruence in a Continental Island System. Insects. 2011; 2(3):369-399. https://doi.org/10.3390/insects2030369
Chicago/Turabian StyleGoldberg, Julia, and Steven A. Trewick. 2011. "Exploring Phylogeographic Congruence in a Continental Island System" Insects 2, no. 3: 369-399. https://doi.org/10.3390/insects2030369
APA StyleGoldberg, J., & Trewick, S. A. (2011). Exploring Phylogeographic Congruence in a Continental Island System. Insects, 2(3), 369-399. https://doi.org/10.3390/insects2030369





