1. Introduction
In social insects, individuals cooperate through division of labor to perform collective tasks such as brood care and defense, with chemosensation playing a central role in regulating recognition and communication among colony members [
1]. Chemical signals are primarily detected by insects through two main chemosensory mechanisms: olfaction, which is responsible for detecting volatile molecules and mediates long-distance communication, and gustation, which recognizes soluble substances and regulates behaviors such as feeding and oviposition [
2]. As one of the most important sensory organs in insects, the antennae carry multiple functions including olfaction, touch, and temperature–humidity perception. Their input signals constitute one of the most fundamental sensory pathways in the central nervous system [
3,
4]. Olfactory receptor neurons (ORNs) can specifically recognize chemical cues related to food, oviposition sites, and mates, thereby activating downstream neural circuits and triggering corresponding behaviors [
5].
Hymenoptera (comprising sawflies, wasps, ants, and bees) represents one of the four major hyperdiverse insect groups. As parasitoids, predators, and pollinators, they play critical roles in terrestrial ecosystems and possess significant economic value [
6]. The antennal sensilla structures in this order often exhibit remarkable sexual dimorphism corresponding to their ecological roles. For instance, in Pteromalidae [
7], differences in sensilla between sexes may be associated with distinct host-location strategies; in Torymidae [
8], the sex-specific differentiation of sensilla provides crucial morphological clues for understanding their host-searching behavior; whereas in Bethylidae [
9], the sensilla on female antennae may be specialized for short-range host detection, while male antennae appear adapted for long-range mate location. Correspondingly, their olfactory systems also demonstrate significant differentiation at the molecular level. A notable example is observed in ants, where distinct caste-specific differences exist in odorant receptor gene expression; in
Camponotus floridanus (Buckley, 1866) (Hymenoptera: Formicidae) and
Harpegnathos saltator (Jerdon, 1851) (Hymenoptera: Formicidae), nearly all olfactory receptors (ORs) are expressed in worker ants, while males express only approximately one-third of these receptors [
10]. Bumblebees (Hymenoptera: Apidae), being truly eusocial and generalist-feeding pollinators of considerable ecological importance, utilize chemosensory mechanisms to mediate both plant–pollinator interactions and intranidal social communication [
11].
Bombus terrestris (Linnaeus, 1758) (Hymenoptera: Apidae), serving as a vital ecological pollinator for numerous crops, has attracted substantial research attention regarding the behavioral and physiological aspects of olfactory systems across different castes (queens, workers, and males) [
12]. However, a systematic comparative analysis of caste-specific differentiation in both antennal sensilla ultrastructure and OR gene expression profiles remains lacking. This knowledge gap hinders comprehensive understanding of the structural and functional integration underlying chemical communication mechanisms in their social behaviors.
To systematically investigate the ultrastructural characteristics of antennal sensilla in different castes of B. terrestris within the context of sex and division of labor variations, this study first employed scanning electron microscopy (SEM) for high-resolution morphological examination of worker, male, and queen antennae. Subsequently, we compared the expression profiles of OR genes in the head tissues (including antennae) across these three castes through high-throughput transcriptome sequencing. By integrating morphological and molecular biological data, this research aims to elucidate the structural and functional adaptations of the olfactory system in B. terrestris under different sexes and social roles, thereby providing new experimental evidence for deeper understanding of caste-specific regulatory mechanisms in olfactory pathways.
2. Materials and Methods
2.1. Test Insects
The B. terrestris (workers, males, and queens) used in this study were provided by the Experimental Apiary of the College of Animal Science, Shanxi Agricultural University.
2.2. Sample Preparation and Observation Methods
Ten healthy and intact workers, males, and queens were randomly collected from healthy and robust colonies of B. terrestris. Antennae from both sides of each individual were included as experimental samples. Under a stereomicroscope, antennae were completely detached at the scape and subsequently fixed with 2.5% glutaraldehyde solution for 24 h. After fixation, the samples were washed three times with PBS phosphate buffer, each wash lasting 10 min. This was followed by sequential dehydration through an ethanol gradient series of 30%, 50%, 70%, 80%, 90%, 95%, and 100%, with each step maintained for 15 min. The 100% ethanol dehydration step was repeated twice. The dehydrated samples were subjected to critical point drying using a JED-320 freeze dryer (JEOL Ltd., Tokyo, Japan). Each sample was mounted on an aluminum stub with conductive carbon adhesive tape and sputter-coated with a 10-nm gold layer using an SBC-12 coating system (KYKY Technology Co., Ltd., Beijing, China) to ensure optimal surface conductivity. The metallized samples were examined under high vacuum conditions with a JSM-7800F field emission scanning electron microscope (JEOL Ltd., Tokyo, Japan), and images were acquired at an accelerating voltage of 5–20 kV.
Additionally, six workers, males, and queens were respectively selected, and their antennae were excised. The antennae were rinsed three times with 75% ethanol, air-dried at room temperature (25 ± 1 °C), and then mounted on glass slides. Panoramic imaging was performed using a KS-X1500S 3D digital depth-of-field microscope (Nanjing Kaishimai Technology Co., Ltd., Nanjing, China).
2.3. Image Processing
SEM images were processed using Microsoft Visio 2024 (Microsoft Corporation, Redmond, WA, USA) to optimize global contrast and unify the background without altering the original morphological features. Antennal parameters were measured using ImageJ software, version 1.54 g (National Institutes of Health, Bethesda, MD, USA).
2.4. Statistical Analysis
The data analysis in this study included both quantitative measurements and qualitative observations. Quantitative analysis focused on sensillum length and total antennal length: the total length of six independent antennae from workers, males, and queens was measured, and for each sensillum type, ten individual structures were randomly measured on different antennal segments to obtain dimensional data. Meanwhile, comparisons of sensillum density were based on qualitative observation of SEM images. One-way ANOVA was performed using SPSS 27 software, version 27.0 (IBM Corp., Armonk, NY, USA), and all quantitative data are presented as the mean ± standard error of the mean (mean ± SEM).
2.5. Total RNA Extraction and Quality Assessment
Thirty workers, males, and queens of B. terrestris were respectively collected. After anesthetizing on ice overlaid with tin foil, their heads (including antennae) were excised, immediately frozen in liquid nitrogen, and stored at −80 °C. Total RNA was extracted from the bumblebee heads using a Trizol reagent kit. Three independent RNA samples were prepared for each caste: each worker sample (designated W_CJ_1 to W_CJ_3) pooled heads from 5 workers, each male sample (D_CJ_1 to D_CJ_3) pooled heads from 5 males, and each queen sample (Q_CJ_1 to Q_CJ_3) pooled heads from 5 queens. RNA purity and concentration were measured using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), while RNA integrity was precisely assessed with an Agilent 2100 Bioanalyzer (Santa Clara, CA, USA). Only samples passing quality control were subjected to transcriptome sequencing.
2.6. Transcriptome Library Construction and Sequencing
High-quality total RNA that passed quality control was used for mRNA enrichment with oligo(dT)-coupled magnetic beads. The enriched mRNA was fragmented, and first-strand cDNA was synthesized using random primers. Second-strand cDNA synthesis was then performed in a reaction system containing buffer, dNTPs, RNase H, and DNA polymerase I. The resulting double-stranded cDNA products were purified using AMPure XP beads (Beckman Coulter, Inc., Brea, CA, USA). The cDNA ends were blunted using the synergistic action of T4 DNA polymerase and Klenow DNA polymerase, followed by 3′ adenylation and adapter ligation. The ligated products were subsequently size-selected with AMPure XP beads. The second cDNA strand was selectively degraded using Uracil-Specific Excision Reagent (USER) enzyme, and the resulting library was amplified via PCR to construct standardized sequencing libraries. After passing quality inspection, the libraries were sequenced on an Illumina NovaSeqTM 6000 platform (San Diego, CA, USA) using a paired-end 150 bp (PE150) read length configuration for data acquisition. The transcriptome sequencing service was provided by Beijing Novogene Co., Ltd. (Beijing, China), and the raw sequencing data have been deposited in the NCBI SRA database under accession number PRJNA1345014.
2.7. Identification of Olfactory Receptor Genes
2.8. Sequence and Phylogenetic Analysis of Olfactory Receptor Genes
To investigate the evolutionary relationships among the screened differentially expressed OR genes in
B. terrestris, the previously annotated OR genes from the bumblebee transcriptome, and the ORs of other insects, this study retrieved amino acid sequences of known ORs from 12 Hymenoptera species (including
B.
terrestris,
B.
affinis,
B.
huntii,
B.
impatiens,
B.
pyrosoma,
B.
vosnesenskii,
B.
bifarius,
B.
pascuorum,
F.
varia,
A.
dorsata,
A.
florea, and
A.
mellifera) from the NCBI database. These downloaded amino acid sequences, along with the
B.
terrestris OR sequences, were comprehensively aligned using MEGA-7.0.26 software (Temple University, Philadelphia, PA, USA). A phylogenetic tree was subsequently constructed with the neighbor-joining (NJ) method in MEGA 7.0. The visualization and refinement of the phylogenetic tree were performed using the iTOL v7 platform (
https://itol.embl.de/, accessed on 26 September 2025).
2.9. Quantitative Real-Time PCR Analysis
Quantitative real-time PCR (qPCR) was performed using the same total RNA samples returned by the sequencing company. cDNA was synthesized by reverse transcription using the PrimeScript
TM RT reagent kit with gDNA Eraser (Perfect Real Time) (Takara Bio Inc., Shiga, Japan, Cat. No. RR047A). This cDNA served as the template for amplification with the TB Green Premix Ex Taq
TM II (Tli RNaseH Plus) (Takara Bio Inc., Shiga, Japan, Cat. No. RR820A). The 10 μL reaction mixture contained: 5 μL of TB Green Premix Ex Taq (Tli RNaseH Plus), 0.5 μL each of forward and reverse primers (10 μmol/L), 500 ng of cDNA, and ddH
2O to a final volume of 10 μL. The thermal cycling protocol was as follows: 95 °C for 3 min; 40 cycles of 95 °C for 5 s, 57 °C for 30 s, and 72 °C for 30 s. The melting curve was generated under the following conditions: 95 °C for 10 s, followed by 52 °C for 5 s. The bumblebee
s18 Ribosomal gene (XM_003400778.3) was selected as the internal reference gene for this study [
13]. Each sample was analyzed with three biological replicates and three technical replicates. The primer sequences used are listed in
Table 1.
4. Discussions
This study integrated the micromorphological characteristics of antennae from three castes of B. terrestris with the expression profiles of OR genes from head transcriptomes. By bridging sensory structures and molecular mechanisms, we systematically revealed the multi-level adaptive evolution of their olfactory system in relation to social division and reproductive strategies.
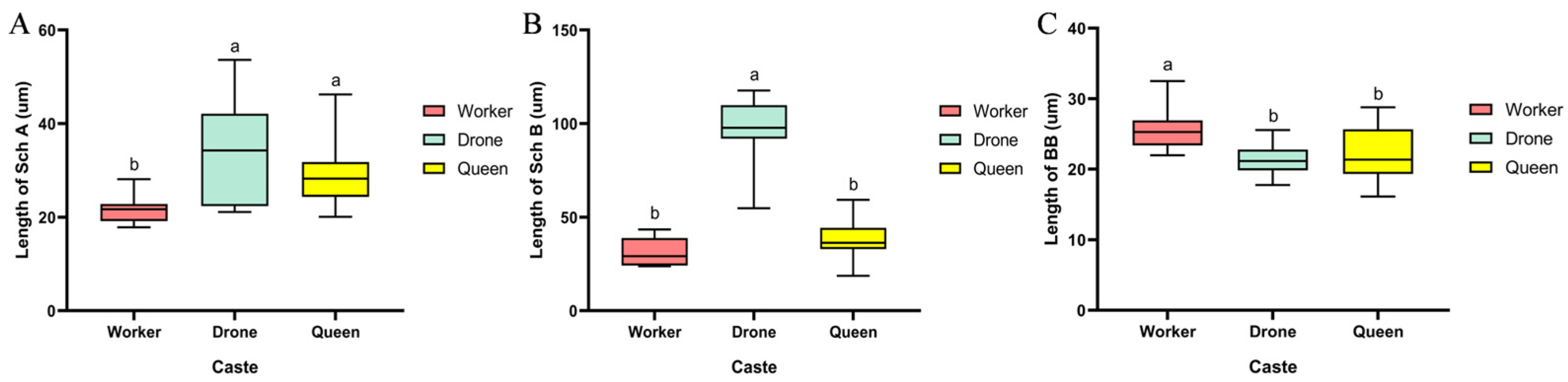
Our study found that the total antennal length of both workers and queens was significantly greater than that of drones, with queens possessing the longest antennae. Previous research has shown that under identical conditions, antennal length is proportional to body size, suggesting that the longer antennae of queens may be partially attributed to their larger body size [
21]. However, this morphological difference is more likely closely associated with their behavioral complexity. Workers are required to perform multiple tasks, including in-nest activities such as brood care and nest maintenance, as well as out-of-nest duties like foraging and collection. Queens, on the other hand, need to execute reproductive behaviors including mating and oviposition. These complex behaviors are highly dependent on a sophisticated sensory system to accurately perceive both internal and external environmental cues. Longer antennae may imply a larger surface area for sensilla distribution and more acute chemosensory capabilities, thereby enhancing the accuracy of behavioral decision-making. In contrast, the primary role of drones is limited to mating with virgin queens, resulting in relatively simpler sensory requirements. This likely has driven the simplification of their antennal structure, reflecting an evolutionary strategy characterized by a “function-structure” trade-off.
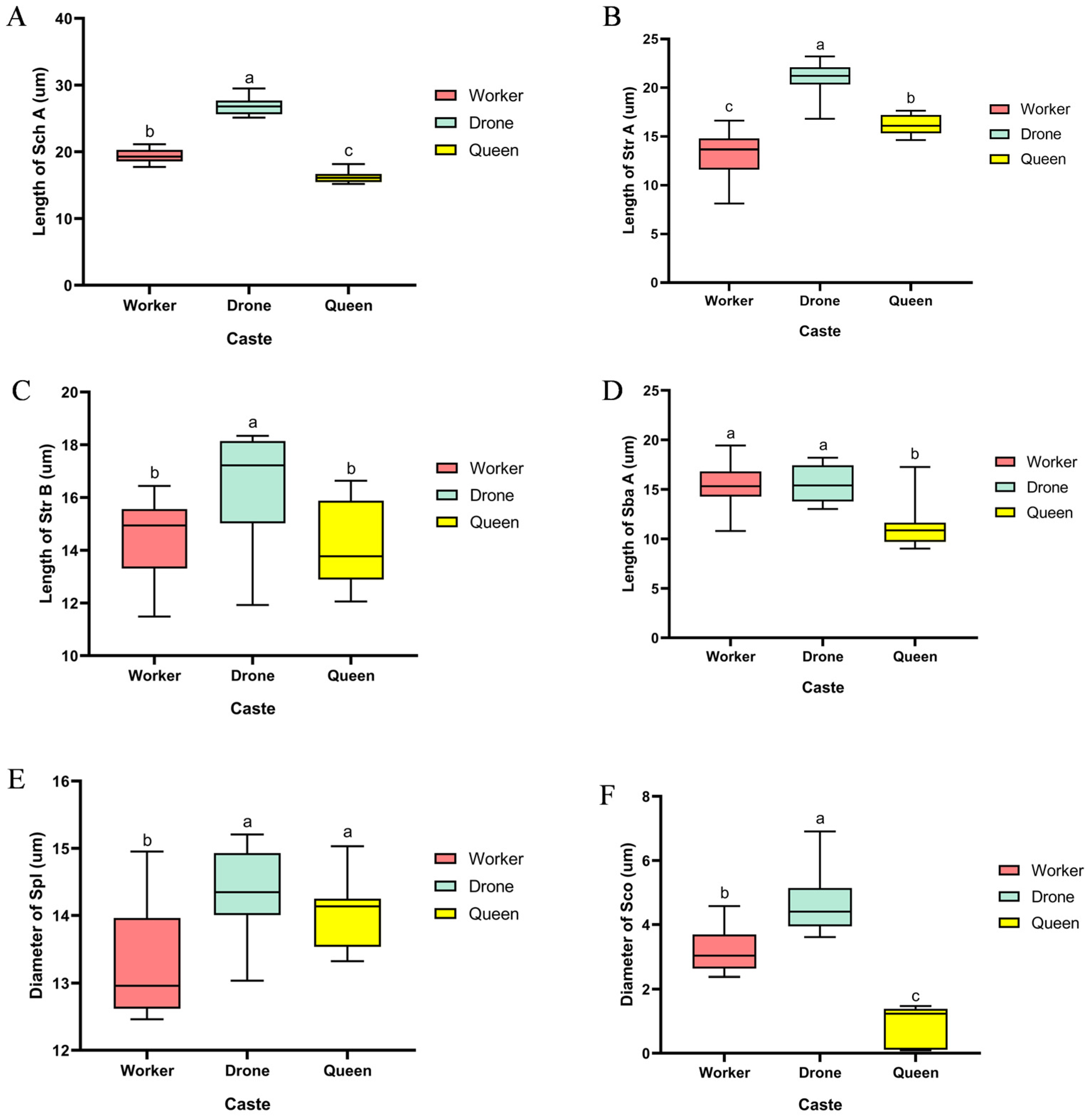
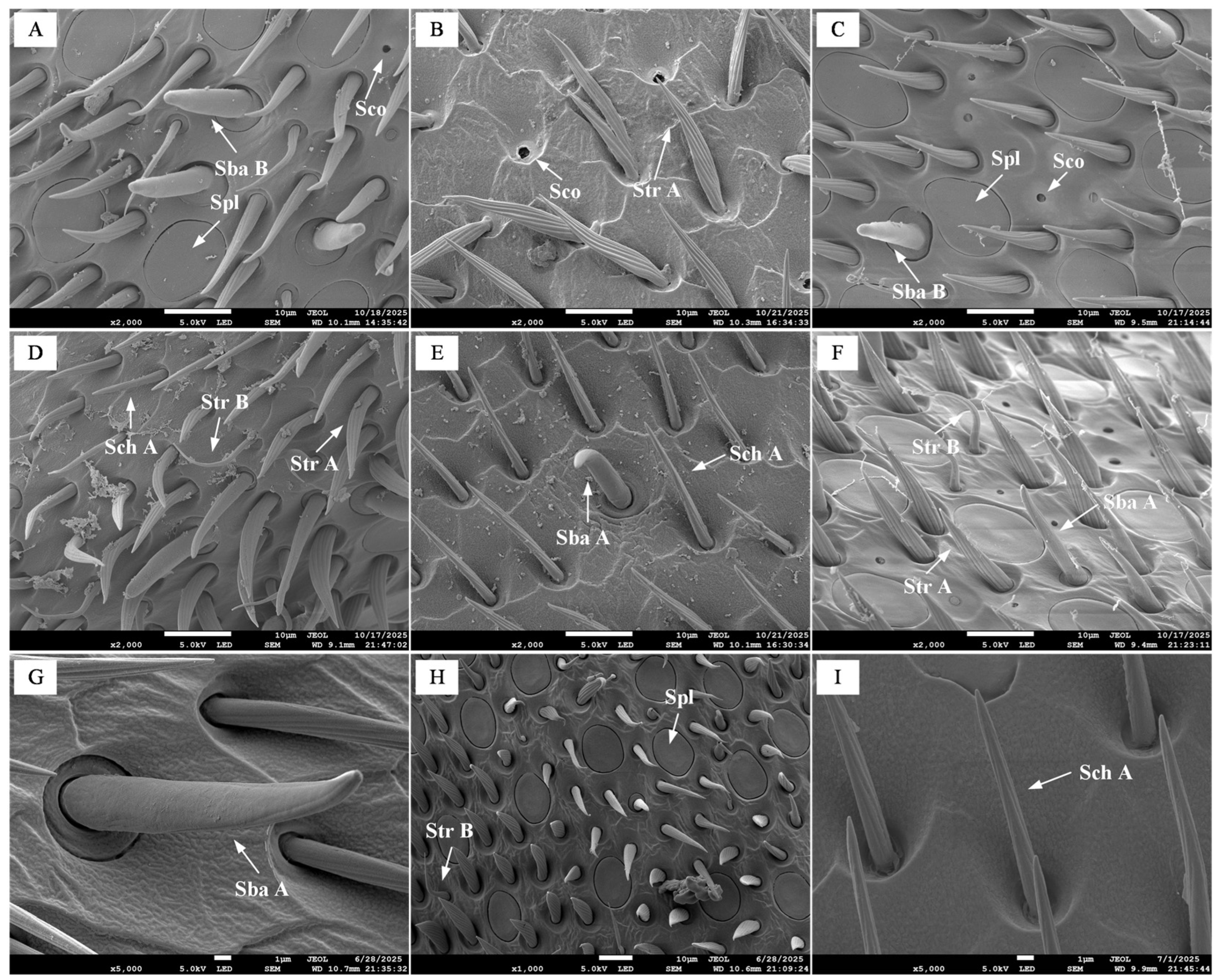
Sch were identified on the scape, pedicel, and flagellum of workers, drones, and queens. Previous studies indicate that this type of sensilla possesses dual functionality in both mechanoreception and contact chemoreception [
14]. Our study revealed that the length of Sch on all antennal segments of drones was significantly greater than that of workers and queens. This morphological difference likely reflects their heightened need for perceiving chemical or mechanical signals during antennal contact, potentially functioning in mating behavior or the recognition of queen cuticular cues. Compared to the scape, BB were present on the pedicel of all three castes, with workers exhibiting significantly greater bristle length than drones and queens. Previous research has established that BB function as proprioceptors, sensing antennal spatial position and regulating its movement [
15]. Consequently, the elongated bristles in workers may enhance their precision in perceiving antennal posture, supporting the more complex flight control demands required for navigating within the nest and hovering between flowers. This indirectly reflects their superior flight capability compared to drones and queens. Str were the most numerous sensilla type on the antennae of
B. terrestris, distributed exclusively on the flagellum and comprising two subtypes, Str A and Str B. This study found that both subtypes were significantly longer in drones than in workers and queens (
p < 0.05). These sensilla have been demonstrated to perceive semiochemicals such as sex pheromones and terpenoids [
19]. Therefore, the marked elongation of Str in drones increases the sensory surface area, likely enhancing the capture efficiency of queen-emitted sex pheromones. This provides a crucial sensory structural foundation for the precise localization of mates during nuptial flights. Sba sensilla, which are sensitive to general odorants such as plant compounds and pheromones [
22], are distributed exclusively on the flagellar segments of the antennae and differ significantly among castes. The length of Sba A was significantly greater in drones than in workers and queens (
p < 0.05), and no Sba B sensilla were detected in drones. This structural pattern suggests a potential functional specialization in the drone olfactory system: the elongated Sba A sensilla may enhance the detection of long-distance, low-concentration odorant molecules (e.g., queen sex pheromones), while the absence of Sba B could streamline the perception pathways for non-reproduction-related odors, thereby sharpening the olfactory focus on key chemical signals associated with reproduction. In contrast, the presence of Sba B sensilla in workers and queens likely facilitates the perception of a diverse array of odors, such as floral scents, nest odors, or social pheromones, accommodating their broader range of behavioral requirements. Spl have been confirmed to be highly sensitive to queen pheromone components, with drones exhibiting significantly higher behavioral sensitivity to these substances than workers in behavioral experiments [
19]. This study found that the diameter of Spl showed no significant difference between drones and queens, but was significantly larger in both castes compared to workers. This structural advantage provides a morphological basis for their high sensitivity to queen pheromones, further supporting the structure-function co-evolution of their olfactory system in fulfilling reproductive tasks. In summary, the caste-specific differentiation in the morphology and quantity of antennal sensilla not only reveals, at the microstructural level, the divergence in olfactory strategies corresponding to “mating specialization” versus “multitask adaptation” in
B. terrestris, but also provides tangible morphological evidence for the co-evolution of sensory systems and reproductive division of labor in social insects. Across the three castes, the Sco shows complementary adaptive traits: drones display the largest Sco diameter, followed by workers, whereas queens exhibit the smallest diameter but the highest spatial density. Studies indicate that the grooved Sco wall, lacking the middle and inner cuticular layers, may constitute an entry path for odor molecules [
23]; moreover, Sco are thought to mediate the detection of olfactory stimuli from long range [
20]. Based on these findings, we speculate that the enlarged Sco diameter in drones may enhance the capture efficiency and effective detection distance of key unitary signals such as queen sex pheromones, which could reflect a specialized sensory adaptation strategy. In contrast, queens and workers may adopt a “small-aperture, high-density” design, where numerous Sco are packed into the limited antennal surface to potentially form a high-throughput olfactory surveillance network. This configuration could allow parallel processing of diverse chemical cues—such as floral scents, nest odors, and social pheromones—thereby potentially fitting their more complex and variable behavioral niches.
High-throughput sequencing (RNA-seq) has become a standard method for analyzing gene expression profiles across species and conditions [
24]. This study performed transcriptome sequencing on the heads (including antennae) of
B. terrestris workers, drones, and queens, systematically characterizing the expression profiles of olfactory receptor genes under different sexes and social roles. qRT-PCR validation of eight significantly differentially expressed OR genes showed complete consistency with the sequencing data, confirming data reliability. Pairwise comparisons among the three groups identified 7151 DEGs, from which eight significantly differentiated OR genes were pinpointed. Their expression patterns reflect three distinct biological differentiations: the differences between workers and queens primarily represent task specialization within the same sex; the differences between drones and queens reflect the combined effects of both sex and social role; while the differences between workers and drones highlight fundamental physiological disparities driven by sex-specific reproductive strategies, despite their shared out-of-nest foraging behavior.
In the worker-queen comparison, the significant upregulation of
BterOR3,
BterOR5, and
BterOR7 may suggest that workers could have evolved more refined olfactory regulatory capabilities to potentially adapt to efficient out-of-nest foraging. GO enrichment analysis revealed “extracellular region” as the top term in the cellular component category, directly corresponding to the initial stage of olfactory perception—where odorant-binding proteins (OBPs) transport odor molecules across the sensillum lymph and activate ORs on the neuronal membrane to initiate neural signaling [
25]. This indicates the coordinated enhancement of genes associated with the extracellular region, establishing a precondition for efficient chemical perception. The concurrent enrichment of “heme binding” and the “Drug metabolism-cytochrome P450” pathway further demonstrates that while enhancing olfaction, workers also upregulate the P450 detoxification system [
26] to cope with plant secondary metabolites in nectar and pollen, as well as potential pesticide residues. This integrated adaptation enables a seamless “perception-intake-detoxification” process.
In the drone-queen comparison, a total of seven OR genes (
BterOR3,
BterOR4,
BterOR5,
BterOR6,
BterOR7,
BterOR8, and
BterOR9) were upregulated in drones, representing the highest number of upregulated genes among all comparison groups. This pattern may suggest a high degree of specialization in their olfactory system, which could align closely with the drone’s primary biological mission of mating [
27]. Unlike queens, which remain in the nest throughout their lives, drones must detect the extremely low concentrations of sex pheromones released by virgin queens in open outdoor environments [
28]. Consequently, this large-scale upregulation of olfactory receptor genes could be interpreted as an adaptive evolutionary response at the olfactory level to potentially maximize their reproductive success. The most significantly enriched term in the GO molecular function category, “transmembrane signaling receptor activity,” indicates that the large-scale upregulation of olfactory receptor genes in drones is not an isolated event, but rather a concentrated manifestation of adaptive evolution within the entire transmembrane signaling perception system. The ultimate objective is to achieve efficient and precise recognition of mating signals. Conversely, the low expression of olfactory receptor genes in queens may correspond to their ecological niche as “in-nest egg-laying machines”. Studies have shown that increased larval pheromones within the nest prompt worker bees to extend their feeding time to the queen, thereby enhancing her oviposition rate [
29]. Within this social structure, which heavily relies on workers for information transfer, the queen has no need to maintain a complex olfactory receptor system for direct perception of external odors. Therefore, the down-regulation of OR expression likely represents an energy-saving adaptation to the queen’s caste-specific role as an in-hive “egg-laying specialist”: by minimizing the metabolic cost of maintaining an extensive olfactory receptor repertoire, she can reallocate resources to oogenesis and oviposition, while relying on worker-mediated information transfer for within-nest chemical communication, thereby maximizing overall reproductive efficiency. Within the drone upregulation profile, the expression levels of
BterOR3 and
BterOR4 were the most prominent. It is hypothesized that these two receptors possess ultra-high affinity for key components of the queen sex pheromone, serving as core molecules mediating the capture of long-distance, low-concentration signals. This represents a crucial evolutionary strategy for drones to maximize their reproductive success.
Although both workers and drones exhibit “out-of-nest” behavior, the underlying motivations are fundamentally distinct: workers leave the nest to forage for nectar sources [
30], whereas drones do so to mate with queens and reproduce [
31]. This fundamental difference drives their olfactory systems toward opposite evolutionary poles: “generalization” versus “specialization.” In the worker-drone comparison, three olfactory receptor genes (
BterOR3,
BterOR4, and
BterOR10) were significantly upregulated in drones. Notably, the expression levels of
BterOR3 and
BterOR4 in drones were significantly higher than those in both workers and queens. This suggests that drones may concentrate their perceptual resources on a limited number of highly specific receptors, forming a “minimalist yet specialized” sex pheromone detection module. In contrast, workers must utilize the same olfactory “hardware” to perform diverse tasks such as brood care, nest maintenance, and foraging from various floral sources. Consequently, their OR expression profile must maintain a “broad and plastic” balanced state [
32]. Behavioral polymorphism has been confirmed as a major driver of brain gene expression in bees [
33]. The multi-task, multi-stage behavioral division of labor in workers necessitates that their olfactory receptor expression profile retains a degree of “balance” and “plasticity” to flexibly respond to varying sensory demands. Therefore, workers maintain only basal expression levels of specific pheromone receptors, while drones adopt an “overexpression–ultra-high affinity” strategy to amplify mating signals. This contrast in molecular-level expression mechanistically explains how the same out-of-nest behavior, driven by divergent survival objectives, can shape fundamentally distinct evolutionary trajectories in the olfactory system.
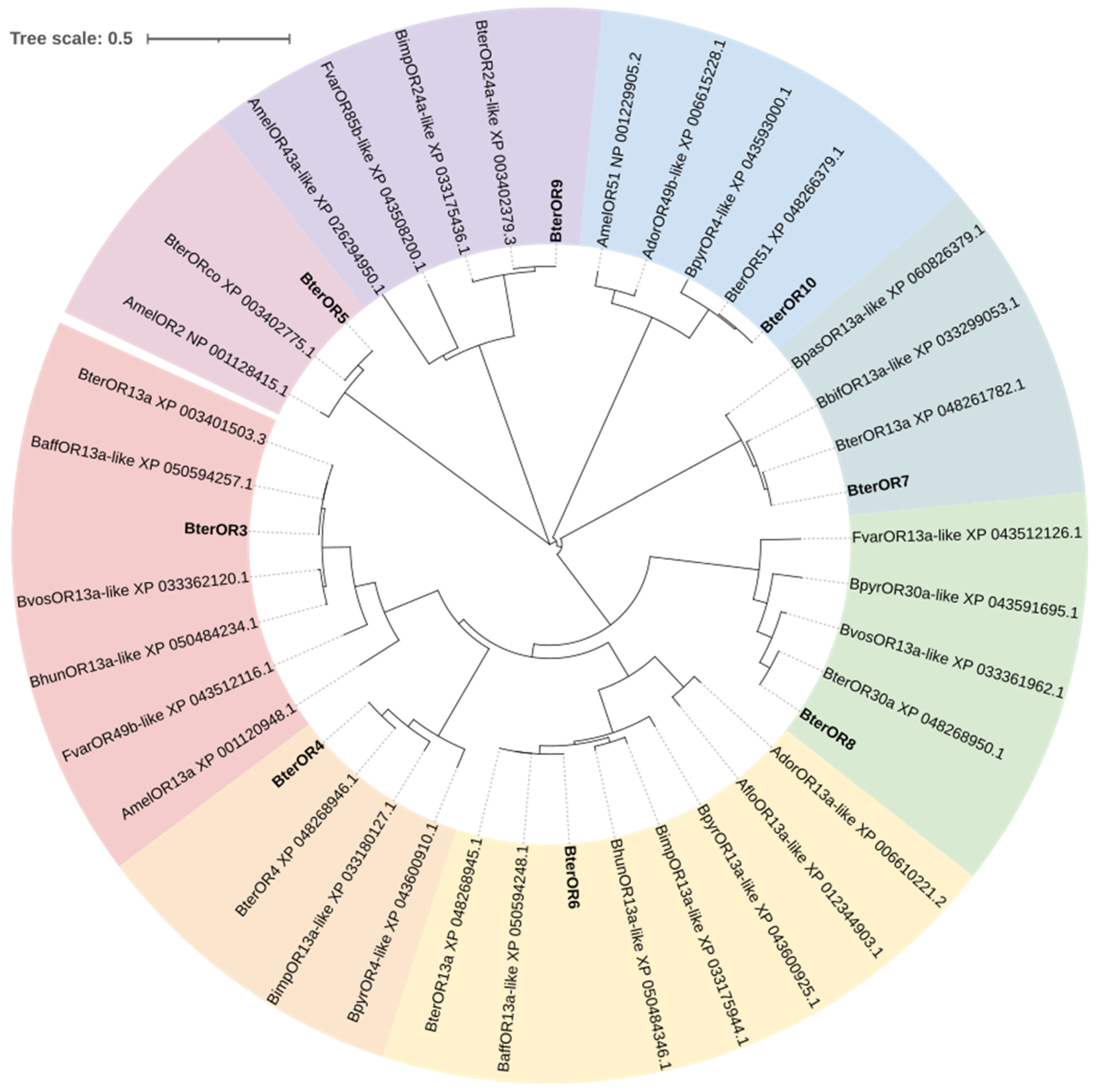
The phylogenetic tree clusters BterOR5 within the same clade as AmelOR2 from
Apis mellifera (100% bootstrap support) and BterORco (99% support), suggesting its potential role as a conserved co-receptor in
B. terrestris (
Figure 7). Previous studies have established that AmelOR2, the ortholog of DmelOR83b, functions as an essential auxiliary subunit in functional olfactory receptor complexes and must be co-expressed with the specific receptor AmelOR1 to mediate sensitive responses to the queen pheromone component 9-ODA [
34]. Although BterOR5 shows high sequence similarity with AmelOR2 (91.63% identity), and thus may serve an analogous co-receptor role in pheromone detection, it should be noted that their expression patterns in homologous olfactory neurons remain unverified. Based on this phylogenetic conservation, we hypothesize that key pheromone receptors in
B. terrestris may similarly require assembly with BterOR5 into heteromeric complexes to achieve full functionality, positioning this gene as a candidate target for future “function-ligand” screening studies. BterOR4 showed 100% identity with its reference sequence, confirming the reliability of its annotation. BterOR7, BterOR8, and BterOR9 clustered with BterOR13, BterOR30a, and BterOR24a-like, respectively, with extremely high bootstrap support. However, their amino acid sequence identity was below 85%, suggesting that they originated from recent gene duplication events followed by rapid divergence. This pattern is consistent with the “birth-and-death” evolutionary model observed in insect olfactory receptor (OR) families [
35,
36]. BterOR3 and BterOR6 formed two distinct, deeply branching clades (with 99% and 100% bootstrap support, respectively) with different accessions of BterOR13a and BaffOR13a-like. This topology reveals that they diverged synchronously following ancestral gene duplication during the radiation of the
Bombus genus, representing a typical “radiation-retention” evolutionary pattern [
37]. BterOR10 and BterOR51 clustered together with moderate support within a highly supported branch containing BpyrOR4-like, suggesting they belong to a conserved OR subfamily within
Bombus. Their functions may exhibit overlap or redundancy. We therefore hypothesize that the function of BterOR10 is likely associated with other members of this subfamily.
Although this study provides comprehensive evidence for caste-specific olfactory adaptation in B. terrestris, several limitations should be noted. First, the association between upregulated BterOR3/5/7 expression and worker foraging behavior is primarily based on cross-caste comparative evidence. While this research design effectively elucidates the influences of sex and social roles, incorporating more multidimensional validation in the future would be more persuasive. Second, due to sampling constraints, this study does not include transcriptomic dynamic data from different developmental stages of worker bees. Subsequent research incorporating temporal expression profiles of newly emerged workers and mature foragers would more clearly reveal the developmental temporal characteristics of OR gene expression regulation. Furthermore, the scanning electron microscopy analysis has certain limitations: the assessment of sensilla density in this study was primarily based on qualitative observation, lacking systematic quantitative statistical analysis. Future research should employ image-based morphometric methods to accurately quantify sensilla density through standardized counting and spatial distribution analysis. Additionally, although BterOR5 and AmelOR2 show high sequence similarity, the conservation of their expression patterns in olfactory neurons still requires direct validation through experiments such as in situ hybridization. These limitations indicate directions for future research, including functional validation through gene knockdown, establishment of gene expression profiles at different behavioral stages, and comparative studies of co-receptor expression patterns across species.
Integrating antennal ultrastructure with high-throughput expression data, this study constructs a complete chain of evidence for “morphology-molecule” co-evolution in B. terrestris. Drones adopt a “quality-priority” olfactory strategy—characterized by longer sensilla trichodea and basiconica, larger-diameter sensilla coeloconica, and the high-level expression of seven OR genes—to form a long-range, high-sensitivity detection system specialized for the singular signal of queen sex pheromones. In contrast, workers and queens follow a “quantity-breadth priority” pathway. By retaining a complete repertoire of sensilla types (e.g., Sba B), increasing the density of sensilla coeloconica, and maintaining a relatively balanced OR expression profile, they construct a high-throughput chemical monitoring network capable of simultaneously recognizing floral scents, nest odors, and social pheromones. The high congruence between sensilla morphology and receptor expression across castes reveals, for the first time, how the olfactory system of B. terrestris undergoes coordinated differentiation at both structural and genetic levels. This precise adaptation aligns with their distinct ecological roles—specialized mating versus complex social tasks—thereby establishing a new multi-level paradigm for understanding sensory-behavioral co-evolution in social insects.