Spatiotemporal Population Genomics of the Invasive Whitefly Bemisia tabaci MED in China: Implications for Surveillance and Sustainable Control
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
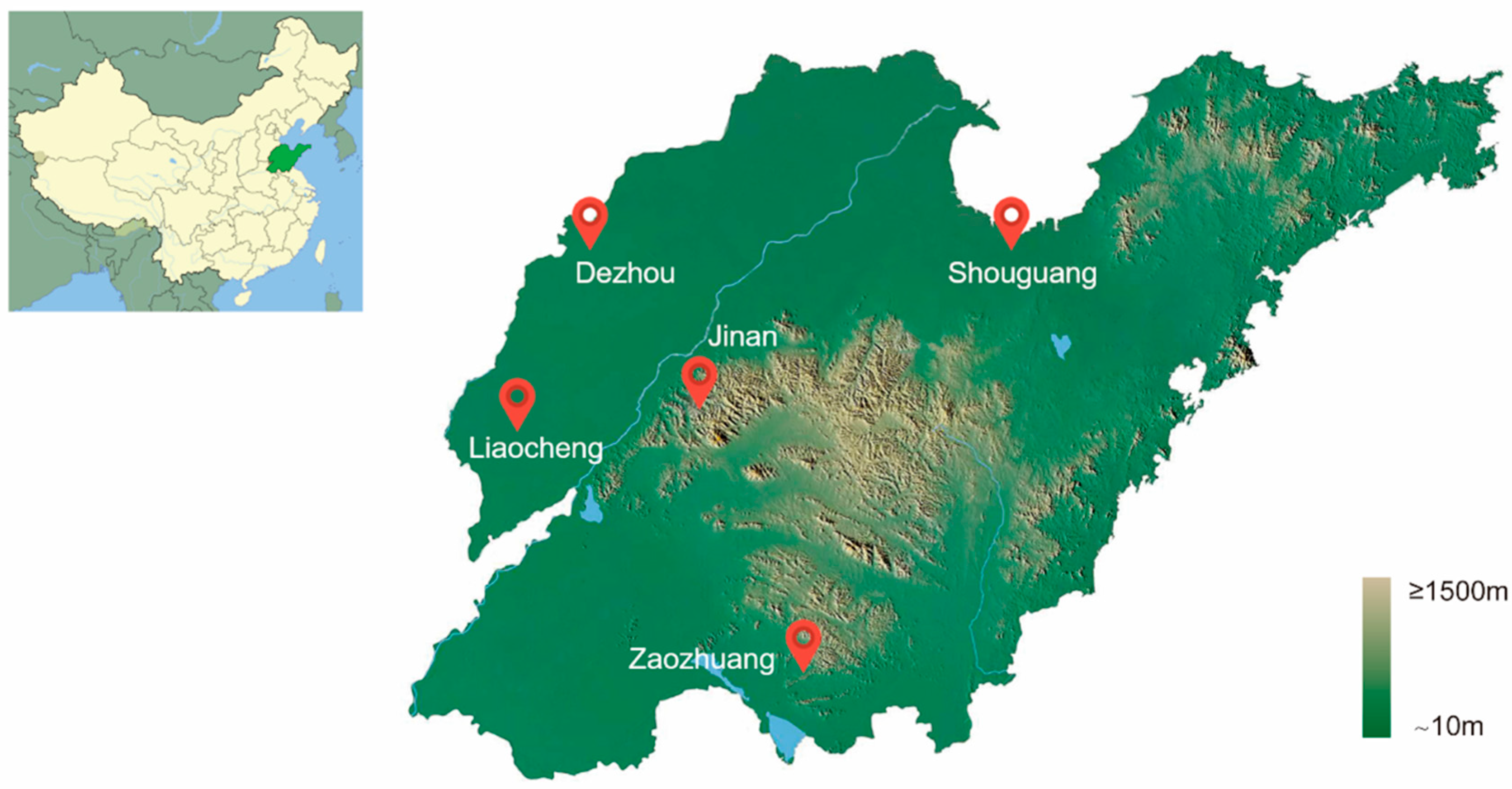
2.1. Sample Collection of Bemisia tabaci MED in Shandong, China
2.2. 2b-RAD Sequencing and Genotyping of Bemisia Tabaci MED
2.3. SNP Filtering and Genetic Diversity Analyses
2.4. Analysis of Molecular Variance and Mantel Test
2.5. Analyses of Population Structure
3. Results
3.1. Genetic Variation Within and Among Bemisia tabaci MED Populations in Shandong Province of China
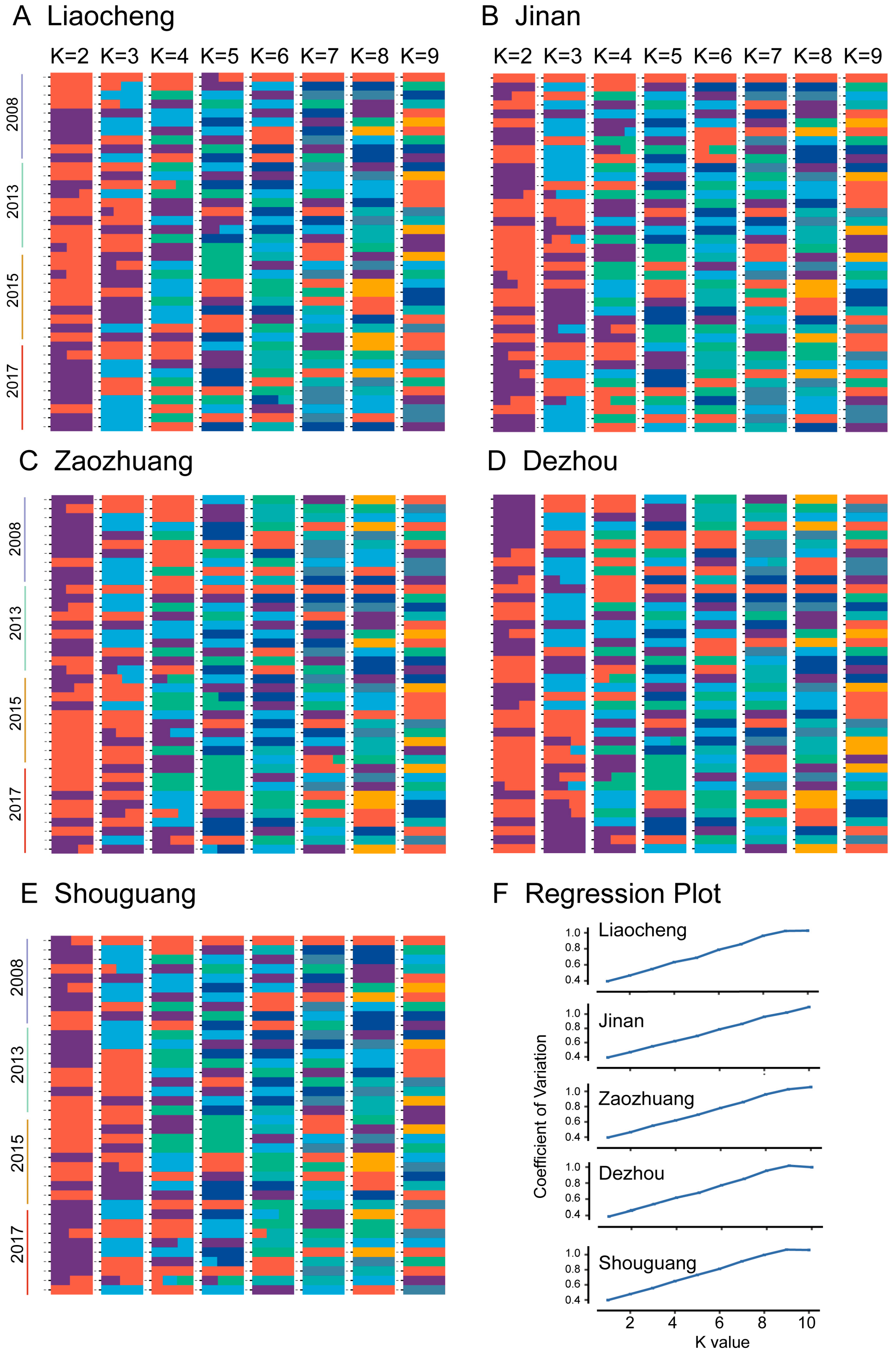
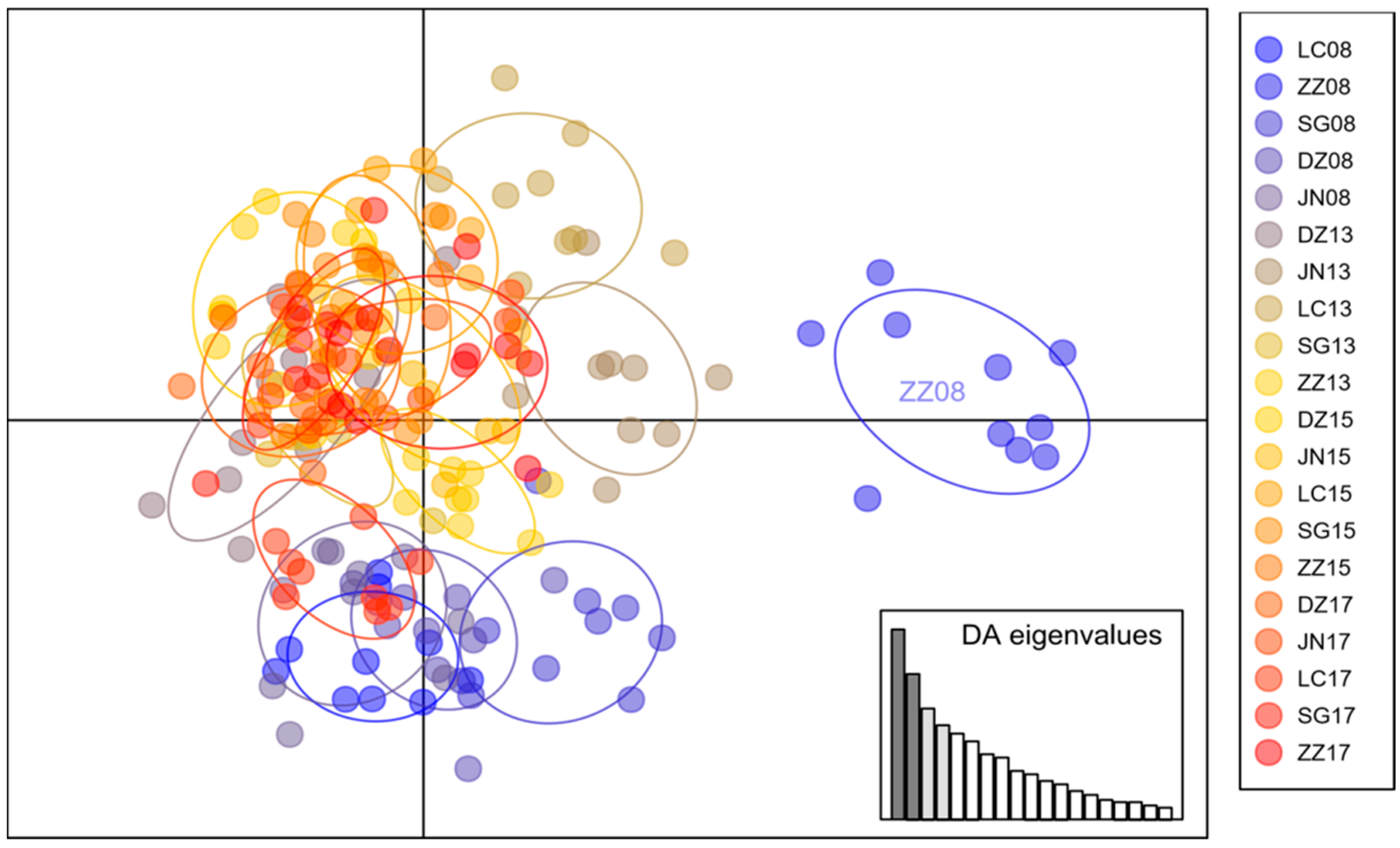
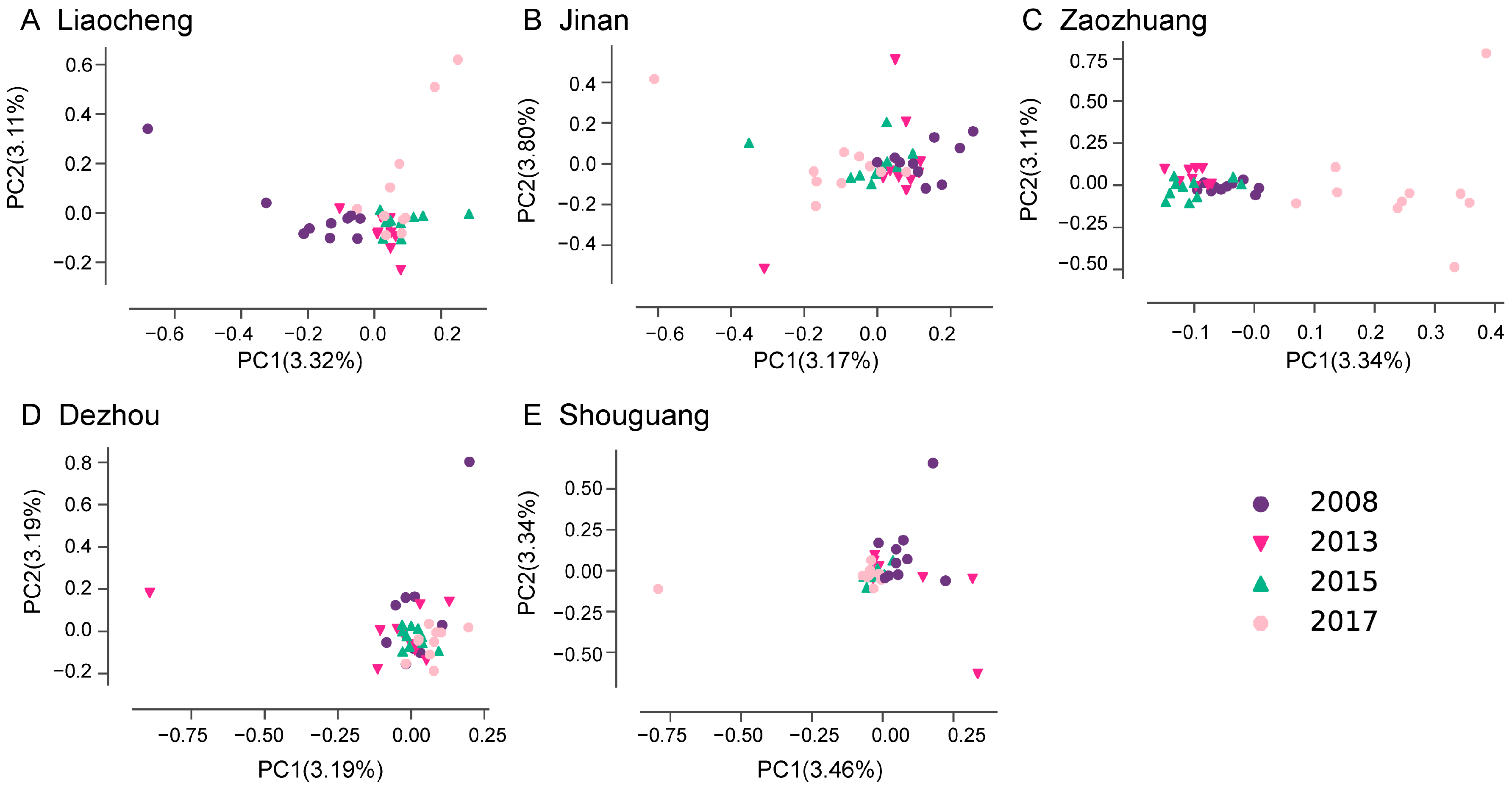
3.2. Population Structure of Bemisia tabaci MED
4. Discussion
4.1. Temporal Genetic Heterogeneity Reveals Bemisia tabaci Med Invasion History
4.2. Spatial Genetic Heterogeneity Driven by Anthropogenic and Ecological Actors
4.3. Implications of Spatiotemporal Heterogeneity for Pest Management
4.4. Limitations and Future Directions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wright, S. The Genetical Structure of Populations. Ann. Eugen. 1949, 15, 323–354. [Google Scholar] [CrossRef]
- Roderick, G.K. Geographic Structure of Insect Populations: Gene Flow, Phylogeography, and Their Uses. Annu. Rev. Entomol. 1996, 41, 325–352. [Google Scholar] [CrossRef]
- Holderegger, R.; Wagner, H.H. A brief guide to Landscape Genetics. Landsc. Ecol. 2006, 21, 793–796. [Google Scholar] [CrossRef]
- Sakai, A.K.; Allendorf, F.W.; Holt, J.S.; Lodge, D.M.; Molofsky, J.; With, K.A.; Baughman, S.; Cabin, R.J.; Cohen, J.E.; Ellstrand, N.C.; et al. The Population Biology of Invasive Species. Annu. Rev. Ecol. Syst. 2001, 32, 305–332. [Google Scholar] [CrossRef]
- Men, Q.-L.; Chen, M.-H.; Zhang, Y.-L.; Feng, J.-N. Genetic structure and diversity of a newly invasive species, the codling moth, Cydia pomonella (L.) (Lepidoptera: Tortricidae) in China. Biol. Invasions 2012, 15, 447–458. [Google Scholar] [CrossRef]
- Wei, S.-J.; Shi, B.-C.; Gong, Y.-J.; Jin, G.-H.; Chen, X.-X.; Meng, X.-F.; Crandall, K.A. Genetic Structure and Demographic History Reveal Migration of the Diamondback Moth Plutella xylostella (Lepidoptera: Plutellidae) from the Southern to Northern Regions of China. PLoS ONE 2013, 8, e59654. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.E. Evolutionary genetics of invasive species. Trends Ecol. Evol. 2002, 17, 386–391. [Google Scholar] [CrossRef]
- Joshi, J.; Schmid, B.; Caldeira, M.C.; Dimitrakopoulos, P.G.; Good, J.; Harris, R.; Hector, A.; Huss-Danell, K.; Jumpponen, A.; Minns, A.; et al. Local adaptation enhances performance of common plant species. Ecol. Lett. 2001, 4, 536–544. [Google Scholar] [CrossRef]
- Chu, D.; Zhang, Y.-J.; Brown, J.K.; Cong, B.; Xu, B.-Y.; Wu, Q.-J.; Zhu, G.-R. The introduction of the exotic q biotype of Bemisia tabaci from the mediterranean region into China on ornamental crops. Fla. Èntomol. 2006, 89, 168–174. [Google Scholar] [CrossRef]
- Verhoeven, K.J.F.; Macel, M.; Wolfe, L.M.; Biere, A. Population admixture, biological invasions and the balance between local adaptation and inbreeding depression. Proc. R. Soc. B Biol. Sci. 2010, 278, 2–8. [Google Scholar] [CrossRef]
- Cao, L.; Wang, Z.; Gong, Y.; Zhu, L.; Hoffmann, A.A.; Wei, S. Low genetic diversity but strong population structure reflects multiple introductions of western flower thrips (Thysanoptera: Thripidae) into China followed by human-mediated spread. Evol. Appl. 2017, 10, 391–401. [Google Scholar] [CrossRef]
- Tang, X.-T.; Cai, L.; Shen, Y.; Xu, L.-L.; Du, Y.-Z. Competitive Displacement between Bemisia tabaci MEAM1 and MED and Evidence for Multiple Invasions of MED. Insects 2019, 11, 35. [Google Scholar] [CrossRef]
- Rowe, G.; Beebee, T.J.C. Defining population boundaries: Use of three Bayesian approaches with microsatellite data from British natterjack toads (Bufo calamita). Mol. Ecol. 2007, 16, 785–796. [Google Scholar] [CrossRef]
- Gould, F. Sustainability of Transgenic Insecticidal Cultivars: Integrating Pest Genetics and Ecology. Annu. Rev. Èntomol. 1998, 43, 701–726. [Google Scholar] [CrossRef]
- Denholm, I.; Cahill, M.; Dennehy, T.J.; Horowitz, A.R.; Pickett, J.A.; Devonshire, A.L. Challenges with managing insecticide resistance in agricultural pests, exemplisfied by the whitefly Bemisia tabaci. Philos. Trans. R. Soc. B Biol. Sci. 1998, 353, 1757–1767. [Google Scholar] [CrossRef]
- Naveen, N.C.; Chaubey, R.; Kumar, D.; Rebijith, K.B.; Rajagopal, R.; Subrahmanyam, B.; Subramanian, S. Insecticide resistance status in the whitefly, Bemisia tabaci genetic groups Asia-I, Asia-II-1 and Asia-II-7 on the Indian subcontinent. Sci. Rep. 2017, 7, 40634. [Google Scholar] [CrossRef]
- De Barro, P.J.; Liu, S.-S.; Boykin, L.M.; Dinsdale, A.B. Bemisia tabaci: A Statement of Species Status. Annu. Rev. Èntomol. 2011, 56, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.-S.; De Barro, P.J.; Xu, J.; Luan, J.-B.; Zang, L.-S.; Ruan, Y.-M.; Wan, F.-H. Asymmetric Mating Interactions Drive Widespread Invasion and Displacement in a Whitefly. Science 2007, 318, 1769–1772. [Google Scholar] [CrossRef]
- Chu, D.; Zhang, Y.J.; Wan, F.H. Cryptic Invasion of the Exotic Bemisia tabaci Biotype Q Occurred Widespread in Shandong Province of China. Fla. Èntomol. 2010, 93, 203–207. [Google Scholar] [CrossRef]
- Hu, J.; De Barro, P.; Zhao, H.; Wang, J.; Nardi, F.; Liu, S.-S.; Roberts, R.G. An Extensive Field Survey Combined with a Phylogenetic Analysis Reveals Rapid and Widespread Invasion of Two Alien Whiteflies in China. PLoS ONE 2011, 6, e16061. [Google Scholar] [CrossRef] [PubMed]
- Chu, D.; Pan, H.-P.; Li, X.-C.; Guo, D.; Tao, Y.-L.; Liu, B.-M.; Zhang, Y.-J.; Sotka, E. Spatial Genetic Heterogeneity in Populations of a Newly Invasive Whitefly in China Revealed by a Nation-Wide Field Survey. PLoS ONE 2013, 8, e79997. [Google Scholar] [CrossRef]
- Dinsdale, A.; Schellhorn, N.; De Barro, P.; Buckley, Y.; Riginos, C. Rapid genetic turnover in populations of the insect pest Bemisia tabaci Middle East: Asia Minor 1 in an agricultural landscape. Bull. Èntomol. Res. 2012, 102, 539–549. [Google Scholar] [CrossRef]
- Chu, D.; Guo, D.; Tao, Y.; Jiang, D.; Li, J.; Zhang, Y. Evidence For Rapid Spatiotemporal Changes in Genetic Structure of an Alien Whitefly During Initial Invasion. Sci. Rep. 2014, 4, 4396. [Google Scholar] [CrossRef] [PubMed]
- Lainhart, W.; Bickersmith, S.A.; Nadler, K.J.; Moreno, M.; Saavedra, M.P.; Chu, V.M.; Ribolla, P.E.; Vinetz, J.M.; Conn, J.E. Evidence for temporal population replacement and the signature of ecological adaptation in a major Neotropical malaria vector in Amazonian Peru. Malar. J. 2015, 14, 1–17. [Google Scholar] [CrossRef]
- Byrne, D. Migration and dispersal by the sweet potato whitefly, Bemisia tabaci. Agric. For. Meteorol. 1999, 97, 309–316. [Google Scholar] [CrossRef]
- Byrne, D.N.; Rathman, R.J.; Orum, T.V.; Palumbo, J.C. Localized migration and dispersal by the sweet potato whitefly, Bemisia tabaci. Oecologia 1996, 105, 320–328. [Google Scholar] [CrossRef] [PubMed]
- EFSA Panel on Plant Health (PLH) Scientific Opinion on the risks to plant health posed by Bemisia tabaci species complex and viruses it transmits for the EU territory. EFSA J. 2013, 11, 3162. [CrossRef]
- Li, H.; Wang, J.; Peng, Y.; Guo, C.; Qu, W.; Yang, N.; Zhu, Y.; Jeong, I.; Li, X.; Ghanim, M.; et al. Invasion genomics uncover complex introduction patterns of the globally invasive whitefly, Bemisia tabaci MED. Divers. Distrib. 2023, 29, 1172–1189. [Google Scholar] [CrossRef]
- Barbanti, A.; Torrado, H.; Macpherson, E.; Bargelloni, L.; Franch, R.; Carreras, C.; Pascual, M. Helping decision making for reliable and cost-effective 2b-RAD sequencing and genotyping analyses in non-model species. Mol. Ecol. Resour. 2020, 20, 795–806. [Google Scholar] [CrossRef]
- Dahms, C.; Kemppainen, P.; Zanella, L.N.; Zanella, D.; Carosi, A.; Merilä, J.; Momigliano, P. Cast away in the Adriatic: Low degree of parallel genetic differentiation in three-spined sticklebacks. Mol. Ecol. 2021, 31, 1234–1253. [Google Scholar] [CrossRef]
- Pauletto, M.; Carraro, L.; Babbucci, M.; Lucchini, R.; Bargelloni, L.; Cardazzo, B. Extending RAD tag analysis to microbial ecology: A comparison between MultiLocus Sequence Typing and 2b-RAD to investigate Listeria monocytogenes genetic structure. Mol. Ecol. Resour. 2015, 16, 823–835. [Google Scholar] [CrossRef]
- Wang, S.; Meyer, E.; McKay, J.K.; Matz, M.V. 2b-RAD: A simple and flexible method for genome-wide genotyping. Nat. Methods 2012, 9, 808–810. [Google Scholar] [CrossRef]
- Catchen, J.M.; Amores, A.; Hohenlohe, P.; Cresko, W.; Postlethwait, J.H. Stacks: Building and Genotyping Loci De Novo From Short-Read Sequences. G3 Genes Genomes Genet. 2011, 1, 171–182. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Peakall, R.O.D.; Smouse, P.E. GenAlEx 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Rousset, F. genepop’007: A complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol. Ecol. Notes 2007, 7, 574–578. [Google Scholar] [CrossRef]
- Earl, D.A.; vonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Jakobsson, M.; Rosenberg, N.A. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007, 23, 1801–1806. [Google Scholar] [CrossRef]
- Rosenberg, N.A. Distruct: A program for the graphical display of population structure. Mol. Ecol. Notes 2004, 4, 137–138. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.r-project.org/ (accessed on 4 February 2024).
- Jombart, T. adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef]
- Jombart, T.; Devillard, S.; Balloux, F. Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genet. 2010, 11, 94. [Google Scholar] [CrossRef] [PubMed]
- Beaumont, M.A.; Nichols, R.A. Evaluating loci for use in the genetic analysis of population structure. Proc. R. Soc. B Biol. Sci. 1996, 263, 1619–1626. [Google Scholar] [CrossRef]
- Boggs, C.L. The fingerprints of global climate change on insect populations. Curr. Opin. Insect Sci. 2016, 17, 69–73. [Google Scholar] [CrossRef]
- Ovčarenko, I.; Kapantaidaki, D.E.; Lindström, L.; Gauthier, N.; Tsagkarakou, A.; Knott, K.E.; Vänninen, I. Agroecosystems shape population genetic structure of the greenhouse whitefly in Northern and Southern Europe. BMC Evol. Biol. 2014, 14, 165. [Google Scholar] [CrossRef] [PubMed]
- Ffrench-Constant, R.H.; Daborn, P.J.; Le Goff, G. The genetics and genomics of insecticide resistance. Trends Genet. 2004, 20, 163–170. [Google Scholar] [CrossRef]
- Pélissié, B.; Crossley, M.S.; Cohen, Z.P.; Schoville, S.D. Rapid evolution in insect pests: The importance of space and time in population genomics studies. Curr. Opin. Insect Sci. 2018, 26, 8–16. [Google Scholar] [CrossRef]
- Haasl, R.J.; Payseur, B.A. Multi-locus inference of population structure: A comparison between single nucleotide polymorphisms and microsatellites. Heredity 2010, 106, 158–171. [Google Scholar] [CrossRef]
- Yang, K.; Qin, P.; Yuan, M.; Chen, L.; Zhang, Y.; Chu, D. Infection density pattern of Cardinium affects the responses of bacterial communities in an invasive whitefly under heat conditions. Insect Sci. 2022, 30, 1149–1164. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Li, C.-R.; Qin, P.-H.; Yuan, M.-Y.; Chu, D.; Wong, A.C. Conditional effects of Cardinium on microbiota in an invasive whitefly under different ecological factors. Microbiol. Spectr. 2025, 13, e0224024. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, Y.; Wei, X.; Pan, H.; Zhang, Y.; Zhou, X.; Chu, D. Heterogeneous distribution of Cardiniumin whitefly populations is associated with host nuclear genetic background. Insect Sci. 2023, 30, 1701–1712. [Google Scholar] [CrossRef]
- Yang, K.; Yuan, M.; Liu, Y.; Guo, C.; Liu, T.; Zhang, Y.; Chu, D. First evidence for thermal tolerance benefits of the bacterial symbiont Cardinium in an invasive whitefly, Bemisia tabaci. Pest Manag. Sci. 2021, 77, 5021–5031. [Google Scholar] [CrossRef]
- Kennedy, T.; Rinker, D.; Broadie, K. Genetic background mutations drive neural circuit hyperconnectivity in a fragile X syndrome model. BMC Biol. 2020, 18, 1–21. [Google Scholar] [CrossRef] [PubMed]

| Sampling Year | Sampling Location | Population ID | %Poly | I | Ho | He | FIS |
|---|---|---|---|---|---|---|---|
| 2008 | Liaocheng | LC08 | 84.45 | 0.341 | 0.194 | 0.198 | 0.006 |
| Zaozhuang | ZZ08 | 85.84 | 0.352 | 0.193 | 0.207 | 0.036 | |
| Shouguang | SG08 | 86.96 | 0.376 | 0.194 | 0.217 | 0.077 | |
| Dezhou | DZ08 | 86.37 | 0.396 | 0.175 | 0.226 | 0.179 | |
| Jinan | JN08 | 86.85 | 0.376 | 0.192 | 0.217 | 0.085 | |
| 2013 | Liaocheng | LC13 | 85.03 | 0.366 | 0.176 | 0.211 | 0.117 |
| Zaozhuang | ZZ13 | 85.52 | 0.377 | 0.167 | 0.214 | 0.172 | |
| Shouguang | SG13 | 81.67 | 0.339 | 0.175 | 0.199 | 0.073 | |
| Dezhou | DZ13 | 85.94 | 0.350 | 0.185 | 0.203 | 0.060 | |
| Jinan | JN13 | 85.52 | 0.376 | 0.172 | 0.216 | 0.159 | |
| 2015 | Liaocheng | LC15 | 86.85 | 0.380 | 0.182 | 0.218 | 0.126 |
| Zaozhuang | ZZ15 | 87.33 | 0.369 | 0.187 | 0.213 | 0.092 | |
| Shouguang | SG15 | 87.60 | 0.389 | 0.174 | 0.223 | 0.176 | |
| Dezhou | DZ15 | 87.28 | 0.356 | 0.190 | 0.206 | 0.050 | |
| Jinan | JN15 | 86.91 | 0.380 | 0.178 | 0.217 | 0.138 | |
| 2017 | Liaocheng | LC17 | 86.75 | 0.383 | 0.180 | 0.220 | 0.147 |
| Zaozhuang | ZZ17 | 86.53 | 0.368 | 0.185 | 0.212 | 0.097 | |
| Shouguang | SG17 | 87.33 | 0.407 | 0.163 | 0.233 | 0.248 | |
| Dezhou | DZ17 | 87.44 | 0.389 | 0.176 | 0.221 | 0.166 | |
| Jinan | JN17 | 87.07 | 0.364 | 0.191 | 0.211 | 0.061 |
| Group Comparison | Average Fst | P-Value |
|---|---|---|
| Among all populations | 0.0042 | 0.006 |
| Among years (Global) | 0.0032 | <0.001 |
| 2008 vs. 2013 | 0.0031 | <0.001 |
| 2008 vs. 2015 | 0.0052 | <0.001 |
| 2008 vs. 2017 | 0.0041 | <0.001 |
| 2013 vs. 2015 | 0.0011 | 0.112 |
| 2013 vs. 2017 | 0.0010 | 0.145 |
| 2015 vs. 2017 | 0.0015 | 0.087 |
| Within same years (Spatial) | −0.0001–0.0020 | >0.05 |
| Group of Populations | Source of Variation | df (Degree of Freedom) | Sum of Squares | Variance Components | Percentage Variation (%) | P-Value |
|---|---|---|---|---|---|---|
| Global | Among populations | 19 | 5219.287 | 0.899 | 0.42 | 0.006 |
| Within populations | 178 | 45,729.188 | 43.553 | 20.33 | 0.001 | |
| Within individuals | 198 | 33,620.500 | 169.801 | 79.25 | 0.001 | |
| Group by geographic | Among geographic groups | 4 | 1111.149 | 0.049 | 0.02 | 0.361 |
| Among yearly populations within geographic groups | 15 | 4108.138 | 0.858 | 0.40 | 0.021 | |
| Within populations | 178 | 45,729.188 | 43.553 | 20.33 | 0.011 | |
| Within individuals | 198 | 33,620.500 | 169.801 | 79.25 | 0.001 | |
| Group by years | Among yearly groups | 3 | 909.059 | 0.339 | 0.16 | 0.051 |
| Among geographic populations within yearly groups | 16 | 4311.158 | 0.634 | 0.30 | 0.044 | |
| Within populations | 178 | 45,729.188 | 43.553 | 20.32 | 0.006 | |
| Within individuals | 198 | 33,620.500 | 169.801 | 79.23 | 0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, K.; Li, H.; Guo, D.; Sun, Z.; Li, F.; Chu, D. Spatiotemporal Population Genomics of the Invasive Whitefly Bemisia tabaci MED in China: Implications for Surveillance and Sustainable Control. Insects 2025, 16, 975. https://doi.org/10.3390/insects16090975
Yang K, Li H, Guo D, Sun Z, Li F, Chu D. Spatiotemporal Population Genomics of the Invasive Whitefly Bemisia tabaci MED in China: Implications for Surveillance and Sustainable Control. Insects. 2025; 16(9):975. https://doi.org/10.3390/insects16090975
Chicago/Turabian StyleYang, Kun, Hongran Li, Dong Guo, Zuowen Sun, Fujun Li, and Dong Chu. 2025. "Spatiotemporal Population Genomics of the Invasive Whitefly Bemisia tabaci MED in China: Implications for Surveillance and Sustainable Control" Insects 16, no. 9: 975. https://doi.org/10.3390/insects16090975
APA StyleYang, K., Li, H., Guo, D., Sun, Z., Li, F., & Chu, D. (2025). Spatiotemporal Population Genomics of the Invasive Whitefly Bemisia tabaci MED in China: Implications for Surveillance and Sustainable Control. Insects, 16(9), 975. https://doi.org/10.3390/insects16090975







