1. Introduction
Due to the global warming, unexpected high temperatures have been frequent in these years. Insects are generally small and are sensitive to heat stress. A recent study used 102 insect species for meta-analyses and revealed that insects exhibit a limited capacity to acclimate their thermal tolerance to elevated temperatures, which renders them more susceptible to the impacts of global warming than previously estimated [
1]. They can be influenced by high temperature in terms of their development, phenotype, reproduction, survival, and population dynamic [
2,
3,
4]. For example, In
Coccophagus japonicus, heat stress shortened adult lifespan, reduced egg production, and altered developmental duration [
3]. Similarly, in
Spodoptera frugiperda, heat stress decreased survival and reduced pupal weight, potentially affecting long-term population dynamics [
4]. A current survey showed that the diversity of pollinating bees has decreased rapidly due to high temperatures [
5]. Therefore, to understand heat resistance in some special insect species is increasingly important to help other species that are highly sensitive to high temperatures.
A few insect species have good heat resistance. For example, the Saharan silver ant (
Cataglyphis bombycina) has developed a unique thermoregulatory adaptation through specialized reflective hairs that minimize solar radiation absorption, enabling activity during peak desert heat with an air temperature over 50 °C [
6,
7]. The male desert locust (
Schistocerca gregaria) extends legs to lift its body off the hot ground and positioned itself parallel to the sun’s rays, minimizing radiative heating [
8]. Some desert beetles (such as
Tenebrionidae and
Chrysomelidae) are among the most successful animals in adapting to the extremely high-temperature habitats of the desert [
9].
The small hive beetle,
Aethina tumida, is a beetle species belonging to the family
Nitidulidae, native to sub-Saharan Africa. It is also well known as a significant pest of honeybee colonies and poses a major threat to apiculture worldwide [
10]. Differing from the above Saharan silver ants and desert beetles that are mainly adapted to the dry and extreme high-temperature desert environment, this small hive beetle is well-adapted to various environmental habitats and is widespread across Africa, America, Europe, Australia, and Asia [
11]. It can cause severe damage to honeybee colonies by feeding on honey, pollen, and brood, leading to the fermentation of hive products and ultimately colony collapse, resulting in significant economic losses for beekeepers due to reduced honey production and the abandonment of hives by bees [
10]. Understanding the heat tolerance of the small hive beetle is important for protecting honeybee health. As a heat-resistant invasive pest, it poses a growing threat to colonies, especially under climate change. Studying its thermal tolerance can reveal how it survives in extreme conditions and help assess potential health risks to bees.
Noor-ul-Ane and Jung (2020) showed that this small hive beetle has good heat resistance and predicted that the high developmental threshold temperatures for this beetle would be 40.4 to 46.5 °C [
12], though high temperature could affect the development of its larvae [
13]. Therefore, although this beetle does not appear to possess specialized strategies for heat resistance, it still exhibits notable thermal tolerance. Investigating the underlying molecular mechanisms could help us to understand the heat resistance ability of this pest and offer suggestions for its management and control. It may also be important for heat-vulnerable insects, rather than for species adapted to extreme heat stress, such as the Saharan silver ant.
The heat resistance molecular mechanisms of small insects are still not well understood. King and MacRae (2015) reviewed the key role of heat-shock protein (
Hsp) genes in insect heat resistance, including
Hsp60,
Hsp70, and
Hsp90 and some small heat shock proteins (
sHsps) [
14].
Hsps facilitate numerous essential molecular processes in insects, such as protein folding, localization, and degradation, helping maintain protein homeostasis under thermal stress [
14]. Heat stress induces a high expression of
Hsps in insects, which bind misfolded proteins and aid refolding. For example, heat stress induces the upregulation of
Hsp90 and
Hsp70 in apple maggot fly (
Rhagoletis pomonella), enhancing its thermal tolerance [
15]. Similarly,
sHsp genes such as
sHsp19.9 and
sHsp20.4 from silkworm (
Bombyx mori) are highly expressed in the larval fat body, testis, and ovary under thermal stress [
16].
Hsp40,
Hsp20,
Hsp70,
Hsp90, and mitochondrial
Hsp60 play an important role in both heat and cold tolerance in two leafminers (
Liriomyza sativa and
Liriomyza huidobrensis) [
17].
In response to heat stress, insects rely not only on heat shock proteins but also on a broad array of other molecular mechanisms. Genes or proteins involved in the oxidative stress response are increasingly recognized for their roles in insect thermotolerance. For instance, superoxide dismutase, catalase and glutathione S-transferase enzymes contribute to cellular protection by mitigating heat-induced oxidative damage [
18]. Moreover, enzymes or related genes involved in energy metabolism and chitin synthase are also involved into insect heat resistance. For example, the potato aphid raises the concentrations of enzymes for ATP generation and circular protein biosynthesis during heat stress [
19]. Therefore, the
Hsp family genes and other related genes are possibly regulated in a coordinated manner, forming a complex and integrated network that underlies the insect’s adaptive response to thermal stress. However, the molecular mechanisms underlying insect heat resistance remain incompletely understood and may involve more interacting pathways than currently documented.
Based on previous evidence that HSPs act as primary molecular chaperones under thermal stress and that other genes contribute to cellular protection, we hypothesized that some key genes enriched in specific pathways, together with HSPs, may constitute the core responders to heat stress in A. tumida. Here, we used A. tumida as a model to investigate the molecular basis of insect thermotolerance. We reared the small hive beetle under 25 °C, 38 °C, 42 °C, and 46 °C, and their survival ability was evaluated. We also sequenced RNA samples from insects in the above conditions by RNA-seq and qPCR to explore A. tumida gene expression profiles related to thermal tolerance, which may contribute significantly to enriching the theoretical research on insect heat tolerance mechanisms.
2. Materials and Methods
2.1. Insects
Thousands of small hive beetles were provided by eight honeybee apiaries in Haikou city, China, and were reared in 10 plastic boxes with a diet consisting of 40% honey, 40% rapeseed pollen, and 20% yeast extract powder (purity 90%, Beijing Hongrun Baoshun Technology Co., Ltd., Beijing, China) under constant room temperature of 30 °C in an incubator (YT-SPX-150BE, Shandong Yuntang Intelligent Technology Co., Ltd., Weifang, China). Totally, 560 newly emerged healthy beetles with similar body size were randomly harvested from the 10 plastic boxes on the same day. For the survival test, 400 beetles were divided into four groups and placed at 25 °C, 38 °C, 42 °C, and 46 °C. Each group had 100 beetles. For the RNA-Seq and qPCR analyses, 160 beetles were divided into four groups as above.
2.2. Survival Test on the Small Hive Beetle
The small hive beetles of each group were transferred into a stainless-steel container (diameter: 10 cm), which was subsequently placed in a water bath (SHJ-4AB, Changzhou Jintan Liangyou Instrument Co., Ltd., Changzhou, China) for heat stress treatment at 25 °C, 38 °C, 42 °C, and 46 °C. The beetles were subjected to heat stress without access to food or water, as the presence of food or water could potentially buffer ambient temperature fluctuations and compromise the stability of the heat stress treatment. We inspected and counted the dead beetles every 0.5 h, and LD50 and LD95 were recorded.
For RNA-Seq and qPCR, the remaining 160 beetles were divided and treated under four temperatures as above but only heated for 0.5 h, an exposure we deemed optimal for assessing heat resistance while minimizing the potential effects of starvation on gene expression. The beetles were then harvested for RNA-Seq and qPCR. Each sample contained four beetles for RNA-Seq or qPCR, and each treatment had three biological replicates. The beetles in a sample were placed into a 1.5 mL microcentrifuge tube and directly immersed in liquid nitrogen for cryopreservation and storage. In total, 12 samples for RNA-Seq and another 12 samples for qPCR were analyzed.
2.3. RNA Extraction and Sequencing
The total RNA of each sample was extracted using Invitrogen RNA extraction kits (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s instructions. RNA concentration and purity were measured using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
In total, 1 µg of each RNA sample with high quality (RIN > 7) was used for library preparation for RNA-Seq. mRNA was enriched from total RNA using oligo(dT) magnetic beads and subsequently fragmented into short fragments. First-strand cDNA was synthesized using random hexamer primers and reverse transcriptase, followed by second-strand cDNA synthesis. The resulting cDNA products were purified using the AMPure XP magnetic bead system and then end-repaired, A-tailed, and ligated to sequencing adapters. Size selection of DNA fragments was conducted using AMPure XP beads. After purification and PCR amplification, the final cDNA libraries were quantified and validated for size distribution using the bioanalyzer. The qualified libraries were then sequenced on an Illumina NovaSeq 6000 platform (Illumina, San Diego, CA, USA) using the paired-end 150 bp (PE150) strategy to generate high-throughput sequencing data.
2.4. Data Processing and Expression Quantification
To ensure the accuracy and reliability of the downstream analyses, the raw sequencing reads were first subjected to a comprehensive quality control process using fastp (version 0.23.2). This procedure included the removal of adapter sequences and reads originating from adapter self-ligation events that failed to capture the target fragments. Bases with Phred quality scores below 20 at both ends of the reads were trimmed, and reads containing bases with a quality score below 10 after trimming were discarded. Additionally, reads containing ambiguous nucleotides (“N”) or those shorter than 30 bp following quality filtering were excluded. After quality control, clean reads were retained for further analysis and quality reassessment.
2.5. Gene Expression Analysis
Low-quality reads were filtered out, and only those with a sequencing error rate below 1% (Q20 > 98%) were retained. The clean reads were mapped to the genome of
A. tumida (icAetTumi1.1). Transcript and gene expression levels were quantified using Cufflinks, employing the Cuffquant and Cuffnorm modules [
20]. Expression abundance was calculated based on the alignment of reads to annotated gene features and normalized as fragments per kilobase of exon per million mapped fragments (FPKM), enabling accurate cross-sample comparisons of the expression profiles. Gene expression after the four treatments was compared using edgeR [
21], and DEGs were identified based on
p value < 0.01 and log2 fold change (Log2FC) ≥ 1.5. Moreover, to assess the overall similarity and variation among the transcriptomic samples, PCA analysis was performed based on the expression profiles of all detected genes from the 12 samples using PCA analysis tools on the platform BMKCloud (
www.biocloud.net, accessed on 15 November 2021).
2.6. Enrichment Analysis of GO and KEGG
All DEGs from three comparisons (38 °C vs. 25 °C, 42 °C vs. 25 °C, and 46 °C vs. 25 °C) were aligned against various protein and nucleotide sequence databases using BLASTX+2.12.0, including the NCBI non-redundant protein (Nr) database, the Swiss-Prot protein database, and the non-redundant nucleotide (Nt) database, with a cut-off E-value of 10
−5. GO enrichment analysis was performed by mapping the DEGs to Gene Ontology (GO) terms, and significantly enriched GO terms were identified using a hypergeometric test (
p < 0.05) [
22].
Subsequently, the DEGs from each of the above comparisons were mapped to the Kyoto Encyclopedia of Genes and Genomes (KEGG) protein database (
http://www.genome.jp/kegg/kegg1.html, accessed on 15 November 2021) using BLAST+2.12.0 (E-value < 10
−5). KEGG pathway enrichment was assessed using the KOBAS 2.0 software [
23], and statistical significance was determined by a hypergeometric test with a Q-value threshold of <0.05.
2.7. qRT-PCR Verification of Eight Genes
Total RNA extracted from the 12 samples was normalized prior to reverse transcription, similar to our previous study [
24]. cDNA was synthesized using MLV reverse transcriptase (Takara, Osaka, Japan) in accordance with the manufacturer’s guidelines. The
β-actin gene was employed as an internal reference gene. Eight genes were randomly selected from the RNA-Seq data for qRT-PCR validation. The primers for these genes were designed using Primer 5.0 software and are presented in
Table S1. qPCR was performed using the ABI 7500 Real-Time PCR System (Applied Biosystems, Rockville, MD, USA) with a 20 μL SYBR Green reaction mixture consisting of SYBR Green master mix 10 μL, forward primer 0.4 μL, reverse primer 0.4 μL, cDNA template 1 μL, and nuclease-free water 8.2 μL. The qPCR cycling conditions were as follows: 94 °C for 2 min, 40 cycles, followed by 94 °C for 15 s, 60 °C for 30 s, and 72 °C for 30 s. The QuantStudio™ 5 System Real-Time-PCR Instrument (Thermo Fisher Scientific Inc., Waltham, MA, USA) was used for qPCR data analysis. Melt curve analysis was conducted for each sample to confirm the specificity of the PCR amplification products. For each gene, three independent biological replicates were analyzed, each with five technical replicates. To minimize inter-plate variability, both reference and target genes from the same sample were assayed on the same qPCR plate. The cycle threshold (Ct) value for each biological replicate was determined by averaging the values from three technical replicates. The relative expression level of each gene from the four treatments was determined using the 2
−ΔΔCt method.
2.8. Data Analysis
The survival data of the small hive beetle from the four treatments (
Figure 1) were compared using Kruskal–Wallis test in Statview 5.0 (SAS Institute Inc., Cary, NC, USA), and a
p value < 0.05 was considered as indicating a significant difference. The relative expression levels of the 9 genes in the qRT-PCR experiment were calculated using the 2
−ΔΔCt format and then analyzed using the ANOVA test followed by Fisher’s LSD test in Statview, and a
p value < 0.05 was considered as indicating a significant difference.
4. Discussion
The small hive beetle is originally native to the deserts of southern Africa and has now spread globally [
11]. This study clearly indicated that
A. tumida possesses a remarkable capacity to endure elevated temperatures. At 38 °C and 42 °C, the beetles exhibited extended survival durations (
Figure 1). Even at higher temperature (46 °C) individuals survived for nearly one hour (
Figure 1). The beetles were exposed to heat stress without access to food or water. Remarkably, they still exhibited high survival rates under these restrictive conditions, suggesting a stronger intrinsic tolerance to heat stress that may reflect their adaptability in natural environments. Another beetle species,
Monochamus alternatus Hope, also showed a similarly high heat resistance [
25]. Therefore, these findings suggest that
A. tumida, like
Monochamus alternatus, possesses an inherently high heat stress tolerance, which may represent a common adaptive trait among certain beetle species. This thermal resilience likely contributes to their survival and ecological success under fluctuating and extreme environmental conditions.
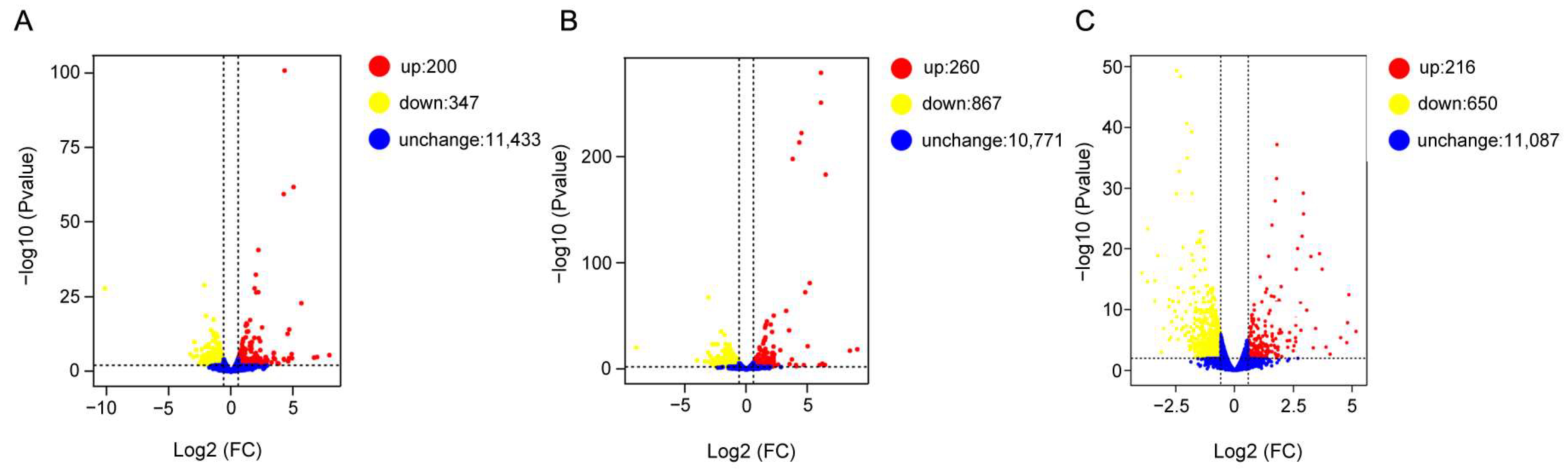
Transcriptome profiling revealed widespread transcriptional remodeling in
A. tumida in response to rising temperatures. A total of 547, 1127, and 866 DEGs were identified at 38 °C, 42 °C, and 46 °C, respectively, in comparison to the control condition at 25 °C (
Figure 3). This temperature-dependent increase in the DEG number up to 42 °C, followed by a slight decrease at 46 °C, and the expression of most heat shock genes showed a similar pattern characterized by an initial upregulation followed by downregulation (
Figure 6). This pattern likely reflects the beetle’s physiological threshold for a transcriptional response before cellular damage becomes irreversible. Among the DEGs, 16, 25, and 5
HSP genes were differentially expressed at 38 °C, 42 °C, and 46 °C, respectively (
Figure 5). Only five
HSP genes were upregulated at 46 °C, indicating that extreme heat may inhibit the induction of protective molecular chaperones or cause transcriptional exhaustion. This aligns with previous reports in
Monochamus alternatus, where the expression of the MaltHSP70-2 protein peaked at 40 °C but declined when the temperature exceeded the cellular tolerance thresholds [
25]. Therefore, these findings suggest that
A. tumida may exhibit transcriptomic plasticity in response to heat stress up to sublethal temperatures, beyond which the transcriptional machinery could become impaired. Additionally, the GO enrichment results revealed distinct biological responses across temperature gradients, which also supports this hypothesis. At 38 °C, the DEGs were mainly enriched in metabolic regulation and stress response genes (
Figure S2A), implying an early-stage adaptation to moderate thermal conditions. At 42 °C, additional enrichment appeared in behavior, detoxification, antioxidant defense, and rhythmic processes genes, reflecting enhanced protective responses (
Figure S2B). At 46 °C, stronger enrichment in immune response, cell junction, oxidative stress, and electron transport genes suggested intensified cellular and systemic responses to severe heat stress (
Figure S2C).
HSP family genes have been widely recognized as key drivers of heat resistance in insect species and act as molecular chaperones that help maintain cellular homeostasis by stabilizing nascent polypeptides, in refolding misfolded proteins, and in preventing the aggregation of denatured proteins under elevated temperatures [
26,
27]. In particular, the
HSP70 genes have been widely implicated in cytoprotection against heat stress, and their expression has been reported to correlate with the survival rates in various insect taxa [
25,
28].
In the present study, 22
HSP genes, mainly the
HSP70,
HSP90, and
HSP20 family genes, were significantly upregulated after the heat stress treatments (
Figure 5). One
HSP70 gene (Loc109602670) was the only DEG consistently upregulated across all temperature treatments. These findings, consistent with previous studies in other insect species [
27], demonstrated that the
HSP family genes play an important role in the heat stress response in insects. Notably, this supports the designation of the
HSP70 gene Loc109602670 as a “core responder” to heat stress and its potential as a molecular biomarker for thermal tolerance in insects.
Beyond
HSPs, one of the most striking observations was the consistent enrichment in lysosome-related pathway genes among the DEGs under all heat stress conditions. This pathway, involved in cellular degradation, recycling of macromolecules, and stress adaptation, likely plays a complementary role in maintaining cellular integrity under thermal insult [
29,
30]. In the present study, dozens of DEGs involved in lysosome pathways were identified, and this key pathway was the top one pathway in all three comparisons, and its proportion increased with the temperature (
Figure 4 and
Figure 5). Notably, key genes in this lysosome pathway such as
Cathepsin L1-like and
Lipase 3-like were upregulated in the heat-stressed groups (
Figure 5C). The
Cathepsin L1 family genes may be able to enhance proteolytic degradation during heat stress, thereby contributing to cellular proteostasis and survival under elevated temperatures [
31].
Lipase 3 is involved in lipid mobilization and energy homeostasis, supporting stress adaptation by facilitating the energy supply during thermal challenges [
32]. Previous studies have reported that the
Lipase 3 gene is also involved in starvation stress in insects [
33]. In the present study, it appears that
Lipase 3 may be simultaneously engaged in both starvation and heat stress responses. The processes in which it participates, namely, lipid mobilization and energy homeostasis, are likely to play important roles in protecting the small hive beetle under both stress conditions. Therefore, these findings suggest that the lysosome pathway in
A. tumida could enhance the proteolytic activity and facilitate the removal of damaged proteins and organelles, thus contributing to cellular homeostasis.
Moreover, lysosomes are central components of the autophagy pathway, which is a key cellular process for recycling damaged proteins and organelles during stress adaptation [
34]. Autophagy has been recognized as an integral part of the heat stress response in both mammals and insects, contributing to proteostasis and survival under elevated temperatures [
30,
35]. The consistent enrichment in lysosome-related genes observed in our study may also indicate the activation of autophagic processes in
A. tumida during heat stress. While we did not directly assess autophagy-specific markers, these transcriptomic signatures suggest that autophagy, in coordination with HSP-mediated protein refolding, could represent an additional protective mechanism underlying the beetle’s thermotolerance. Future studies incorporating autophagy assays will be valuable for confirming this hypothesis. Additionally, starvation can also activate lysosomal pathways associated with autophagy [
36], as well as the expression of
HSP genes such as
HSP90 [
37]. In this study, we sought to minimize the potential starvation-related impacts on gene expression; however, the possible influence of starvation on the expression of lysosomal pathway genes should still be taken into consideration.
The simultaneous upregulation of both
HSPs and lysosomal proteases points toward a coordinated response involving both protein repair and degradation mechanisms. For example,
HSPs coordinate their activity with that of antioxidant enzymes to confer thermal tolerance in
Pardosa pseudoannulata [
38]. In the present study,
HSPs may also coordinate their activity with that of lysosome pathway genes and antioxidant enzymes during heat resistance. While
HSPs act to refold or stabilize misfolded proteins, lysosomes may degrade irreparably damaged cellular components, collectively ensuring cellular survival under prolonged heat exposure. This dual response underscores the complexity of thermotolerance strategies in
A. tumida, suggesting that successful adaptation involves a dynamic balance between cytoprotection and selective degradation. From an applied perspective, understanding the molecular basis of thermal adaptation in invasive pests is critical for developing predictive models of species distribution under climate change. Given the increasing frequency and intensity of heatwaves, species like
A. tumida may gain competitive advantages over native species, leading to shifts in ecosystem dynamics and agricultural impacts. Consequently, molecular markers such as the
HSPs and lysosomal genes identified in this study may serve as early warning indicators or targets for pest control strategies.