Rapid and Accurate Detection of the Most Common Bee Pathogens; Nosema ceranae, Aspergillus flavus, Paenibacillus larvae and Black Queen Cell Virus
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Total Nucleic Acid (TNA) Extraction
2.2. Guanidinium Thiocyanate-Phenol-Chloroform (Trizol) Method
2.3. Manual Cetyl-Rimethylammonium Bromide (CTAB) Protocol
2.4. Automated CTAB-Based Protocol
2.5. Primer Design and Specificity Confirmation
2.6. Amplification Conditions, Primers Sensitivity and Validation
2.7. Statistical Analysis
3. Results
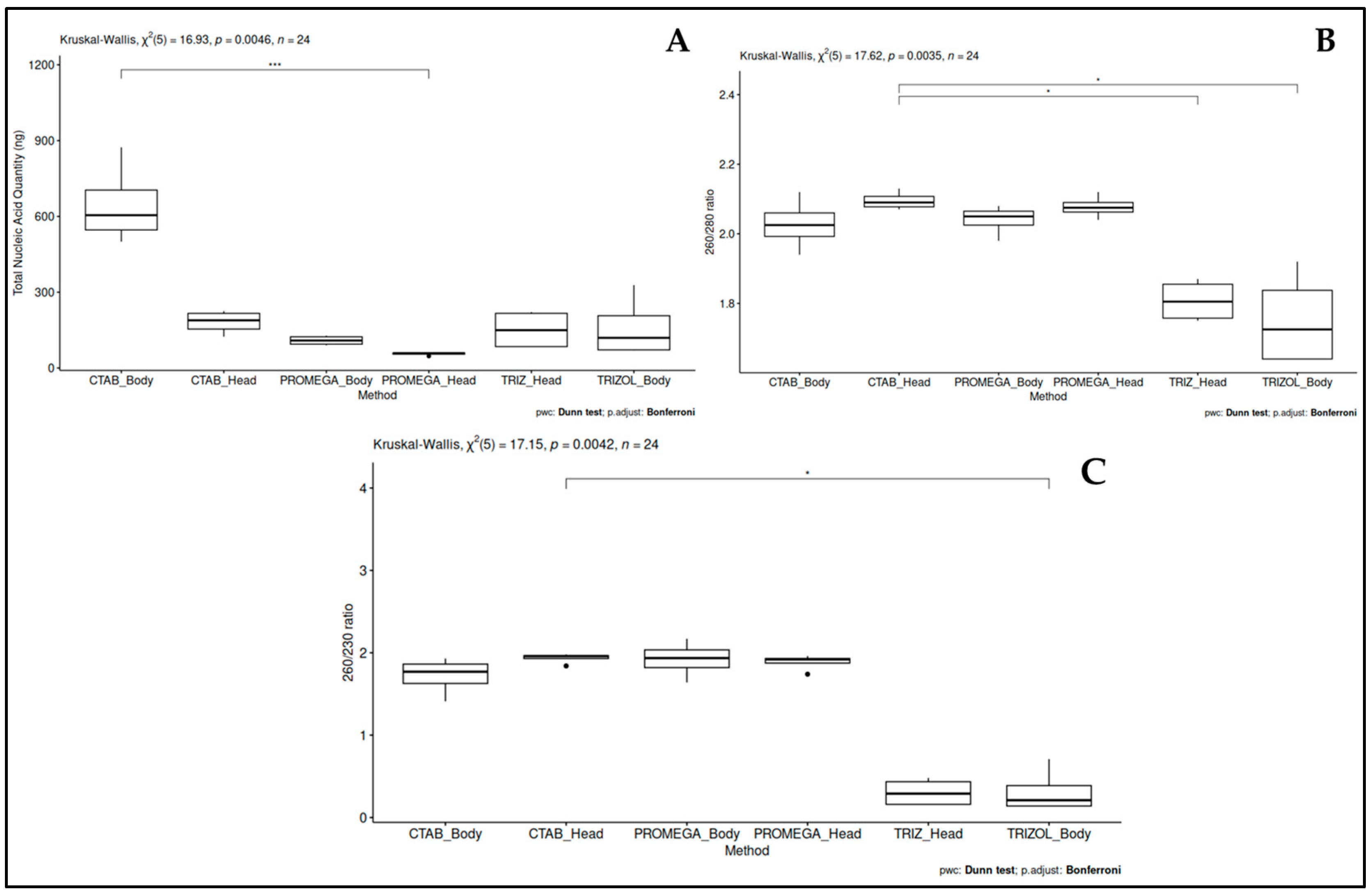
3.1. TNA Extraction Method Selection
3.2. Primer Specificity
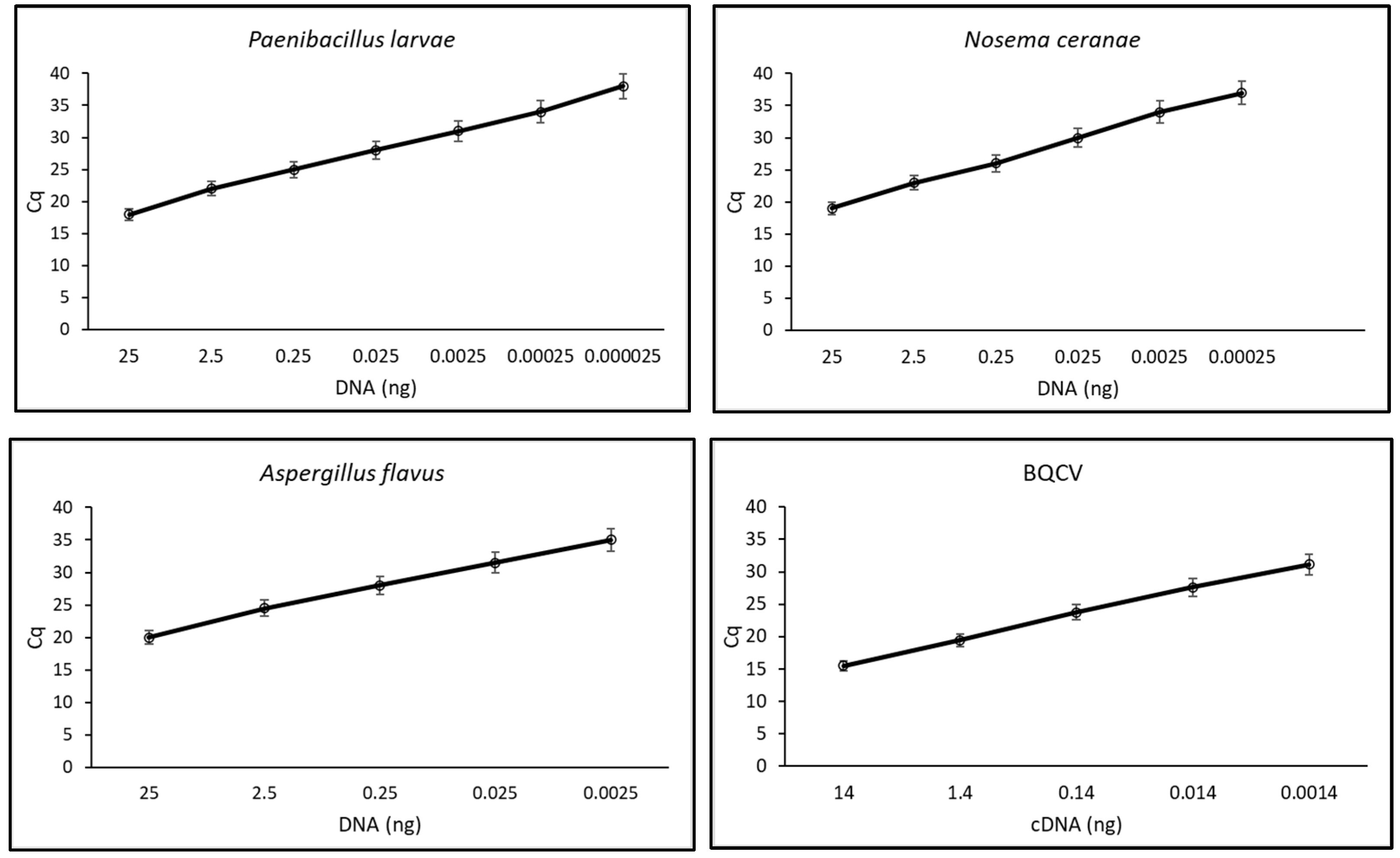
3.3. Assay Sensitivity and Validation
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Evans, J.D.; Schwarz, R.S. Bees brought to their knees: Microbes affecting honey bee health. Trends Microbiol. 2011, 19, 614–620. [Google Scholar] [CrossRef]
- Dainat, B.; Vanengelsdorp, D.; Neumann, P. Colony collapse disorder in Europe. Environ. Microbiol. Rep. 2012, 4, 123–125. [Google Scholar] [CrossRef]
- Vanengelsdorp, D.; Evans, J.D.; Saegerman, C.; Mullin, C.; Haubruge, E.; Nguyen, B.K.; Frazier, M.; Frazier, J.; Cox-Foster, D.; Chen, Y.; et al. Colony collapse disorder: A descriptive study. PLoS ONE 2009, 4, e6481. [Google Scholar] [CrossRef]
- De Graaf, D.; Méroc, E.; Nguyen, B.K.; Roelandt, S.; Roels, S.; Van der Stede, Y.; Tonnersen, T.; Kryger, P.; Jaarma, K.; Kuus, M.; et al. Risk indicators affecting honeybee colony survival in Europe: One year of surveillance. Apidologie 2016, 47, 348–378. [Google Scholar] [CrossRef]
- McMenamin, A.J.; Brutscher, L.M.; Glenny, W.; Flenniken, M.L. Abiotic and biotic factors affecting the replication and pathogenicity of bee viruses. Curr. Opin. Insect Sci. 2016, 16, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Ellis, J.D.; Munn, P.A. The worldwide health status of honey bees. Bee World 2005, 86, 88–101. [Google Scholar] [CrossRef]
- Dainat, B.; Grossar, D.; Ecoffey, B.; Haldemann, C. Triplex real-time PCR method for the qualitative detection of European and American foulbrood in honeybee. J. Microbiol. Methods 2018, 146, 61–63. [Google Scholar] [CrossRef]
- Lee, J.-S.; Luong, G.T.H.; Yoon, B.-S. Development of In-Field-Diagnosis of Aspergillus Flavus by Loop-Mediated Isothermal Amplification in Honeybee. J. Apic. 2016, 31, 25–30. [Google Scholar] [CrossRef]
- Swaileh, K.M.; Abdulkhaliq, A. Analysis of aflatoxins, caffeine, nicotine and heavy metals in Palestinian multifloral honey from different geographic regions. J. Sci. Food Agric. 2013, 93, 2116–2120. [Google Scholar] [CrossRef]
- Chen, Y.; Evans, J.D.; Zhou, L.; Boncristiani, H.; Kimura, K.; Xiao, T.; Litkowski, A.M.; Pettis, J.S. Asymmetrical coexistence of Nosema ceranae and Nosema apis in honey bees. J. Invertebr. Pathol. 2009, 101, 204–209. [Google Scholar] [CrossRef]
- Emsen, B.; De la Mora, A.; Lacey, B.; Eccles, L.; Kelly, P.G.; Medina-Flores, C.A.; Petukhova, T.; Morfin, N.; Guzman-Novoa, E. Seasonality of Nosema ceranae Infections and Their Relationship with Honey Bee Populations, Food Stores, and Survivorship in a North American Region. Vet. Sci. 2020, 7, 131. [Google Scholar] [CrossRef]
- Botías, C.; Martín-Hernández, R.; Barrios, L.; Meana, A.; Higes, M. Nosema spp. infection and its negative effects on honey bees (Apis mellifera iberiensis) at the colony level. Vet. Res. 2013, 44, 25. [Google Scholar] [CrossRef]
- Fürst, M.A.; McMahon, D.P.; Osborne, J.L.; Paxton, R.J.; Brown, M.J.F. Disease associations between honeybees and bumblebees as a threat to wild pollinators. Nature 2014, 506, 364–366. [Google Scholar] [CrossRef] [PubMed]
- de Miranda, J.R.; Dainat, B.; Locke, B.; Cordoni, G.; Berthoud, H.; Gauthier, L.; Neumann, P.; Budge, G.E.; Ball, B.V.; Stoltz, D.B. Genetic characterization of slow bee paralysis virus of the honeybee (Apis mellifera L.). J. Gen. Virol. 2010, 91, 2524–2530. [Google Scholar] [CrossRef] [PubMed]
- Scott, A.; Hassler, E.; Formato, G.; Rünzel, M.A.S.; Wilkes, J.; Hassan, A.; Cazier, J. Data mining hive inspections: More frequently inspected honey bee colonies have higher over-winter survival rates. J. Apic. Res. 2023, 62, 983–991. [Google Scholar] [CrossRef]
- Copley, T.R.; Giovenazzo, P.; Jabaji, S.H. Detection of Nosema apis and N. ceranae in honeybee bottom scraps and frass in naturally infected hives. Apidologie 2012, 43, 753–760. [Google Scholar] [CrossRef]
- Schurr, F.; Tison, A.; Militano, L.; Cheviron, N.; Sircoulomb, F.; Rivière, M.-P.; Ribière-Chabert, M.; Thiéry, R.; Dubois, E. Validation of quantitative real-time RT-PCR assays for the detection of six honeybee viruses. J. Virol. Methods 2019, 270, 70–78. [Google Scholar] [CrossRef]
- Truong, A.-T.; Sevin, S.; Kim, S.; Yoo, M.-S.; Cho, Y.S.; Yoon, B. Rapidly quantitative detection of Nosema ceranae in honeybees using ultra-rapid real-time quantitative PCR. J. Vet. Sci. 2021, 22, e40. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Li, J.; Zhang, Y.; Li, Z.; Evans, J.D.; Rose, R.; Gilligan, T.M.; LeBrun, A.; He, N.; Zheng, T.; et al. A novel method for the detection and diagnosis of virus infections in honey bees. J. Virol. Methods 2021, 293, 114163. [Google Scholar] [CrossRef]
- Benjeddou, M.; Leat, N.; Allsopp, M.; Davison, S. Detection of Acute Bee Paralysis Virus and Black Queen Cell Virus from Honeybees by Reverse Transcriptase PCR. Appl. Environ. Microbiol. 2001, 67, 2384–2387. [Google Scholar] [CrossRef]
- Babin, A.; Schurr, F.; Rivière, M.-P.; Chauzat, M.-P.; Dubois, E. Specific detection and quantification of three microsporidia infecting bees, Nosema apis, Nosema ceranae, and Nosema bombi, using probe-based real-time PCR. Eur. J. Protistol. 2022, 86, 125935. [Google Scholar] [CrossRef] [PubMed]
- Genersch, E. Honey bee pathology: Current threats to honey bees and beekeeping. Appl. Microbiol. Biotechnol. 2010, 87, 87–97. [Google Scholar] [CrossRef]
- Forsgren, E. European foulbrood in honey bees. J. Invertebr. Pathol. 2010, 103, S5–S9. [Google Scholar] [CrossRef] [PubMed]
- de Graaf, D.C.; Vandekerchove, D.; Dobbelaere, W.; Peeters, J.E.; Jacobs, F.J. Influence of the proximity of American foulbrood cases and apicultural management on the prevalence of Paenibacillus larvae spores in Belgian honey. Apidologie 2001, 32, 587–599. [Google Scholar] [CrossRef]
- Evans, J.D.; Schwarz, R.S.; Chen, Y.P.; Budge, G.; Cornman, R.S.; De la Rua, P.; de Miranda, J.R.; Foret, S.; Foster, L.; Gauthier, L.; et al. Standard methods for molecular research in Apis mellifera. J. Apic. Res. 2013, 52, 1–54. [Google Scholar] [CrossRef]
- Foley, K.; Fazio, G.; Jensen, A.B.; Hughes, W.O.H. The distribution of Aspergillus spp. opportunistic parasites in hives and their pathogenicity to honey bees. Vet. Microbiol. 2014, 169, 203–210. [Google Scholar] [CrossRef]
- de Graaf, D.C.; Alippi, A.M.; Antúnez, K.; Aronstein, K.A.; Budge, G.; De Koker, D.; De Smet, L.; Dingman, D.W.; Evans, J.D.; Foster, L.J.; et al. Standard methods for American foulbrood research. J. Apic. Res. 2013, 52, 1–28. [Google Scholar] [CrossRef]
- American Foulbrood of Honey Bees (Infection of Honey Bees with Paenibacillus larvae). Available online: https://www.woah.org/fileadmin/Home/fr/Health_standards/tahm/3.02.02_AMERICAN_FOULBROOD.pdf (accessed on 1 July 2025).
- Rossi, F.; Amadoro, C.; Ruberto, A.; Ricchiuti, L. Evaluation of Quantitative PCR (qPCR) Paenibacillus larvae Targeted Assays and Definition of Optimal Conditions for Its Detection/Quantification in Honey and Hive Debris. Insects 2018, 9, 165. [Google Scholar] [CrossRef]
- Ribani, A.; Utzeri, V.J.; Taurisano, V.; Fontanesi, L. Honey as a Source of Environmental DNA for the Detection and Monitoring of Honey Bee Pathogens and Parasites. Vet. Sci. 2020, 7, 113. [Google Scholar] [CrossRef] [PubMed]
- Mackay, J.F.; Hewett, R.E.; Smith, N.T.; Waters, T.L.; Scandrett, J.S. The Foster method: Rapid and non-invasive detection of clinically significant American Foulbrood disease levels using eDNA sampling and a dual-target qPCR assay, with its potential for other hive pathogens. J. Apic. Res. 2023, 64, 693–700. [Google Scholar] [CrossRef]
- Papić, B.; Žvokelj, L.; Ocepek, M.P.; Hočevar, B.; Kozar, M.; Rus, R.; Zajc, U.; Kušar, D. The Diagnostic Value of qPCR Quantification of Paenibacillus larvae in Hive Debris and Adult Bees for Predicting the Onset of American Foulbrood. Vet. Sci. 2024, 11, 442. [Google Scholar] [CrossRef]
- Han, S.-H.; Lee, D.-B.; Lee, D.-W.; Kim, E.-H.; Yoon, B.-S. Ultra-rapid real-time PCR for the detection of Paenibacillus larvae, the causative agent of American Foulbrood (AFB). J. Invertebr. Pathol. 2008, 99, 8–13. [Google Scholar] [CrossRef]
- Traver, B.E.; Fell, R.D. Prevalence and infection intensity of Nosema in honey bee (Apis mellifera L.) colonies in Virginia. J. Invertebr. Pathol. 2011, 107, 43–49. [Google Scholar] [CrossRef]
- Carra, E.; Galletti, G.; Carpana, E.; Bergamini, F.; Loglio, G.; Bosi, F.; Palminteri, S.; Bassi, S. A Probe-Based qPCR Method, Targeting 16S rRNA Gene, for the Quantification of Paenibacillus larvae Spores in Powdered Sugar Samples. Appl. Sci. 2022, 12, 9895. [Google Scholar] [CrossRef]
- Houbraken, J.; Kocsubé, S.; Visagie, C.M.; Yilmaz, N.; Wang, X.-C.; Meijer, M.; Kraak, B.; Hubka, V.; Bensch, K.; Samson, R.A.; et al. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series and species. Stud. Mycol. 2020, 95, 5–169. [Google Scholar] [CrossRef]
- Sanzani, S.M.; Schena, L.; De Cicco, V.; Ippolito, A. Early detection of Botrytis cinerea latent infections as a tool to improve postharvest quality of table grapes. Postharvest Biol. Technol. 2012, 68, 64–71. [Google Scholar] [CrossRef]
- WOAH. Nosemosis of Honey Bees. WOAH Terrestrial Manual 2024, Chapter 3.2.4. Available online: https://www.woah.org/fileadmin/Home/eng/Health_standards/tahm/3.02.04_NOSEMOSIS.pdf (accessed on 8 July 2025).
- Bourgeois, A.L.; Rinderer, T.E.; Beaman, L.D.; Danka, R.G. Genetic detection and quantification of Nosema apis and N. ceranae in the honey bee. J. Invertebr. Pathol. 2010, 103, 53–58. [Google Scholar] [CrossRef]
- Hamiduzzaman, M.M.; Guzman-Novoa, E.; Goodwin, P.H. A multiplex PCR assay to diagnose and quantify Nosema infections in honey bees (Apis mellifera). J. Invertebr. Pathol. 2010, 105, 151–155. [Google Scholar] [CrossRef]
- Chapter 9.2. Available online: https://www.woah.org/fileadmin/Home/eng/Health_standards/tahc/current/en_chapitre_paenibacillus_larvae.htm?utm_source=chatgpt.com (accessed on 11 July 2025).
- de Miranda, J.R.; Bailey, L.; Ball, B.V.; Blanchard, P.; Budge, G.E.; Chejanovsky, N.; Chen, Y.-P.; Gauthier, L.; Genersch, E.; de Graaf, D.C.; et al. Standard methods for virus research in Apis mellifera. J. Apic. Res. 2013, 52, 1–56. [Google Scholar] [CrossRef]
- Tantillo, G.; Bottaro, M.; Di Pinto, A.; Martella, V.; Di Pinto, P.; Terio, V. Virus Infections of Honeybees Apis mellifera. Ital. J. Food Saf. 2015, 4, 5364. [Google Scholar] [CrossRef] [PubMed]

| Protocols | Honey Bee Part |
|---|---|
| Automated CTAB-method | Head |
| Body (excluding the head) | |
| Manual CTAB-method | Head |
| Body (excluding the head) | |
| Guanidinium thiocyanate-phenol-chloroform (Trizol) | Head |
| Body (excluding the head) |
| Pathogen | Primer | Sequence (5′-3′) | Annealing T (°C) | Fragment Size (bp) | Region/Gene (Genbank No.) |
|---|---|---|---|---|---|
| N. ceranae | NcerRT-F | GCAGCCGCGGTAATACTTGT | 60 | 201 | ITS (OL966535) |
| NcerRT-F | TCCTGCATTCGACCTCCT | ||||
| A. flavus | Aflav-F | TAGCCGCCATAATTTTATCCAG | 60 | 127 | Calmodulin (OR947226) |
| Aflav-R | TTTTGGCCCAGAGAGCGCAT | ||||
| P. larvae | Plar-F | GGCGACCTTTCAACCCTTGT | 60 | 212 | 16S rDNA (JF423915) |
| Plar-R | TCCTCCGTGTGCTCTTACCA | ||||
| BQCV | BQCV1F | GGGAGTCGCAGAGTTCCAAA | 58 | 205 | Capsid protein gene (MW442614) |
| BQCV1R | CATGAATACAGGGCGGCGTA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanzani, S.M.; Kubaa, R.A.; Jabri, B.-E.; Zaidat, S.A.E.; Addante, R.; Admane, N.; Djelouah, K. Rapid and Accurate Detection of the Most Common Bee Pathogens; Nosema ceranae, Aspergillus flavus, Paenibacillus larvae and Black Queen Cell Virus. Insects 2025, 16, 810. https://doi.org/10.3390/insects16080810
Sanzani SM, Kubaa RA, Jabri B-E, Zaidat SAE, Addante R, Admane N, Djelouah K. Rapid and Accurate Detection of the Most Common Bee Pathogens; Nosema ceranae, Aspergillus flavus, Paenibacillus larvae and Black Queen Cell Virus. Insects. 2025; 16(8):810. https://doi.org/10.3390/insects16080810
Chicago/Turabian StyleSanzani, Simona Marianna, Raied Abou Kubaa, Badr-Eddine Jabri, Sabri Ala Eddine Zaidat, Rocco Addante, Naouel Admane, and Khaled Djelouah. 2025. "Rapid and Accurate Detection of the Most Common Bee Pathogens; Nosema ceranae, Aspergillus flavus, Paenibacillus larvae and Black Queen Cell Virus" Insects 16, no. 8: 810. https://doi.org/10.3390/insects16080810
APA StyleSanzani, S. M., Kubaa, R. A., Jabri, B.-E., Zaidat, S. A. E., Addante, R., Admane, N., & Djelouah, K. (2025). Rapid and Accurate Detection of the Most Common Bee Pathogens; Nosema ceranae, Aspergillus flavus, Paenibacillus larvae and Black Queen Cell Virus. Insects, 16(8), 810. https://doi.org/10.3390/insects16080810







