Comparative Mitogenomic Analysis of Three Chionea Species (Tipulomorpha: Limoniidae): Insights into Phylogenetic Relationships and Selection Pressure
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Preservation
2.2. DNA Isolation, PCR Amplification, and Sanger Sequencing
2.3. Gene Annotation and Sequence Analysis
2.4. Calculation of Genetic Distance
2.5. Phylogenetic Analyses
2.6. Selection Analysis
3. Results
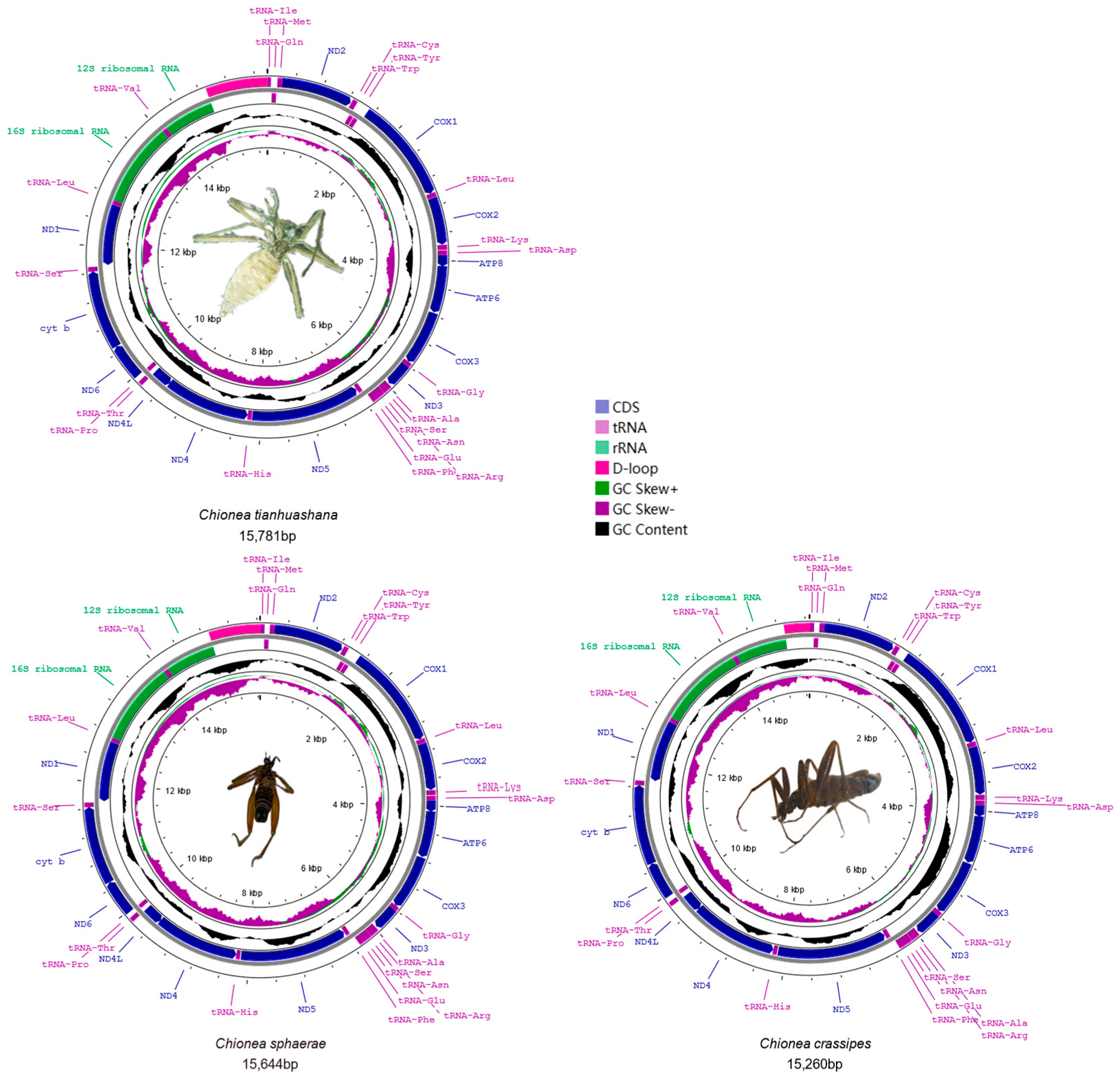
3.1. Gene Structure of Three Mitochondrial Genomes
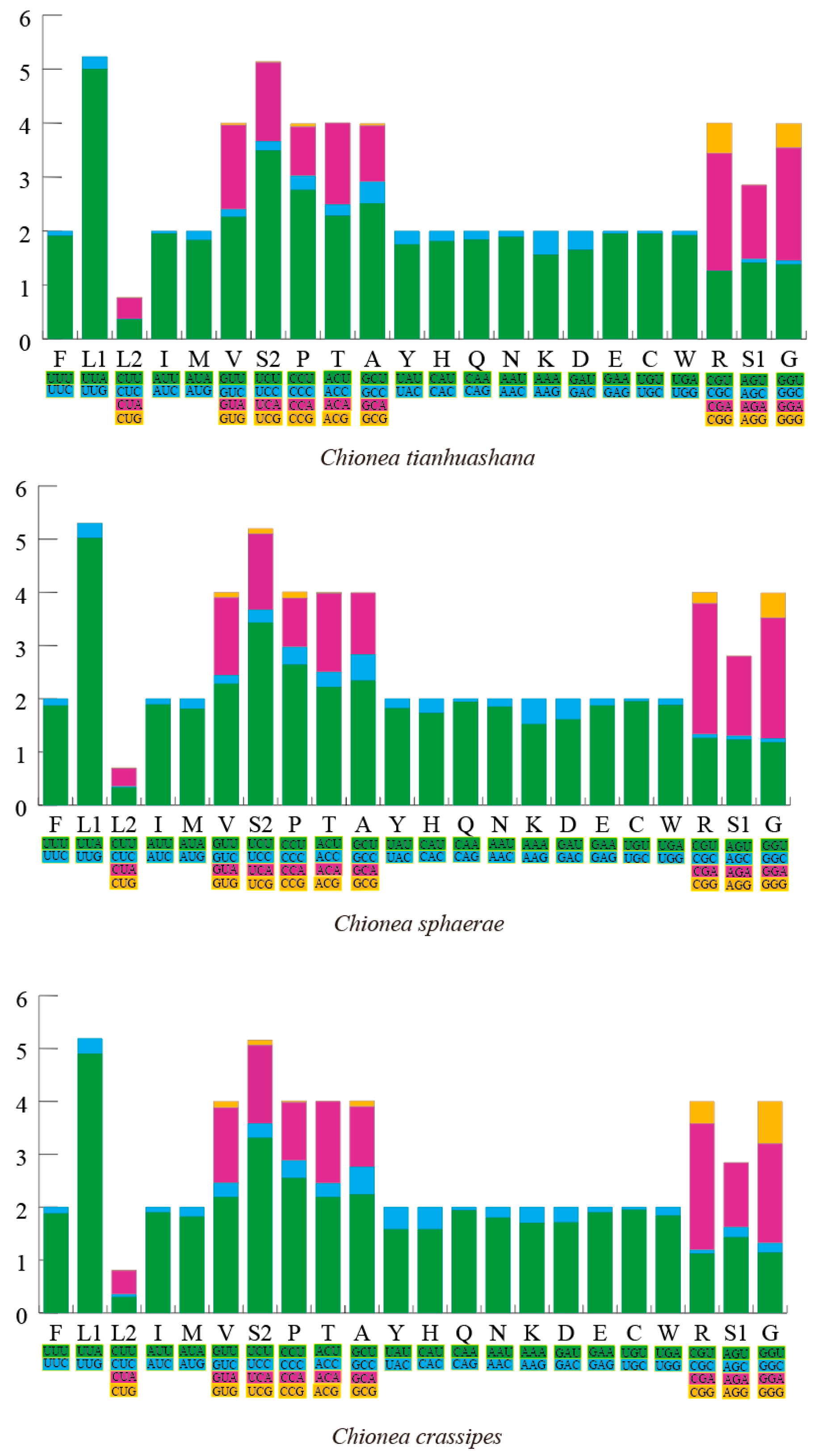
3.2. Utilization of Codons
3.3. Transfer RNA (tRNA) and Ribosomal RNA (rRNA)
3.4. Genetic Distances
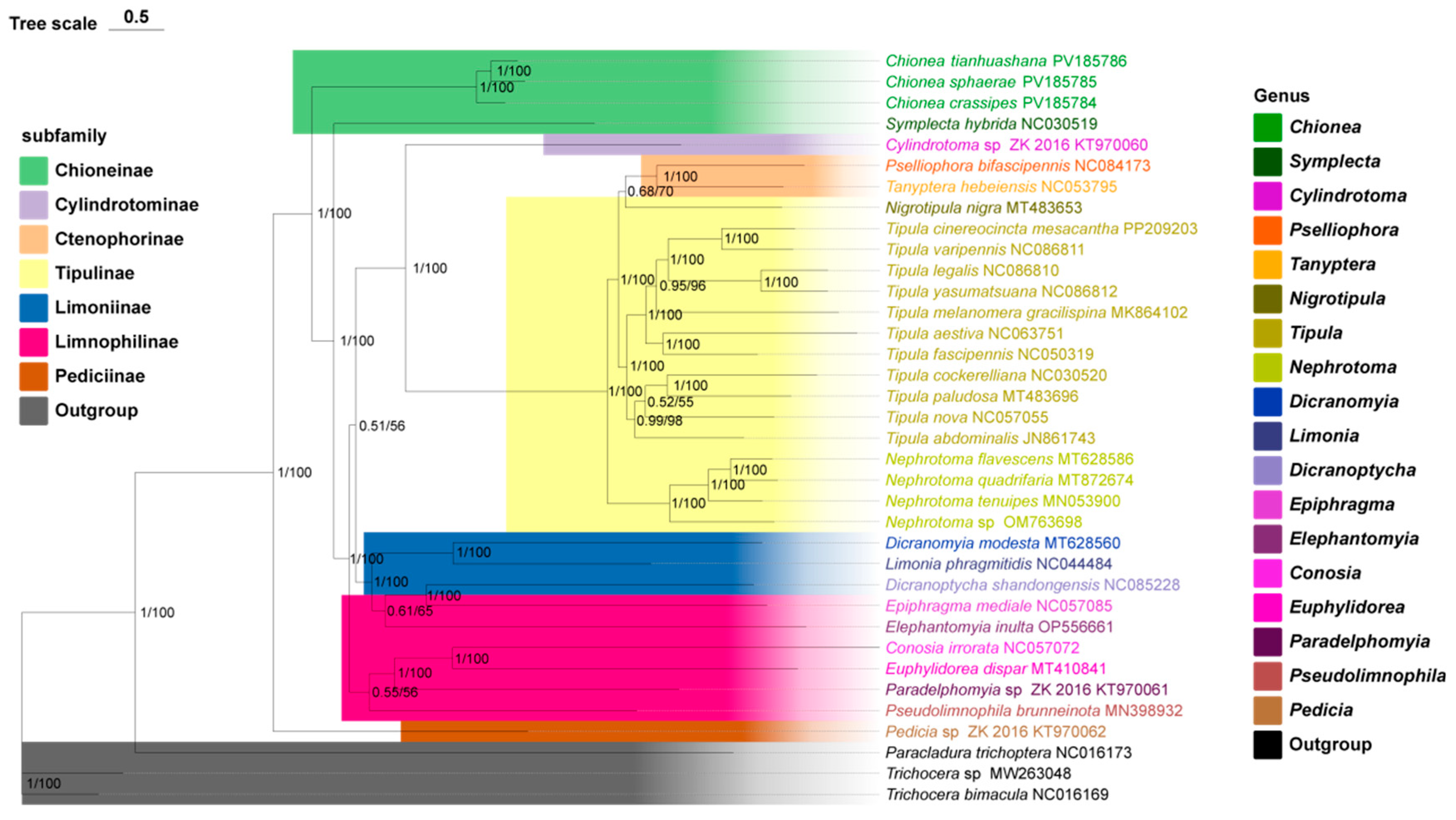
3.5. Phylogenetic Relationship
3.6. Positive Selection
4. Discussion
4.1. Organization of the Mitochondrial Genome
4.2. Genetic Divergence and Evolutionary Relationships
4.3. Selection Analysis of Three Crane Flies
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Jong, H.; Oosterbroek, P.; Gelhaus, J.; Reusch, H.; Young, C. Global diversity of craneflies (Insecta, Diptera: Tipulidea or Tipulidae sensu lato) in freshwater. Freshw. Anim. Divers. Assess. 2008, 595, 457–467. [Google Scholar] [CrossRef]
- Vane-Wright, R. A re-assessment of the genera Holorusia Loew (= Ctenacroscelis Enderlein), Ischnotoma Skuse and Zelandotipula Alexander (Diptera: Tipulidae), with notes on their phylogeny and zoogeography. J. Nat. Hist. 1967, 1, 511–547. [Google Scholar] [CrossRef]
- Wagner, R.; Barták, M.; Borkent, A.; Courtney, G.; Goddeeris, B.; Haenni, J.P.; Knutson, L.; Pont, A.; Rotheray, G.E.; Rozkošný, R. Global diversity of dipteran families (Insecta Diptera) in freshwater (excluding Simulidae, Culicidae, Chironomidae, Tipulidae and Tabanidae). Hydrobiologia 2008, 595, 489–519. [Google Scholar] [CrossRef]
- Vanin, S. Chionea (Chionea) mirabilis n. sp., a new species of snow fly (Insecta, Diptera, Limoniidae) from Korea. Zoosystema 2008, 30, 413–418. [Google Scholar]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef]
- Ballard, J.W.O.; Rand, D.M. The population biology of mitochondrial DNA and its phylogenetic implications. Annu. Rev. Ecol. Evol. Syst. 2005, 36, 621–642. [Google Scholar] [CrossRef]
- Lang, B.F.; Gray, M.W.; Burger, G. Mitochondrial genome evolution and the origin of eukaryotes. Annu. Rev. Genet. 1999, 33, 351–397. [Google Scholar] [CrossRef]
- Bernt, M.; Braband, A.; Schierwater, B.; Stadler, P.F. Genetic aspects of mitochondrial genome evolution. Mol. Phylogenet. Evol. 2013, 69, 328–338. [Google Scholar] [CrossRef]
- Yang, L.; Tan, Z.; Wang, D.; Xue, L.; Guan, M.X.; Huang, T.; Li, R. Species identification through mitochondrial rRNA genetic analysis. Sci. Rep. 2014, 4, 4089. [Google Scholar] [CrossRef]
- Wolf, C.; Rentsch, J.; Hübner, P. PCR− RFLP analysis of mitochondrial DNA: A reliable method for species identification. J. Agric. Food Chem. 1999, 47, 1350–1355. [Google Scholar] [CrossRef]
- Rastogi, G.; Dharne, M.S.; Walujkar, S.; Kumar, A.; Patole, M.S.; Shouche, Y.S. Species identification and authentication of tissues of animal origin using mitochondrial and nuclear markers. Meat Sci. 2007, 76, 666–674. [Google Scholar] [CrossRef] [PubMed]
- Palumbi, A.; Cipriano, F. Species identification using genetic tools: The value of nuclear and mitochondrial gene sequences in whale conservation. J. Hered. 1998, 89, 459–464. [Google Scholar] [CrossRef] [PubMed]
- Salinas-Giegé, T.; Giegé, R.; Giegé, P. tRNA biology in mitochondria. Int. J. Mol. Sci. 2015, 16, 4518–4559. [Google Scholar] [CrossRef]
- Roger, A.J.; Muñoz-Gómez, S.A.; Kamikawa, R. The origin and diversification of mitochondria. Curr. Biol. 2017, 27, R1177–R1192. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Lv, Y.; Wen, Z.; Bian, C.; Zhang, X.; Guo, S.; Shi, Q.; Li, D. The complete mitochondrial genome of the intertidal spider (Desis jiaxiangi) provides novel insights into the adaptive evolution of the mitogenome and the evolution of spiders. BMC Ecol. Evol. 2021, 21, 72. [Google Scholar] [CrossRef]
- Clary, D.O.; Wolstenholme, D.R. The mitochondrial DNA molecule of Drosophila yakuba: Nucleotide sequence, gene organization, and genetic code. J. Mol. Evol. 1985, 22, 252–271. [Google Scholar] [CrossRef]
- Wei, S.J.; Shi, M.; Sharkey, M.J.; van Achterberg, C.; Chen, X.X. Comparative mitogenomics of Braconidae (Insecta: Hymenoptera) and the phylogenetic utility of mitochondrial genomes with special reference to Holometabolous insects. BMC Genomics 2010, 11, 371. [Google Scholar] [CrossRef]
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef]
- Zhang, D.X.; Hewitt, G.M. Insect mitochondrial control region: A review of its structure, evolution and usefulness in evolutionary studies. Biochem. Syst. Ecol. 1997, 25, 99–120. [Google Scholar] [CrossRef]
- Hebert, P.D.; Cywinska, A.; Ball, S.L.; DeWaard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. London. Ser. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef]
- Kang, Z.; Xu, Y.; Wang, G.; Yang, D.; Zhang, X. First mitochondrial genomes of the crane fly tribe Elephantomyiini (Diptera, Tipuloidea, Limoniidae): Comparative analysis and phylogenetic implications. Arthropod. Syst. Phylo. 2023, 81, 731–746. [Google Scholar] [CrossRef]
- Ribeiro, G.C. Homology of the gonostylus parts in crane flies, with emphasis on the families Tipulidae and Limoniidae (Diptera, Tipulomorpha). Zootaxa 2006, 1110, 47–57. [Google Scholar] [CrossRef]
- Petersen, M.J.; Bertone, M.A.; Wiegmann, B.M.; Courtney, G.W. Phylogenetic synthesis of morphological and molecular data reveals new insights into the higher-level classification of Tipuloidea (Diptera). Syst. Entomol. 2010, 35, 526–545. [Google Scholar] [CrossRef]
- Savchenko, E. New or little-known palaearctic species of the genus Paradelphomyia Al. (Diptera, Limoniidae). Zool. Zhurnal 1976, 55, 387–394. [Google Scholar]
- Starý, J. Phylogeny and classification of Tipulomorpha, with special emphasis on the family Limoniidae. Acta Zool. Cracoviensia 1992, 35, 11–36. [Google Scholar]
- Song, N.; Wu, Q.; Lin, X.; Zhang, Y.; Zhao, T. The complete mitochondrial genome of Nephrotoma scalaris parvinotata (Brunetti 1918)(Diptera: Tipulidae) and insights into the phylogeny of Tipulomorpha. J. Asia-Pac. Entomol. 2023, 26, 102133. [Google Scholar] [CrossRef]
- Kania-Kłosok, I.; Nel, A.; Szwedo, J.; Jordan-Stasiło, W.; Krzemiński, W. Phylogeny and biogeography of the enigmatic ghost lineage Cylindrotomidae (Diptera, Nematocera). Sci. Rep. 2021, 11, 13916. [Google Scholar] [CrossRef]
- Kolcsár, L.-P.; Oosterbroek, P.; Olsen, K.M.; Paramonov, N.M.; Gavryushin, D.I.; Pilipenko, V.E.; Polevoi, A.V.; Eiroa, E.; Andersson, M.; Dufour, C. Contribution to the knowledge of Cylindrotomidae, Pediciidae and Tipulidae (Diptera: Tipuloidea): First records of 86 species from various European countries. Diversity 2023, 15, 336. [Google Scholar] [CrossRef]
- Ribeiro, G.C. Phylogeny of the Limnophilinae (Limoniidae) and early evolution of the Tipulomorpha (Diptera). Invertebr. Syst. 2008, 22, 627–694. [Google Scholar] [CrossRef]
- Lukashevich, E.D.; Ribeiro, G.C. Mesozoic fossils and the phylogeny of Tipulomorpha (Insecta: Diptera). J. Syst. Palaeontol. 2019, 17, 635–652. [Google Scholar] [CrossRef]
- Kang, Z.; Zhang, X.; Ding, S.; Tang, C.; Wang, Y.; Jong, H.D.; Cameron, S.L.; Wang, M.; Yang, D. Transcriptomes of three species of Tipuloidea (Diptera, Tipulomorpha) and implications for phylogeny of Tipulomorpha. PLoS ONE 2017, 12, e0173207. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Kang, Z.; Mao, M.; Li, X.; Cameron, S.L.; Jong, H.d.; Wang, M.; Yang, D. Comparative Mt genomics of the Tipuloidea (Diptera: Nematocera: Tipulomorpha) and its implications for the phylogeny of the Tipulomorpha. PLoS ONE 2016, 11, e0158167. [Google Scholar] [CrossRef] [PubMed]
- Bertram, J.; Gomez, K.; Masel, J. Predicting patterns of long-term adaptation and extinction with population genetics. Evolution 2017, 71, 204–214. [Google Scholar] [CrossRef] [PubMed]
- Kryazhimskiy, S.; Plotkin, J.B. The population genetics of dN/dS. PLoS Genet. 2008, 4, e1000304. [Google Scholar] [CrossRef]
- Bomblies, K.; Peichel, C.L. Genetics of adaptation. Proc. Natl. Acad. Sci. USA 2022, 119, e2122152119. [Google Scholar] [CrossRef]
- Zong, S.B.; Li, Y.L.; Liu, J.X. Genomic architecture of rapid parallel adaptation to fresh water in a wild fish. Mol. Biol. Evol. 2021, 38, 1317–1329. [Google Scholar] [CrossRef]
- Ballard, J.W.O.; Kreitman, M. Is mitochondrial DNA a strictly neutral marker? Trends. Ecol. Evol. 1995, 10, 485–488. [Google Scholar] [CrossRef]
- Xu, X.D.; Guan, J.Y.; Zhang, Z.Y.; Cao, Y.R.; Cai, Y.Y.; Storey, K.B.; Yu, D.N.; Zhang, J.Y. Insight into the phylogenetic relationships among three subfamilies within Heptageniidae (Insecta: Ephemeroptera) along with low-temperature selection pressure analyses using mitogenomes. Insects 2021, 12, 656. [Google Scholar] [CrossRef]
- Lane, N. Mitochondria and the W chromosome: Low variability on the W chromosome in birds is more likely to indicate selection on mitochondrial genes. Heredity 2008, 100, 444–445. [Google Scholar] [CrossRef]
- James, J.E.; Piganeau, G.; Eyre-Walker, A. The rate of adaptive evolution in animal mitochondria. Mol. Ecol. 2016, 25, 67–78. [Google Scholar] [CrossRef]
- Ning, T.; Xiao, H.; Li, J.; Hua, S.; Zhang, Y. Adaptive evolution of the mitochondrial ND6 gene in the domestic horse. Genet. Mol. Res. 2010, 9, 144–150. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Xu, S.; Xu, J.; Guo, Y.; Yang, G. Adaptive evolution of mitochondrial energy metabolism genes associated with increased energy demand in flying insects. PLoS ONE 2014, 9, e99120. [Google Scholar] [CrossRef] [PubMed]
- Illustrator, A. Adobe Illustrator. 2021. Available online: https://adobe.hmzsya.com (accessed on 12 January 2025).
- Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Zhang, J.Y.; Zhou, C.F.; Gai, Y.H.; Song, D.X.; Zhou, K.Y. The complete mitochondrial genome of Parafronurus youi (Insecta: Ephemeroptera) and phylogenetic position of the Ephemeroptera. Gene 2008, 424, 18–24. [Google Scholar] [CrossRef]
- Lalitha, S. Primer premier 5. Biotech Softw. Internet Rep. Comput. Softw. J. Sci. 2000, 1, 270–272. [Google Scholar] [CrossRef]
- Burland, T.G. DNASTAR’s Lasergene sequence analysis software. Bioinform. Methods Protoc. 1999, 132, 71–91. [Google Scholar]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Grant, J.R.; Stothard, P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, W181–W184. [Google Scholar] [CrossRef]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE Online: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Krzemińska, E. A new subgenus and two new species of the genus Trichocera Meigen, 1803 (Diptera: Trichoceridae). Ann. Zool. 2002, 52, 201–205. [Google Scholar]
- Beckenbach, A.T. Mitochondrial genome sequences of Nematocera (lower Diptera): Evidence of rearrangement following a complete genome duplication in a winter crane fly. Genome Biol. Evol. 2012, 4, 89–101. [Google Scholar] [CrossRef]
- Kolcsár, L.P.; Ivković, M.; Ternjej, I. New records of Limoniidae and Pediciidae (Diptera) from Croatia. ZooKeys 2015, 513, 23–37. [Google Scholar]
- Starý, J.; Brodo, F. Arctic species of the subgenus Symplecta sensu stricto (Diptera: Limoniidae). Can. Entomol. 2009, 141, 1–30. [Google Scholar] [CrossRef]
- Zhang, B.; Gao, S.; Yang, D. The mitochondrial genome of Conosia irrorata (Diptera: Limoniidae). Mitochondrial DNA B 2019, 4, 2367–2368. [Google Scholar] [CrossRef]
- Dahl, C.; Alexander, C.P. A world catalogue of Trichoceridae Kertész, 1902*(Diptera). Insect. Syst. Evol. 1976, 7, 7–18. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Kück, P.; Meid, S.A.; Groß, C.; Wägele, J.W.; Misof, B. AliGROOVE–visualization of heterogeneous sequence divergence within multiple sequence alignments and detection of inflated branch support. BMC Bioinform. 2014, 15, 294. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, R.; Calcott, B.; Ho, S.Y.; Guindon, S. PartitionFinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 2012, 29, 1695–1701. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 214. [Google Scholar] [CrossRef]
- Gao, F.; Chen, C.; Arab, D.A.; Du, Z.; He, Y.; Ho, S.Y.J.E. EasyCodeML: A visual tool for analysis of selection using CodeML. Ecol. Evol. 2019, 9, 3891–3898. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Fouquet, A.; Marinho, P.; Réjaud, A.; Carvalho, T.R.; Caminer, M.A.; Jansen, M.; Rainha, R.N.; Rodrigues, M.T.; Werneck, F.P.; Lima, A.P. Systematics and biogeography of the Boana albopunctata species group (Anura, Hylidae), with the description of two new species from Amazonia. Syst. Biodivers. 2021, 19, 375–399. [Google Scholar] [CrossRef]
- Xu, T.J.; Cheng, Y.Z.; Sun, Y.N.; Shi, G.; Wang, R.X. The complete mitochondrial genome of bighead croaker, Collichthys niveatus (Perciformes, Sciaenidae): Structure of control region and phylogenetic considerations. Mol. Biol. Rep. 2011, 38, 4673–4685. [Google Scholar] [CrossRef] [PubMed]
- Prabhu, V.R.; Singha, H.S.; Kumar, R.G.; Gopalakrishnan, A.; Nagarajan, M. Characterization of the complete mitochondrial genome of Barilius malabaricus and its phylogenetic implications. Genomics 2020, 112, 2154–2163. [Google Scholar] [CrossRef] [PubMed]
- Behura, S.K.; Severson, D.W. Comparative analysis of codon usage bias and codon context patterns between dipteran and hymenopteran sequenced genomes. PLoS ONE 2012, 7, e43111. [Google Scholar] [CrossRef] [PubMed]
- Behura, S.K.; Severson, D.W. Coadaptation of isoacceptor tRNA genes and codon usage bias for translation efficiency in Aedes aegypti and Anopheles gambiae. Insect Mol. Biol. 2011, 20, 177–187. [Google Scholar] [CrossRef]
- Vicario, S.; Moriyama, E.N.; Powell, J.R. Codon usage in twelve species of Drosophila. BMC Evol. Biol. 2007, 7, 226. [Google Scholar] [CrossRef]
- Yuan, Y.N.; Zhang, L.H.; Li, K.; Hong, Y.H.; Storey, K.B.; Zhang, J.Y.; Yu, D.N. Nine mitochondrial genomes of Phasmatodea with two novel mitochondrial gene rearrangements and phylogeny. Insects 2023, 14, 485. [Google Scholar] [CrossRef]
- Guan, J.Y.; Zhang, Z.Y.; Cao, Y.R.; Xu, X.D.; Storey, K.B.; Yu, D.N.; Zhang, J.Y. The complete mitochondrial genome of Choroterpes (Euthralus) yixingensis (Ephemeroptera: Leptophlebiidae) and its mitochondrial protein-coding gene expression under imidacloprid stress. Gene 2021, 800, 145833. [Google Scholar] [CrossRef]
- Bickford, D.; Lohman, D.J.; Sodhi, N.S.; Ng, P.K.; Meier, R.; Winker, K.; Ingram, K.K.; Das, I. Cryptic species as a window on diversity and conservation. Trends. Ecol. Evol. 2007, 22, 148–155. [Google Scholar] [CrossRef]
- Oosterbroek, P. The western palaearctic species of Nephrotoma Meigen, 1803 (Diptera, Tipulidae). Part 5: Phylogeny and Biogeography. Beaufortia 1980, 29, 311–385. [Google Scholar]
- Ren, J.; Zhang, C.; Chang, W.; Yang, D. The mitochondrial genome of Nephrotoma tenuipes (Diptera Tipulidae). Mitochondrial DNA B 2019, 4, 3092–3093. [Google Scholar] [CrossRef]
- Hong, Y.H.; Huang, H.M.; Wu, L.; Storey, K.B.; Zhang, J.Y.; Zhang, Y.P.; Yu, D.N. Characterization of two mitogenomes of Hyla sanchiangensis (Anura: Hylidae), with phylogenetic relationships and selection pressure analyses of Hylidae. Animals 2023, 13, 1593. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.Q.; Gao, Y.J.; Chen, Y.X.; Zhan, L.M.; Storey, K.B.; Yu, D.N.; Zhang, J.Y. Comparative mitogenome of phylogenetic relationships and divergence time analysis within Potamanthidae (Insecta: Ephemeroptera). Insects 2024, 15, 357. [Google Scholar] [CrossRef] [PubMed]
- De Jong, H.; Ciliberti, P. How cold-adapted flightless flies dispersed over the northern hemisphere: Phylogeny and biogeography of the snow fly genus Chionea Dalman (Diptera: Limoniidae). Syst. Entomol. 2014, 39, 563–589. [Google Scholar] [CrossRef]
- Saraste, M. Oxidative phosphorylation at the fin de siecle. Science 1999, 283, 1488–1493. [Google Scholar] [CrossRef]
- Dotson, E.M.; Beard, C.B. Sequence and organization of the mitochondrial genome of the Chagas disease vector, Triatoma dimidiata. Insect Mol. Biol. 2001, 10, 205–215. [Google Scholar] [CrossRef]
- McKenzie, M.; Chiotis, M.; Pinkert, C.A.; Trounce, I.A. Functional respiratory chain analyses in murid xenomitochondrial cybrids expose coevolutionary constraints of cytochrome b and nuclear subunits of complex III. Mol. Biol. Evol. 2003, 20, 1117–1124. [Google Scholar] [CrossRef]
- Calvaruso, M.A.; Willems, P.; van den Brand, M.; Valsecchi, F.; Kruse, S.; Palmiter, R.; Smeitink, J.; Nijtmans, L. Mitochondrial complex III stabilizes complex I in the absence of NDUFS4 to provide partial activity. Hum. Mol. Genet. 2012, 21, 115–120. [Google Scholar] [CrossRef]
- Da Fonseca, R.R.; Johnson, W.E.; O’Brien, S.J.; Ramos, M.J.; Antunes, A. The adaptive evolution of the mammalian mitochondrial genome. BMC Genomics 2008, 9, 119. [Google Scholar] [CrossRef]
- Yu, D.N.; Zhang, J.Y.; Li, P.; Zheng, R.Q.; Shao, C. Do cryptic species exist in Hoplobatrachus rugulosus? An examination using four nuclear genes, the Cyt b gene and the complete MT genome. PLoS ONE 2015, 10, e0124825. [Google Scholar] [CrossRef]
- Bai, Y.; Shakeley, R.M.; Attardi, G. Tight control of respiration by NADH dehydrogenase ND5 subunit gene expression in mouse mitochondria. Mol. Cell Biol. 2000, 20, 805–815. [Google Scholar] [CrossRef]
- Deng, J.H.; Li, Y.; Park, J.S.; Wu, J.; Hu, P.; Lechleiter, J.; Bai, Y. Nuclear suppression of mitochondrial defects in cells without the ND6 subunit. Mol. Cell Biol. 2006, 26, 1077–1086. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fu, X.; Jiang, X.; Chen, X.; Zhu, L.; Zhang, G. The Differential Expression of Mitochondrial Function-Associated Proteins and Antioxidant Enzymes during Bovine Herpesvirus 1 Infection: A Potential Mechanism for Virus Infection-Induced Oxidative Mitochondrial Dysfunction. Mediat. Inflamm. 2019, 2019, 7072917. [Google Scholar] [CrossRef] [PubMed]
- Marienfeld, J.R.; Reski, R.; Abel, W.O. The first analysed archegoniate mitochondrial gene (COX3) exhibits extraordinary features. Curr. Genet. 1991, 20, 319–329. [Google Scholar] [CrossRef]
- Zhang, J.; Nielsen, R.; Yang, Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 2005, 22, 2472–2479. [Google Scholar] [CrossRef] [PubMed]
- Li, X.D.; Jiang, G.F.; Yan, L.Y.; Li, R.; Mu, Y.; Deng, W.A. Positive selection drove the adaptation of mitochondrial genes to the demands of flight and high-altitude environments in grasshoppers. Front. Genet. 2018, 9, 605. [Google Scholar] [CrossRef]
| Family | Subfamily | Genus | Species | Length (bp) | GenBank Accession Number |
|---|---|---|---|---|---|
| Cylindrotomidae | Cylindrotominae | Cylindrotoma | Cylindrotoma sp. ZK 2016 | 15,913 | KT970060 |
| Tipulidae | Ctenophorinae | Pselliophora | Pselliophora bifascipennis | 15,821 | NC084173 |
| Tanyptera | Tanyptera hebeiensis | 15,888 | NC053795 | ||
| Tipulinae | Nigrotipula | Nigrotipula nigra | 15,400 | MT483653 | |
| Tipula | Tipula cinereocincta mesacantha | 15,907 | PP209203 | ||
| Tipula varipennis | 15,772 | NC086811 | |||
| Tipula legalis | 15,625 | NC086810 | |||
| Tipula yasumatsuana | 15,735 | NC086812 | |||
| Tipula melanomera gracilispina | 14,575 | MK864102 | |||
| Tipula aestiva | 16,083 | NC063751 | |||
| Tipula fascipennis | 15,701 | NC050319 | |||
| Tipula cockerelliana | 14,453 | NC030520 | |||
| Tipula paludosa | 15,121 | MT483696 | |||
| Tipula nova | 15,668 | NC057055 | |||
| Tipula abdominalis | 14,566 | JN861743 | |||
| Nephrotoma | Nephrotoma flavescens | 15,269 | MT628586 | ||
| Nephrotoma quadrifaria | 16,579 | MT872674 | |||
| Nephrotoma tenuipes | 14,851 | MN053900 | |||
| Nephrotoma sp. | 17,862 | OM763698 | |||
| Limoniidae | Limoniinae | Dicranomyia | Dicranomyia modesta | 15,311 | MT628560 |
| Limonia | Limonia phragmitidis | 15,924 | NC044484 | ||
| Dicranoptycha | Dicranoptycha shandongensis | 16,157 | NC085228 | ||
| Limnophilinae | Epiphragma | Epiphragma mediale | 14,858 | NC057085 | |
| Elephantomyia | Elephantomyia inulta | 14,551 | OP556661 | ||
| Conosia | Conosia irrorata | 14,634 | NC057072 | ||
| Euphylidorea | Euphylidorea dispar | 16,069 | MT410841 | ||
| Paradelphomyia | Paradelphomyia sp. ZK 2016 | 14,639 | KT970061 | ||
| Pseudolimnophila | Pseudolimnophila brunneinota | 15,985 | MN398932 | ||
| Chioneinae | Chionea | Chionea tianhuashana | 15,781 | PV185786 | |
| Chionea sphaerae | 15,644 | PV185785 | |||
| Chionea crassipes | 15,260 | PV185784 | |||
| Symplecta | Symplecta hybrida | 15,811 | NC030519 | ||
| Pediciidae | Pediciinae | Pedicia | Pedicia sp. ZK 2016 | 14,605 | KT970062 |
| Trichoceridae | Paracladura | Paracladura trichoptera | 16,143 | NC016173 | |
| Trichocera | Trichocera sp. | 16,094 | MW263048 | ||
| Trichocera bimacula | 16,140 | NC016169 |
| 1 | 2 | 3 | 4 | |
|---|---|---|---|---|
| 1. S. hybrida | ||||
| 2. C. tianhuashana | 0.153 | |||
| 3. C. sphaerae | 0.154 | 0.050 | ||
| 4. C. crassipes | 0.154 | 0.058 | 0.061 |
| Site model (SM) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Model | np | Ln L | Estimates of Parameters | Model Compared | LRT P-Value | Positive Sites | |||
| M3 | 70 | −142,014.2418 | p: | 0.6129 | 0.2908 | 0.0963 | M0 vs. M3 | 0.0000 | [] |
| ω: | 0.0035 | 0.0659 | 0.2489 | ||||||
| M0 | 66 | −149,292.6487 | ω0: | 0.0389 | Not Allowed | ||||
| M2a | 69 | −146,919.6452 | p: | 0.9345 | 0.0233 | 0.0422 | M1a vs. M2a | 1.0000 | [] |
| ω: | 0.0344 | 1.0000 | 1.0000 | ||||||
| M1a | 67 | −146,919.6452 | p: | 0.9345 | 0.0655 | Not Allowed | |||
| ω: | 0.0344 | 1.0000 | |||||||
| M8 | 69 | −142,234.6699 | p0 = 0.9862 | p = 0.3307 | q = 4.7220 | M7 vs. M8 | 0.0000 | 270 E 0.556, 273 N 0.798, 2830 R 0.573, 3381 K 0.619, 3497 N 0.703, 3501 I 0.590 | |
| (p1 = 0.0138) | ω = 1.0000 | ||||||||
| M7 | 67 | −142,437.4345 | p = 0.3481 | q = 4.0661 | Not Allowed | ||||
| M8a | 68 | −141,844.7323 | p0 = 0.9940 | p = 0.2520 | q = 4.6615 | M8a vs. M8 | 0.0000 | Not Allowed | |
| (p1 = 0.0060) | ω = 1.0000 | ||||||||
| Branch Model (BM) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Model | np | Ln L | Estimates of Parameters | Model Compared | LRT P-Value | Omega for Foreground Branch | ||
| Two-Ratio Model 2 | 67 | −149,287.6863 | ω: | ω0 = 0.0387 | ω1 = 0.05670 | Model 0 vs. Two-Ratio Model 2 | 0.0016 | ω1 = 0.0570 |
| Model 0 | 66 | −149,292.6488 | ω = 0.0389 | |||||
| Genes | Positive Selection Sites | Amino Acids | BEB Value | Feature Key | |
|---|---|---|---|---|---|
| Foreground | Background | ||||
| COX3 | 1247 | N/T | L/S/T | 0.910 | Domain |
| ND5 | 3489 | S/M | Q/Y/F | 0.953 * | Domain |
| ND6 | 3666 | T/S | N/S | 0.900 | Domain |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, Y.; Cen, W.; Storey, K.B.; Liu, L.; Yu, D.; Zhang, J. Comparative Mitogenomic Analysis of Three Chionea Species (Tipulomorpha: Limoniidae): Insights into Phylogenetic Relationships and Selection Pressure. Insects 2025, 16, 720. https://doi.org/10.3390/insects16070720
Feng Y, Cen W, Storey KB, Liu L, Yu D, Zhang J. Comparative Mitogenomic Analysis of Three Chionea Species (Tipulomorpha: Limoniidae): Insights into Phylogenetic Relationships and Selection Pressure. Insects. 2025; 16(7):720. https://doi.org/10.3390/insects16070720
Chicago/Turabian StyleFeng, Yufeng, Wei Cen, Kenneth B. Storey, Lingjuan Liu, Danna Yu, and Jiayong Zhang. 2025. "Comparative Mitogenomic Analysis of Three Chionea Species (Tipulomorpha: Limoniidae): Insights into Phylogenetic Relationships and Selection Pressure" Insects 16, no. 7: 720. https://doi.org/10.3390/insects16070720
APA StyleFeng, Y., Cen, W., Storey, K. B., Liu, L., Yu, D., & Zhang, J. (2025). Comparative Mitogenomic Analysis of Three Chionea Species (Tipulomorpha: Limoniidae): Insights into Phylogenetic Relationships and Selection Pressure. Insects, 16(7), 720. https://doi.org/10.3390/insects16070720






