Laboratory Test Indirectly Reveals the Unreliability of RNA-Dependent 16S rRNA Amplicon Sequences in Detecting the Gut Bacterial Diversity of Delia antiqua
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Rearing and Microbial Culture Media
2.2. Larval Gut Sample Collection
2.3. DNA- and RNA-Dependent 16S rRNA Amplicon Sequence
DNA/RNA Extraction and Amplicon Sequencing
2.4. Isolation and Identification of D. antiqua Gut Bacteria
2.5. SynComs Constructed According to DNA/RNA-Dependent Amplicon Sequencing
2.6. Effects of SynComs on Larval Survival and Growth
2.7. Data Analysis
3. Results
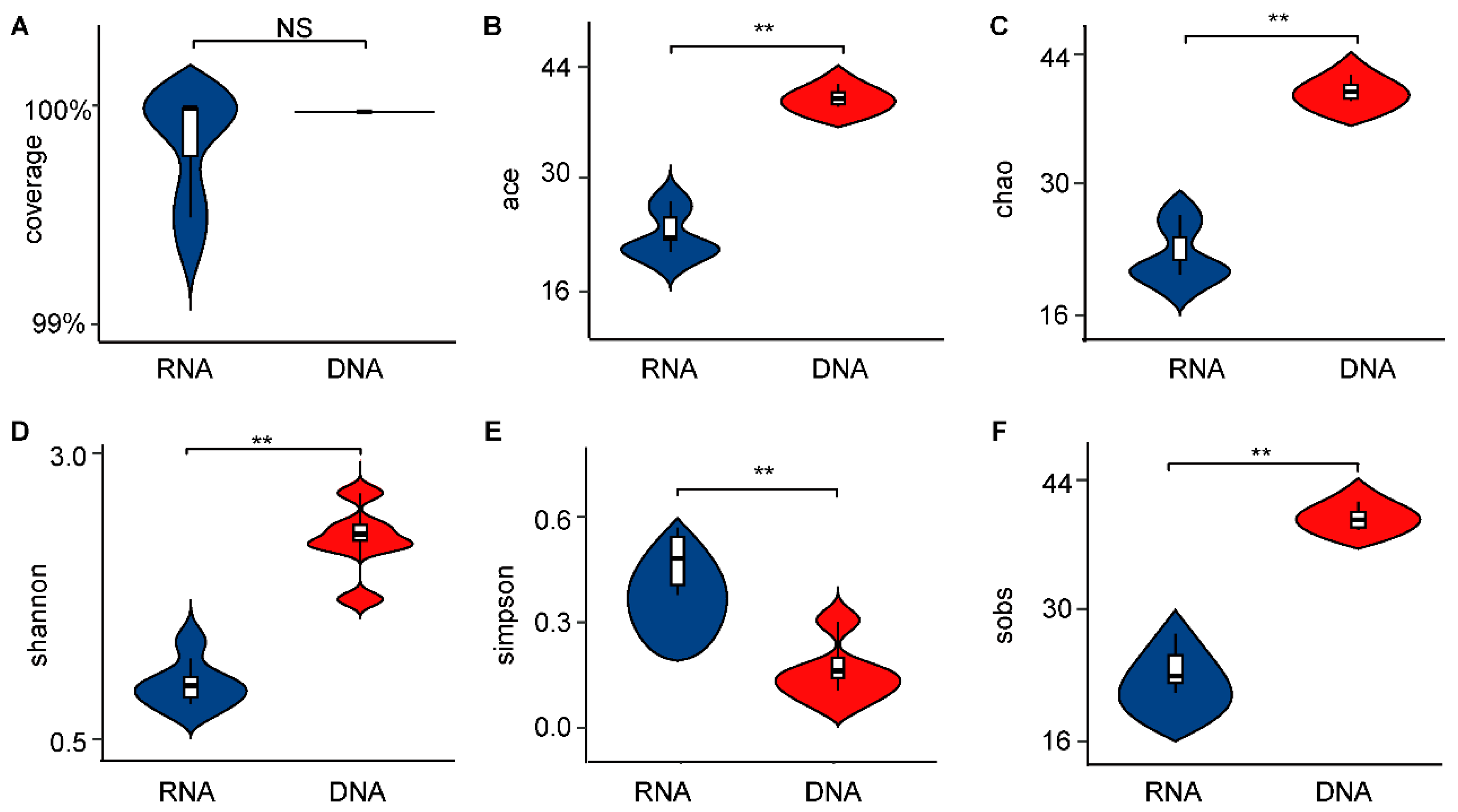
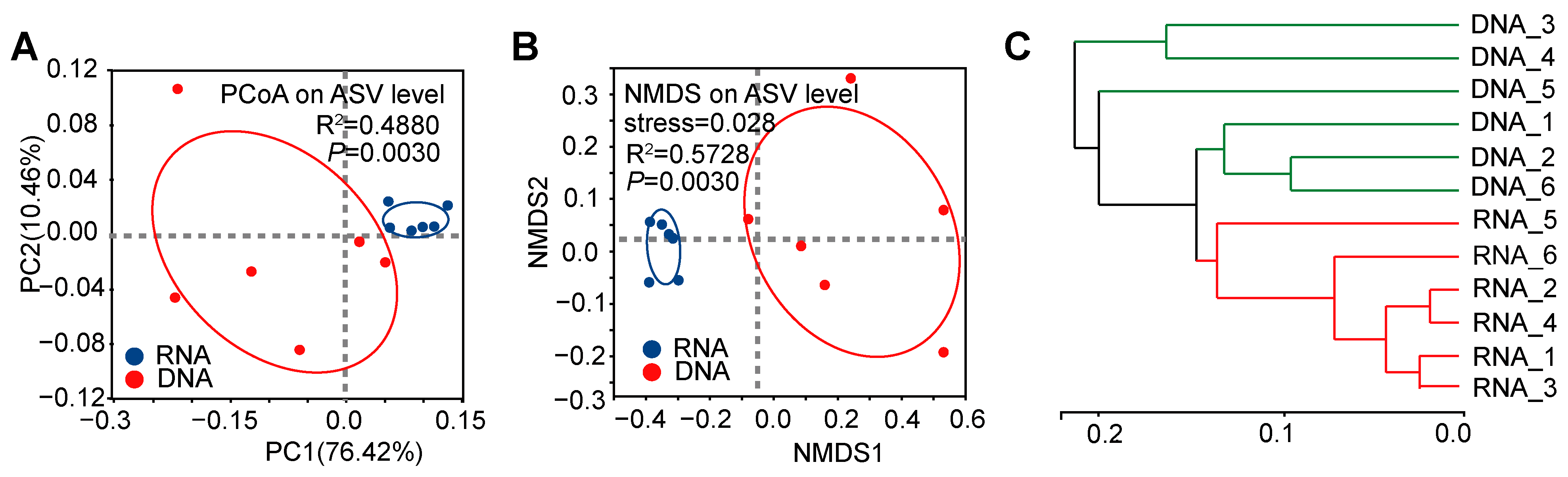
3.1. Significant Differences in α-Diversity and β-Diversity of Bacterial Communities Between DNA- and RNA-Dependent Sequence Samples
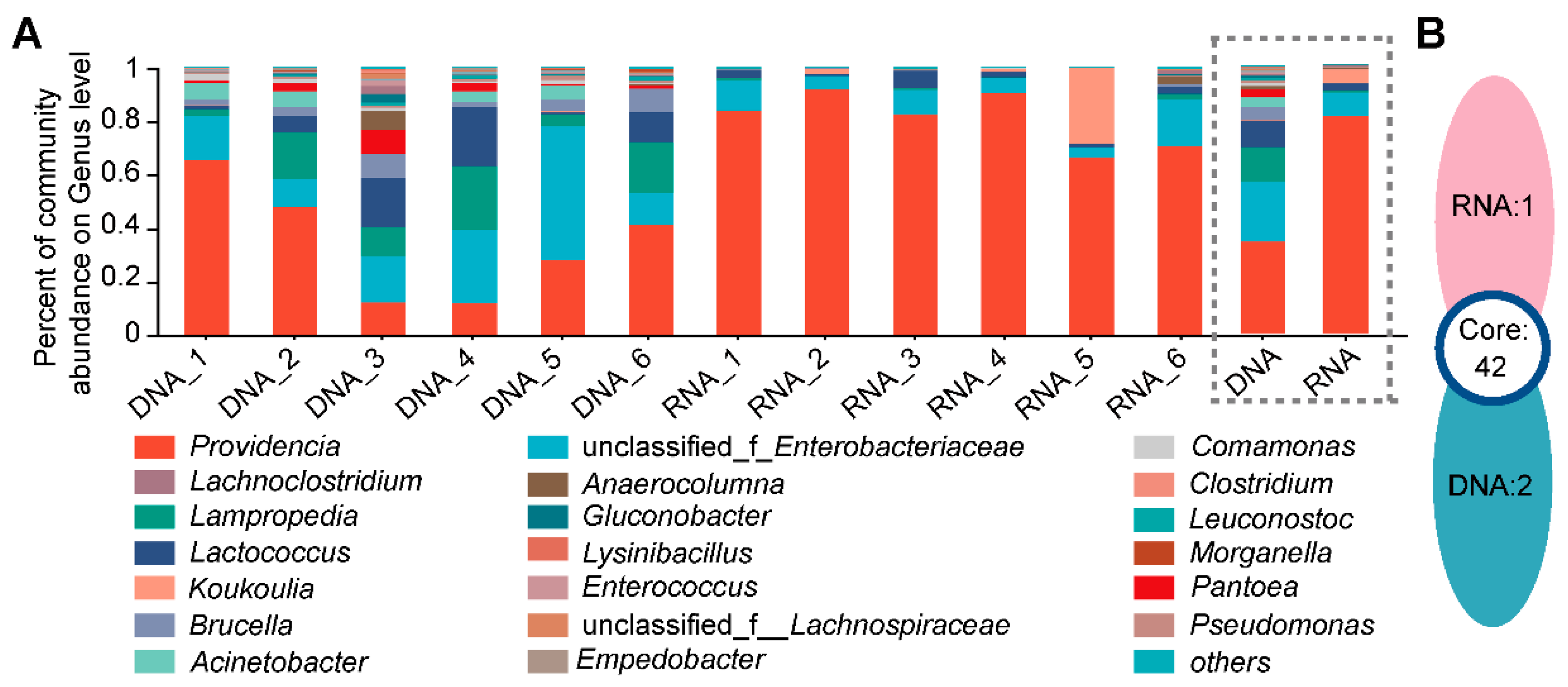
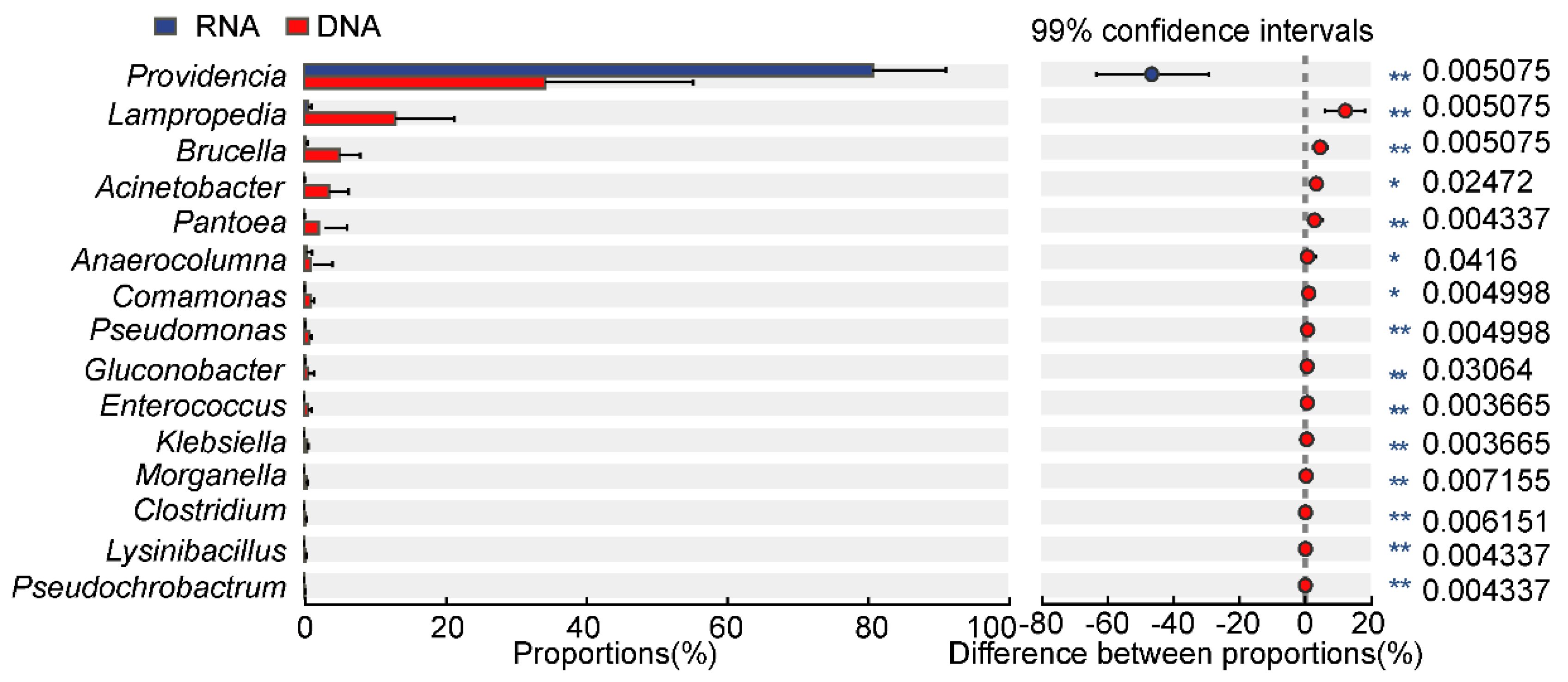
3.2. Different Bacterial Species Composition and Genus Abundance in the DNA- and RNA-Dependent Sequence Samples
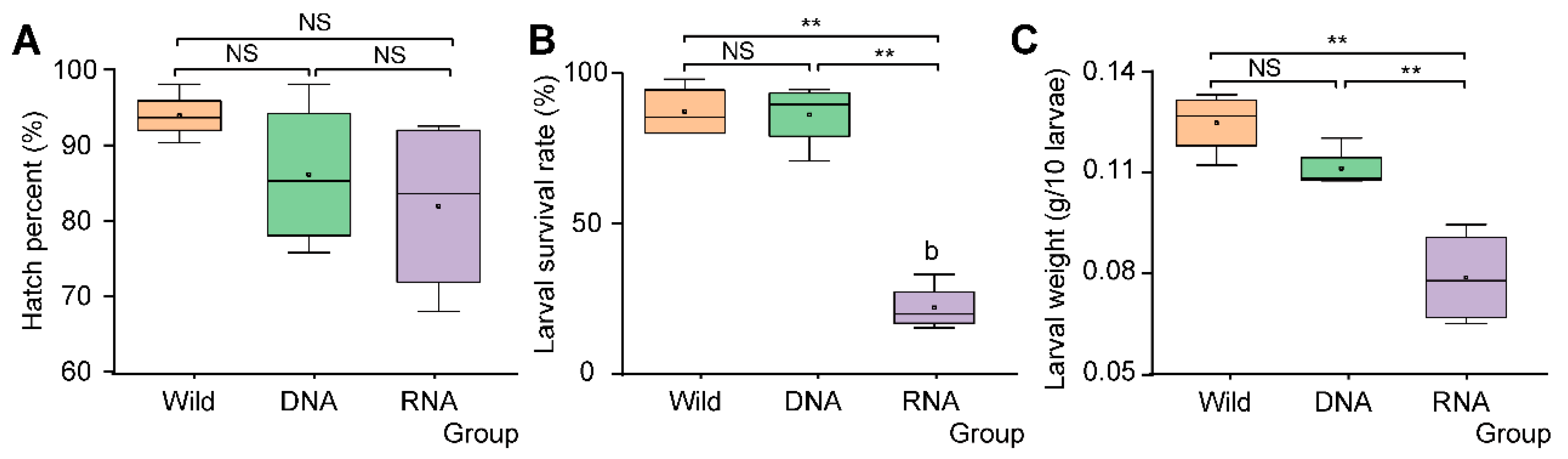
3.3. SynComs Constructed According to RNA-Dependent 16S rRNA Amplicon Sequencing Showed Inhibition Effects on D. antiqua Larvae
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Douglas, A.E. Multiorganismal insects: Diversity and function of resident microorganisms. Annu. Rev. Entomol. 2015, 60, 17–34. [Google Scholar] [CrossRef] [PubMed]
- Nahar, K.; Sultana, S.; Akter, T.; Begum, S. Biological traits and susceptibility of Delia antiqua (Meigen, 1826) (Diptera: Anthomyiidae) in onion. Bangladesh J. Zool. 2019, 47, 325–332. [Google Scholar] [CrossRef]
- Erica, M.; Kyle, W.; Brian, N. Environmental factors and crop management that affect Delia antiqua damage in onion fields. Agric. Ecosyst. Environ. 2021, 314, 107420. [Google Scholar] [CrossRef]
- Alejandro, T.M.; Guillermo, R.A.; Patricio, I.C.; Agustín, H. Incidence of Delia platura (Meigen) (Diptera: Anthomyiidae) in onion and scallion crops in Mexico. Fla. Entomol. 2022, 105, 319–320. [Google Scholar] [CrossRef]
- Akman, L.; Yamashita, A.; Watanabe, H.; Oshima, K.; Shiba, T.; Hattori, M.; Aksoy, S. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat. Genet. 2002, 32, 402–407. [Google Scholar] [CrossRef]
- Zhou, F.; Xu, L.; Wu, X.; Zhao, X.; Liu, M.; Zhang, X. Symbiotic bacterium-derived organic acids protect Delia antiqua larvae from entomopathogenic fungal Infection. mSystems 2020, 5, e00778-20. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yan, Y.; Thompson, K.N.; Bae, S.; Accorsi, E.K.; Zhang, Y.; Shen, J.; Vlamakis, H.; Hartmann, E.M.; Huttenhower, C. Whole microbial community viability is not quantitatively reflected by propidium monoazide sequencing approach. Microbiome 2021, 9, 17. [Google Scholar] [CrossRef]
- Shen, J.; McFarland, A.G.; Young, V.B.; Hayden, M.K.; Hartmann, E.M. Toward accurate and robust environmental surveillance using metagenomics. Front. Genet. 2021, 12, 600111. [Google Scholar] [CrossRef]
- Rackaityte, E.; Halkias, J.; Fukui, E.M.; Mendoza, V.F.; Hayzelden, C.; Crawford, E.D.; Fujimura, K.E.; Burt, T.D.; Lynch, S.V. Viable bacterial colonization is highly limited in the human intestine in utero. Nat. Med. 2020, 26, 599–607. [Google Scholar] [CrossRef]
- Ting, Y.; Hyder, Q.I.; Jinhao, L.; Peijia, Y.; Hongyu, Y.; Xueyin, Z.; Weili, L.; Jiping, L. Analysis of changes in bacterial diversity in healthy and bacterial wilt mulberry samples using metagenomic sequencing and culture-dependent approaches. Front. Plant Sci. 2023, 14, 1206691. [Google Scholar] [CrossRef]
- Bowsher, A.W.; Kearns, P.J.; Shade, A. 16S rRNA/rRNA gene ratios and cell activity staining reveal consistent patterns of microbial activity in plant-associated soil. mSystems 2019, 4, e00003-19. [Google Scholar] [CrossRef] [PubMed]
- Schostag, M.; Stibal, M.; Jacobsen, C.S.; Bælum, J.; Taş, N.; Elberling, B.; Jansson, J.K.; Semenchuk, P.; Priemé, A. Distinct summer and winter bacterial communities in the active layer of Svalbard permafrost revealed by DNA- and RNA-based analyses. Front. Microbiol. 2015, 6, 399. [Google Scholar] [CrossRef] [PubMed]
- Wutkowska, M.; Vader, A.; Mundra, S.; Cooper, E.J.; Eidesen, P.B. Dead or Alive; or does it really matter? level of congruency between trophic modes in total and active fungal communities in high arctic soil. Front. Microbiol. 2018, 9, 3243. [Google Scholar] [CrossRef]
- Blazewicz, S.J.; Barnard, R.L.; Daly, R.A.; Firestone, M.K. Evaluating rRNA as an indicator of microbial activity in environmental communities: Limitations and uses. ISME J. 2013, 7, 2061–2068. [Google Scholar] [CrossRef]
- Ishikawa, Y.; Yamashita, T.; Nomura, M. Characteristics of summer diapause in the onion maggot, Delia antiqua (Diptera: Anthomyiidae). J. Insect Physiol. 2000, 46, 161–167. [Google Scholar] [CrossRef]
- Xin, C.; Liang, Q.; Li, M.; Wu, X.; Fan, S.; Zhang, X.; Zhou, F.; Zhao, Z. Rearing axenic Delia antiqua with half-fermented sterile diets. J. Vis. Exp. 2023, 202, e66259. [Google Scholar] [CrossRef]
- Zhou, F.Y.; Wu, X.Q.; Fan, S.S.; Zhao, X.; Li, M.; Song, F.; Huang, Y.; Zhang, X. Detoxification of phoxim by a gut bacterium of Delia antiqua. Sci. Total Environ. 2024, 943, 173866. [Google Scholar] [CrossRef]
- Fangyuan, Z.; Qingxia, L.; Xiaoyan, Z.; Xiaoqing, W.; Susu, F.; Xinjian, Z. Comparative metaproteomics reveal co-contribution of onion maggot and its gut microbiota to phoxim resistance. Ecotoxicol. Environ. Saf. 2023, 267, 115649. [Google Scholar] [CrossRef]
- Fangyuan, Z.; Yunxiao, G.; Mei, L.; Letian, X.; Xiaoqing, W.; Xiaoyan, Z.; Xinjian, Z. Bacterial Inhibition on beauveria bassiana contributes to microbiota stability in Delia antiqua. Front. Microbiol. 2021, 12, 710800. [Google Scholar] [CrossRef]
- Fangyuan, Z.; Xiaoqing, W.; Letian, X.; Shuhai, G.; Guanhong, C.; Xinjian, Z. Repressed beauveria bassiana Infections in Delia antiqua due to associated microbiota. Pest Manag. Sci. 2018, 75, 170–179. [Google Scholar] [CrossRef]
- Cao, X.; Li, M.; Wu, X.; Fan, S.; Lin, L.; Xu, L.; Zhang, X.; Zhou, F. Gut fungal diversity across different life stages of the onion fly Delia antiqua. Microb. Ecol. 2024, 87, 115. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Zhao, X.; Li, X.; Wu, X.; Zhou, H.; Gao, Y.; Zhang, X.; Zhou, F. Antagonistic effects of Delia antiqua (Diptera: Anthomyiidae)-associated bacteria against four phytopathogens. J. Econ. Entomol. 2021, 114, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Jingying, Z.; Xin, L.Y.; Xiaoxuan, G.; Yuan, Q.; Ruben, G.O.; Paul, S.L.; Yang, B. High-throughput cultivation and identification of bacteria from the plant root microbiota. Nat. Protoc. 2021, 16, 988–1012. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Stöver, B.C.; Müller, K.F. TreeGraph 2: Combining and visualizing evidence from different phylogenetic analyses. BMC Bioinf. 2010, 11, 7. [Google Scholar] [CrossRef]
- Fakruddin, M.; Mannan, S.B. Methods for analyzing diversity of microbial communities in natural environments. Ceylon J. Sci. Biol. Sci. 2013, 42, 19. [Google Scholar] [CrossRef]
- Rabee, A.E.; Forster, R.J.; Elekwachi, C.O.; Kewan, K.Z.; Sabra, E.; Mahrous, H.A.; Khamiss, O.A.; Shawket, S.M. Composition of bacterial and archaeal communities in the rumen of dromedary camel using cDNA-amplicon sequencing. Int. Microbiol. 2020, 23, 137–148. [Google Scholar] [CrossRef]
- Adams, R.I.; Lymperopoulou, D.S.; Misztal, P.K.; De Cassia Pessotti, R.; Behie, S.W.; Tian, Y.; Goldstein, A.H.; Lindow, S.E.; Nazaroff, W.W.; Taylor, J.W.; et al. Microbes and associated soluble and volatile chemicals on periodically wet household surfaces. Microbiome 2017, 5, 128. [Google Scholar] [CrossRef]
- Kajla, M.; Gupta, K.; Gupta, L.; Kumar, S. A Fine-Tuned management between physiology and immunity maintains the gut microbiota in insects. Biochem. Physiol.Open Access 2015, 4, 1000182. [Google Scholar] [CrossRef]
- Lax, S.; Cardona, C.; Zhao, D.; Winton, V.J.; Goodney, G.; Gao, P.; Gottel, N.; Hartmann, E.M.; Henry, C.; Thomas, P.M.; et al. Microbial and metabolic succession on common building materials under high humidity conditions. Nat. Commun. 2019, 10, 1767. [Google Scholar] [CrossRef] [PubMed]
- Fahimipour, A.K.; Hartmann, E.M.; Siemens, A.; Kline, J.; Levin, D.A.; Wilson, H.; Betancourt-Román, C.M.; Brown, G.Z.; Fretz, M.; Northcutt, D.; et al. Daylight exposure modulates bacterial communities associated with household dust. Microbiome 2018, 6, 175. [Google Scholar] [CrossRef] [PubMed]
- Nkongolo, K.K.; Narendrula-Kotha, R. Advances in monitoring soil microbial community dynamic and function. J. Appl. Genet. 2020, 61, 249–263. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Thompson, K.N.; Yan, Y.; Short, M.I.; Zhang, Y.; Franzosa, E.A.; Shen, J.; Hartmann, E.M.; Huttenhower, C. RNA-based amplicon sequencing is ineffective in measuring metabolic activity in environmental microbial communities. Microbiome 2023, 11, 131. [Google Scholar] [CrossRef]
- Tan, S.C.; Yiap, B.C. DNA, RNA, and protein extraction: The past and the present. BioMed Res. Int. 2009, 2009, 574398. [Google Scholar] [CrossRef]
- Ledeker, B.M.; De Long, S.K. The effect of multiple primer-template mismatches on quantitative PCR accuracy and development of a multi-primer set assay for accurate quantification of pcrA gene sequence variants. J. Microbiol. Methods 2013, 94, 224–231. [Google Scholar] [CrossRef]
- Gaby, J.C.; Buckley, D.H. The use of degenerate primers in qPCR analysis of functional genes can cause dramatic quantification bias as revealed by Investigation of nifH primer performance. Microb. Ecol. 2017, 74, 701–708. [Google Scholar] [CrossRef]
- Reid, N.M.; Addison, S.L.; Macdonald, L.J.; Lloyd-Jones, G. Biodiversity of active and inactive bacteria in the gut flora of wood-feeding huhu beetle larvae (Prionoplus reticularis). Appl. Environ. Microbiol. 2011, 77, 7000–7006. [Google Scholar] [CrossRef]
- Mugo Kamiri, L.; Querejeta, M.; Raymond, B.; Herniou, E.A. The effect of diet composition on the diversity of active gut bacteria and on the growth of Spodoptera exigua (Lepidoptera: Noctuidae). J. Insect Sci. 2024, 24, 13. [Google Scholar] [CrossRef]
- Zhang, Y.; Ke, Z.; Xu, L.; Yang, Y.; Chang, L.; Zhang, J. A faster killing effect of plastid-mediated RNA interference on a leaf beetle through induced dysbiosis of the gut bacteria. Plant Commun. 2024, 5, 100974. [Google Scholar] [CrossRef]
- Zeng, T.; Fu, Q.Y.; Luo, F.; Dai, J.; Fu, R.; Qi, Y.; Deng, X.; Lu, Y.; Xu, Y. Lactic acid bacteria modulate the CncC pathway to enhance resistance to β-cypermethrin in the oriental fruit fly. ISME J. 2024, 18, wrae058. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Xu, L.; Wang, S.; Wang, B.; Lou, Q.; Lu, M.; Sun, J. Bacterial volatile ammonia regulates the consumption sequence of d-pinitol and d-glucose in a fungus associated with an invasive bark beetle. ISME J. 2017, 11, 2809–2820. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Cao, X.; Xu, L.; Lin, L.; Wu, X.; Fan, S.; Zhang, X.; Zhou, F. Laboratory Test Indirectly Reveals the Unreliability of RNA-Dependent 16S rRNA Amplicon Sequences in Detecting the Gut Bacterial Diversity of Delia antiqua. Insects 2025, 16, 611. https://doi.org/10.3390/insects16060611
Li M, Cao X, Xu L, Lin L, Wu X, Fan S, Zhang X, Zhou F. Laboratory Test Indirectly Reveals the Unreliability of RNA-Dependent 16S rRNA Amplicon Sequences in Detecting the Gut Bacterial Diversity of Delia antiqua. Insects. 2025; 16(6):611. https://doi.org/10.3390/insects16060611
Chicago/Turabian StyleLi, Miaomiao, Xin Cao, Linfeng Xu, Luyao Lin, Xiaoqing Wu, Susu Fan, Xinjian Zhang, and Fangyuan Zhou. 2025. "Laboratory Test Indirectly Reveals the Unreliability of RNA-Dependent 16S rRNA Amplicon Sequences in Detecting the Gut Bacterial Diversity of Delia antiqua" Insects 16, no. 6: 611. https://doi.org/10.3390/insects16060611
APA StyleLi, M., Cao, X., Xu, L., Lin, L., Wu, X., Fan, S., Zhang, X., & Zhou, F. (2025). Laboratory Test Indirectly Reveals the Unreliability of RNA-Dependent 16S rRNA Amplicon Sequences in Detecting the Gut Bacterial Diversity of Delia antiqua. Insects, 16(6), 611. https://doi.org/10.3390/insects16060611





