Molecular Phylogeny of the Subfamily Notodontinae (Lepidoptera: Noctuoidea: Notodontidae)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Taxon Sampling
2.2. Molecular Experiments
2.3. Phylogenetic Analyses
2.4. Divergence Time Estimation
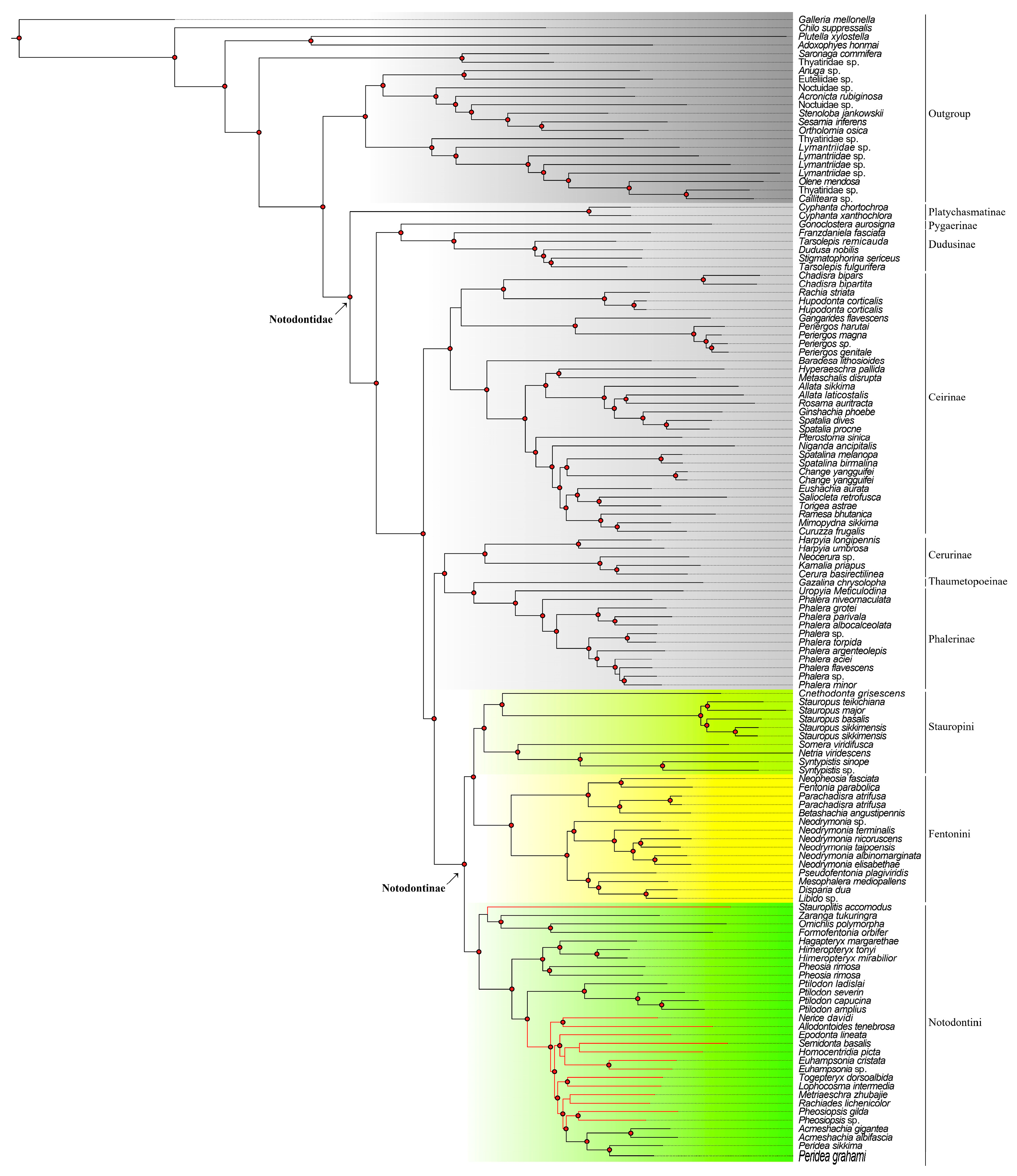
3. Results
3.1. Dataset Construction and Phylogeny
3.1.1. Monophyly of Notodontinae
3.1.2. Stauropini Matsumura, 1925
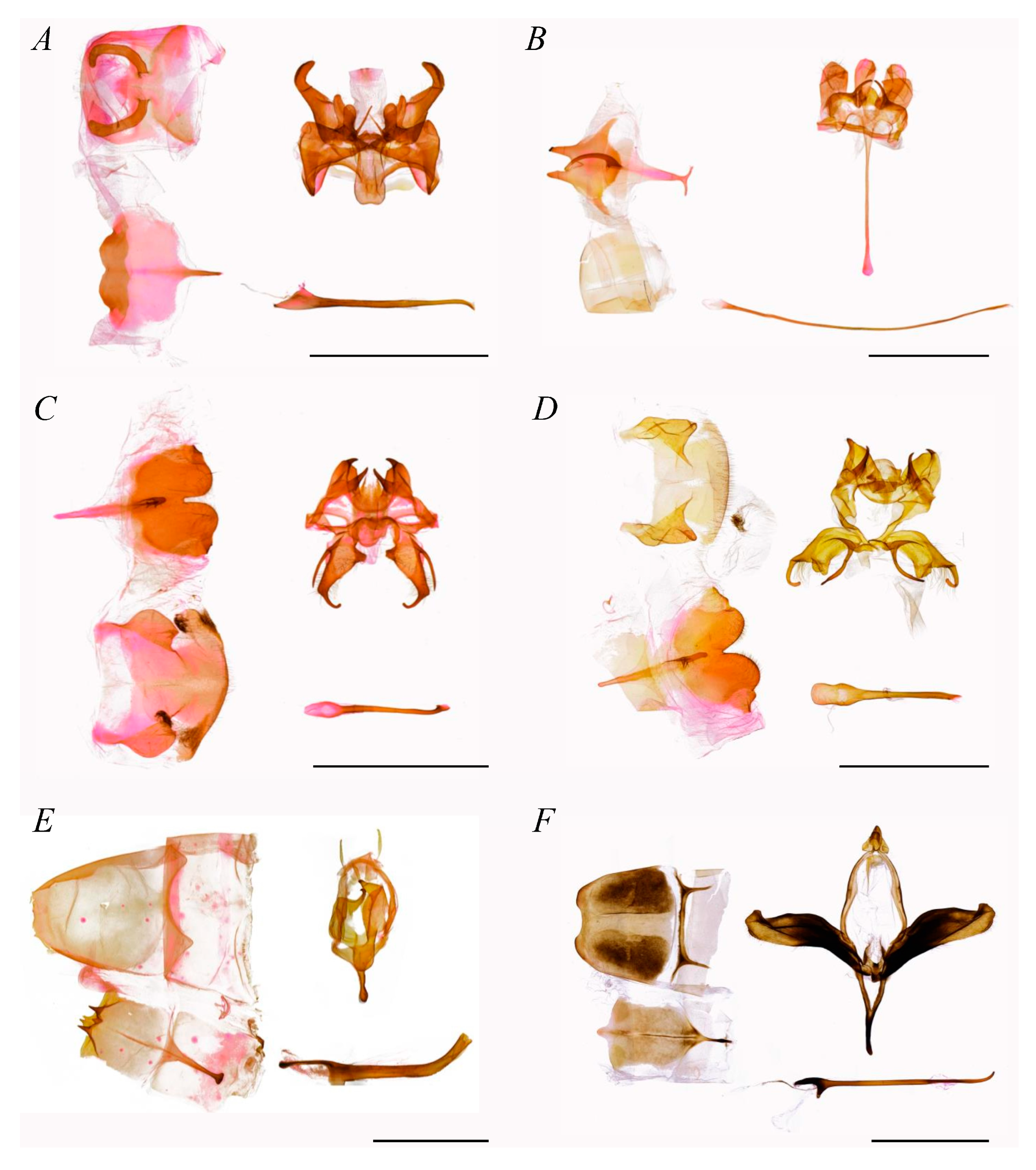
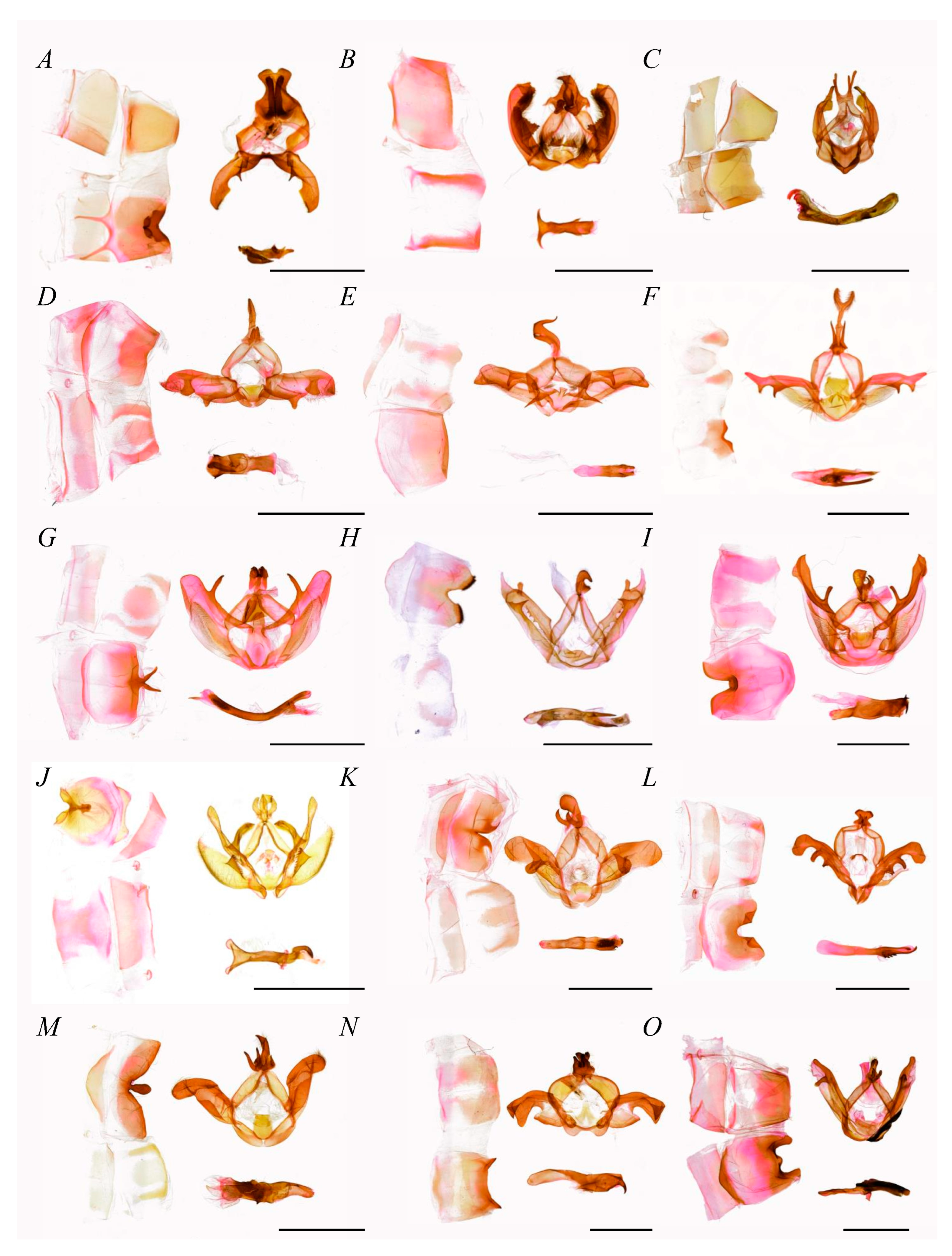
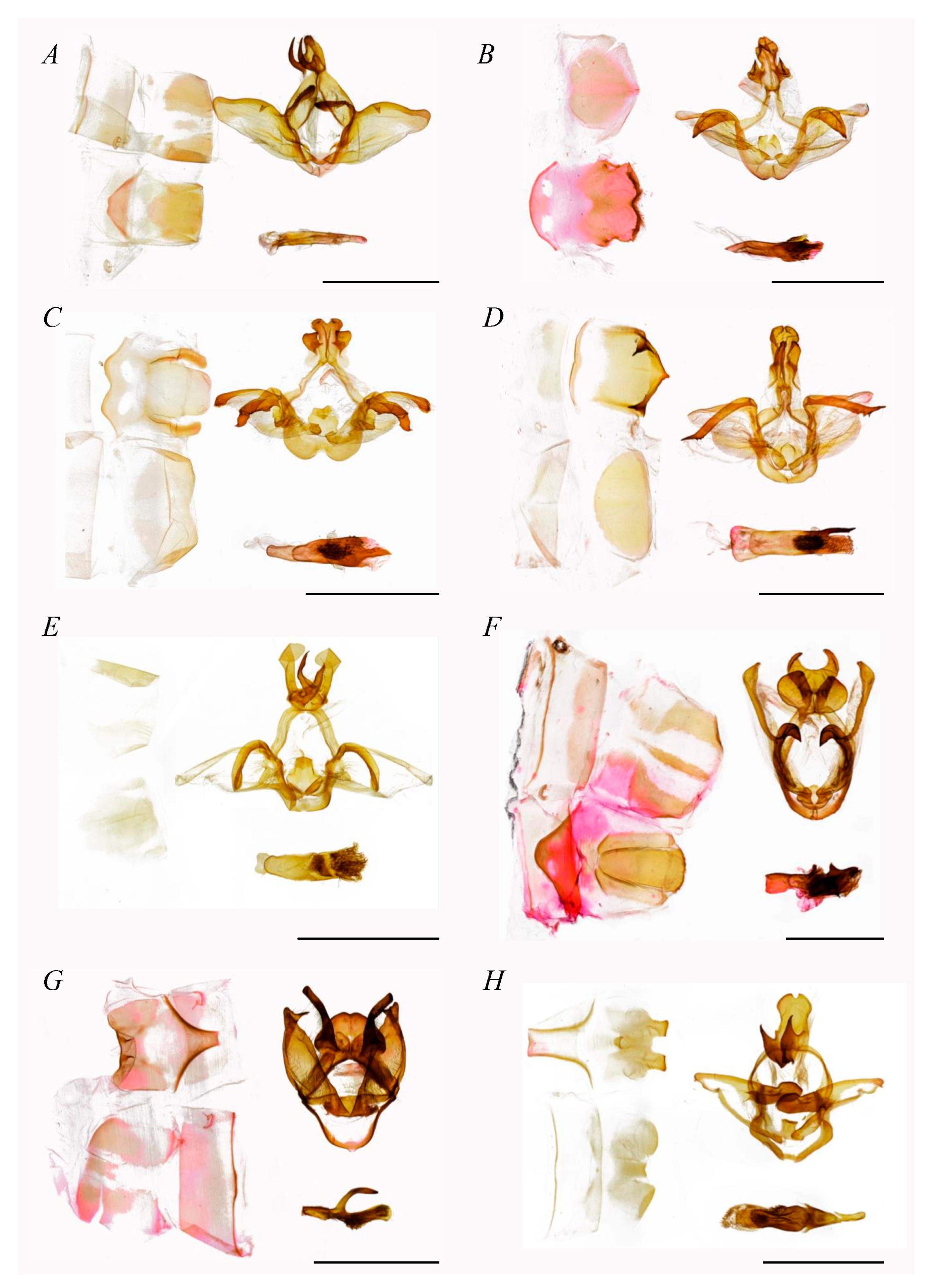
- Diagnosis.
3.1.3. Notodontini Stephens, 1829
- Diagnosis.
3.1.4. Fentoniini Matsumura, 1929
- Diagnosis.
3.2. Divergence Time Estimation
4. Discussion
4.1. Stauropini Matsumura, 1925
4.2. Notodontini Stephens, 1829
4.3. Fentoniini Matsumura, 1929
4.4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| PCGs | protein-coding genes |
| OGs | orthologous genes |
| ML | maximum likelihood |
| CTAB | cetyltrimethylammonium bromide |
| cPAS | combinatorial probe-anchor synthesis |
| MSA | multiple sequence alignment |
Appendix A

References
- Miller, J.S. Cladistics and classification of the Notodontidae (Lepidoptera: Noctuoidea) based on larval and adult morphology. Bull. Am. Mus. Nat. Hist. 1991, 204, 1–230. [Google Scholar]
- Schintlmeister, A. Notodontidae and Oenosandridae (Lepidoptera). In World Catalogue of Insects, Volume II; van Nieukerken, E.J., Ed.; Brill: Leiden, The Netherlands, 2013; p. 605. [Google Scholar]
- Kobayashi, H.; Nonaka, M. Molecular phylogeny of the Notodontidae: Subfamilies inferred from 28S rRNA sequences. Tinea 2016, 23, 213–230. [Google Scholar]
- Basso, A.; Battisti, A.; Roques, A. A total evidence phylogeny for the processionary moths of the genus Thaumetopoea (Lepidoptera: Notodontidae: Thaumetopoeinae). Cladistics 2017, 33, 557–573. [Google Scholar] [CrossRef]
- Schintlmeister, A. Palaearctic Macrolepidoptera Volume 1: Notodontidae; Apollo Books: Stenstrup, Denmark, 2008; pp. 1–400. [Google Scholar]
- Matsumura, S. On the Formosan Notodontidae. Zool. Mag. 1925, 37, 213–230. [Google Scholar]
- Matsumura, S. Generic revision of the Palaearctic Notodontidae. Insecta Matsumurana Ser. Entomol. 1929, 4, 132–136. [Google Scholar]
- Forbes, W.T.M. The Lepidoptera of Barro Colorado Island, Panama. Bull. Mus. Comp. Zool. 1939, 85, 97–322. [Google Scholar]
- Forbes, W.T.M. Lepidoptera of New York and neighboring states. Part 2. Notodontidae. Mem. Cornell Univ. Agric. Exp. Stn. 1948, 274, 1–263. [Google Scholar]
- Neumoegen, B.; Dyar, H.G. A preliminary revision of the lepidopterous family Notodontidae. Trans. Am. Entomol. Soc. 1894, 21, 179–208. [Google Scholar]
- Dyar, H.G. A generic revision of the Ptilodontidae and Melalophidae. Trans. Am. Entomol. Soc. 1897, 24, 1–20. [Google Scholar]
- Packard, A.S. Notodontidae. In Monograph of the Bombycine Moths of America North of Mexico Including Their Transformations and Origin of the Larval Markings and Armature, Volume 7; Dyar, H.G., Ed.; Memoirs of the National Academy of Sciences: Washington, DC, USA, 1895; pp. 1–390. [Google Scholar]
- Draudt, M. Notodontidae. In Macrolepidoptera of the World, Volume 6; Seitz, A., Ed.; Verlag des Seitzschen Werkes: Stuttgart, Germany, 1932; pp. 347–412. [Google Scholar]
- Kiriakoff, S.G. Sur la classification et la phylogenie de la superfamille Notodontoidea (F. d’Almeida) Kiriakoff. Bull. Ann. Soc. R. Entomol. Belg. 1950, 86, 23–255. [Google Scholar]
- Nakamura, M. Pupae of Japanese Notodontidae (Lepidoptera); Japan Lepidopterists’ Society: Tokyo, Japan, 2007; pp. 1–150. [Google Scholar]
- Reineke, A.; Karlovsky, P.; Zebitz, C.P.W. Preparation and purification of DNA from insects for AFLP analysis. Insect Mol. Biol. 1998, 7, 95–99. [Google Scholar] [CrossRef]
- Meng, G.; Li, Y.; Yang, H. MitoZ: A toolkit for animal mitochondrial genome assembly and analysis. Methods Ecol. Evol. 2019, 10, 274–280. [Google Scholar]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo Metazoan Mitochondrial Genome Annotation. Mol. Phylogenetics Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Huang, Y.-H.; Sun, Y.-F.; Li, H.; Li, H.-S.; Pang, H. PhyloAln: A Convenient Reference-Based Tool to Align Sequences and High-Throughput Reads for Phylogeny and Evolution in the Omic Era. Mol. Biol. Evol. 2024, 41, msae150. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Suchard, M.A.; Lemey, P.; Baele, G.; Ayres, D.L.; Drummond, A.J.; Rambaut, A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018, 4, vey016. [Google Scholar] [CrossRef]
- Drummond, A.J.; Nicholls, G.K.; Rodrigo, A.G.; Solomon, W. Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics 2002, 161, 1307–1320. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A. Estimating the rate of molecular evolution: Incorporating non-contemporaneous sequences into maximum likelihood phylogenies. Bioinformatics 2000, 16, 395–399. [Google Scholar] [CrossRef] [PubMed]
- Pybus, O.G.; Rambaut, A. GENIE: Estimating demographic history from molecular phylogenies. Bioinformatics 2002, 18, 1404–1405. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, M.V. Paleontologiya cheshuekrylykh i voprosy filogenii otrayada Papilionida. In Melovoy Biotsenoticheskiy Krizis i Evolyutsiya Nasekomykh; Ponomarenko, A.G., Ed.; Akademiya Nauk SSSR: Moscow, Russia, 1988; pp. 50–75. [Google Scholar]
- Simonsen, T.J. Reassessment of known fossil Pyraloidea (Lepidoptera) with descriptions of the oldest fossil pyraloid and a crambid larva in Baltic amber. Zootaxa 2018, 4483, 101–127. [Google Scholar]
- Kusnezov, N. Oligamatites martynovi, gen. et sp. nn., a fossil amatid lepidopteron from the Oligocene beds of central Asia. Dokl. Akad. Nauk SSSR 1928, 20, 431–436. [Google Scholar]
- Skalski, A.W. Studies on the Lepidoptera from fossil resins part III. Two new genera and species of Tortricidae from the Baltic amber. Ann. Up. Silesian Mus. Entomol. 1992, 3, 137–146. [Google Scholar]
- Kernbach, K. Über die bisher im Pliozän von Willershausen gefundenen Schmetterlings und Raupenreste. Ber. Der Naturhistorischen Ges. Zu Hann. 1967, 111, 103–106. [Google Scholar]
- Schintlmeister, A. The Notodontidae of South Africa Including Swaziland and Lesotho (Lepidoptera: Notodontidae); Museum Witt: Munich, Germany; Vilnius Academy of Arts Press: Vilnius, Lithuania, 2015; pp. 1–289. [Google Scholar]
- Schintlmeister, A. Notodontidae of the Indonesian Archipelago; Brill: Leiden, The Netherlands, 2020; Volume 1, pp. 1–400. [Google Scholar]
- Moore, F. Family Notodontidae. In The Lepidoptera of Ceylon, Volume 2; Moore, F., Ed.; L. Reeve & Co.: London, UK, 1882; pp. 1–120. [Google Scholar]
- Kiriakoff, S.G. Lepidoptera Familia Notodontidae Pars tertia Genera Indo-Australica. In Genera Insectorum, fasc.217 C; Wytsman, P., Ed.; Imprimerie H. Vaillant-Carmanne: Liège, Belgium, 1968; p. 269. [Google Scholar]

| Taxon | Family | PBDB (Ma) | Fossil Type | Location |
|---|---|---|---|---|
| Plutellites tenebricus | Plutellidae | 38.0–33.9 | Baltic Amber | Russia |
| Eopyralis morsae | Pyralidae | 56.0–47.8 | Rock | Denmark |
| Oligamatites martynovi | Erebidae | 28.1–23.0 | Rock | Kazahkstan |
| Tortricibaltia diakonoffi | Tortricidae | 38.0–33.9 | Baltic Amber | Russia |
| Cerurites wagneri | Notodontidae | 3.6–2.588 | Rock | Germany |
| Species Name | Accession No. | Species Name | Accession No. |
|---|---|---|---|
| Adoxophyes honmai | NC_008141 | Cnethodonta grisescens | NC_068541 |
| GCA_005406045.1 | Dudusa sphingiformis | MW788876 | |
| Chilo suppressalis | NC_015612 | Furcula furcula | OU452271 |
| GCA_902850365.2 | Neocerura liturata | NC_062182 | |
| Galleria mellonella | KT750964 | Ochrogaster lunifer | NC_011128 |
| GCA_026898425.1 | Peridea elzet | NC_062111 | |
| Plutella xylostella | NC_025322 | Phalera assimilis | OP784764 |
| GCA_932276165.1 | Pheosia gnoma | FR989925 | |
| Sesamia inferens | NC_015835 | Pheosia rimosa | NC_061645 |
| GCA_037179545.1 | Pheosia tremula | HG995428 | |
| Clostera anachoreta | NC_034740 | Syntypistis chambae | NC_062123 |
| Clostera anastomosis | NC_041140 | Thaumetopoea pityocampa | NC_053256 |
| Stauropini | Notodontini | Fentoniini |
|---|---|---|
| Stauropus, Somera, Cnethodonta, Netria, Syntypistis | Stauroplitis, Zaranga, Omichlis, Formofentonia, Hagapteryx, Himeropteryx, Pheosia, Ptilodon, Homocentridia, Allodontoides, Lophocosma, Semidontoides, Epodonta, Togepteryx, Nerice, Euhampsonia, Rachiades, Notodonta, Metriaeschra, Pheosiopsis, Achmeshachia, Peridea | Neopheosia, Fentonia, Parachadisra, Betashachia, Pseudofentonia, Mesophalera, Disparia, Libido, Neodrymonia |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, M.; Geng, Q.; Zhang, D. Molecular Phylogeny of the Subfamily Notodontinae (Lepidoptera: Noctuoidea: Notodontidae). Insects 2025, 16, 526. https://doi.org/10.3390/insects16050526
Guo M, Geng Q, Zhang D. Molecular Phylogeny of the Subfamily Notodontinae (Lepidoptera: Noctuoidea: Notodontidae). Insects. 2025; 16(5):526. https://doi.org/10.3390/insects16050526
Chicago/Turabian StyleGuo, Muyu, Qingliu Geng, and Dandan Zhang. 2025. "Molecular Phylogeny of the Subfamily Notodontinae (Lepidoptera: Noctuoidea: Notodontidae)" Insects 16, no. 5: 526. https://doi.org/10.3390/insects16050526
APA StyleGuo, M., Geng, Q., & Zhang, D. (2025). Molecular Phylogeny of the Subfamily Notodontinae (Lepidoptera: Noctuoidea: Notodontidae). Insects, 16(5), 526. https://doi.org/10.3390/insects16050526







