1. Introduction
Herbivorous insects, with nearly 500,000 identified species, serve as a major driver of biodiversity and a fundamental component of terrestrial ecosystems [
1,
2]. Over 400 million years of co-evolution, plants and herbivorous insects have been engaged in a continuous arms race, driving the evolution of intricate plant defense mechanisms and corresponding insect adaptations that promote survival [
3,
4,
5]. To counteract herbivory, plants have evolved sophisticated chemical and structural defenses, including toxic secondary metabolites, proteinase inhibitors, and immune signaling pathways [
6,
7,
8]. In turn, herbivorous insects have evolved strategies to circumvent or suppress these defenses, enabling efficient host plant utilization [
9,
10,
11,
12]. During feeding, the insect herbivores deposit oral secretions (OS) onto wounded plant tissues, where they interact with plant signaling pathways and modulate defense responses [
13,
14]. The regurgitant originating from the gut contents is the primary component of OS and carries digestive enzymes, microbial communities, and metabolic compounds that further modulate plant responses [
15]. These secretions facilitate digestion, detoxification, and immune modulation, ultimately promoting insect adaptation to host plants [
16,
17].
Previous findings highlight the essential roles of effectors in overcoming plant defenses and facilitating insect adaptation [
18,
19,
20]. For example, Glucose oxidase (GOX), a well-characterized effector secreted by the labial glands of caterpillars such as
Helicoverpa zea, suppresses nicotine biosynthesis and other defense-related compounds in tobacco while attenuating the jasmonic acid (JA) signaling pathway through the upregulation of salicylic acid (SA) signaling [
18]. Notably,
H. zea larvae feeding on tomato (
Solanum lycopersicum) induce the expression of the proteinase inhibitor gene
Pin2 and elevate JA levels, whereas feeding on tobacco (
Nicotiana tabacum) increases SA levels. This differential response indicates that GOX, as a salivary-gland-derived effector, may exert distinct functions depending on the host plant species [
19]. Interestingly, GOX has also been identified in the regurgitant of
P. xylostella, where it catalyzes the oxidation of plant-derived glucose, leading to the production of H
2O
2. The accumulation of H
2O
2 activates the SA pathway while suppressing the JA pathway, thereby attenuating JA-mediated defense responses against chewing insects [
20]. However, the transcription and enzymatic activity of GOX can be suppressed by specific bacterial strains isolated from the regurgitant of
P. xylostella, thereby impairing its suppression of plant defense responses and negatively influencing the insect’s adaptation to host-induced immunity [
21]. Another representative effector,
Helicoverpa armigera R-like protein 1 (HARP1), identified in the OS of
H. armigera, is structurally homologous to an R-like protein found in the venom glands of
Nasonia vitripennis (parasitoid wasp). HARP1 infiltrates host cells through feeding-induced plant wounds, where it interacts with JASMONATE-ZIM-domain (JAZ) proteins to suppress JA signaling and attenuate plant defenses. Notably,
harp1 transcripts are highly expressed in the foregut and midgut rather than in the labial glands, and the protein is detectable in both tissues, underscoring its function in host plant adaptation through regurgitation [
22]. These findings have also been shown in other insect species [
23], further highlighting the role of regurgitant-associated effectors in enhancing the adaptation of chewing insects to host plants.
Although well-established effectors such as GOX and HARP1 have been extensively studied for their role in modulating plant defenses and facilitating insect adaptation to host plants, heat shock proteins (HSPs) have received comparatively less attention as potential effectors in insect–plant interactions. As molecular chaperones, HSPs facilitate protein folding, stabilization, and transmembrane transport, thus contributing to cellular adaptation to both biotic and abiotic stressors [
24,
25]. HSP70 is particularly noteworthy due to its high evolutionary conservation, widespread presence across various organisms, and diverse functional roles [
26,
27,
28]. Traditionally known for their role in maintaining cellular homeostasis and mediating stress responses [
29,
30,
31], HSP70s are increasingly acknowledged for their potential role in modulating plant immunity during insect feeding. Notably, studies have shown that insect HSP70s, including
Nilaparvata lugens NlHSC70-3, have been shown to suppress plant immune responses by downregulating defense-related genes and reducing reactive oxygen species (ROS) accumulation [
32]. In contrast,
NlDNAJB9, an HSP40 family member in
N. lugens, triggers ROS bursts and activates immune responses through its DNAJ domain, thereby enhancing plant resistance to both insect herbivory and pathogens [
33]. These findings highlight the dual potential of HSPs in plant–insect interactions, where they can both suppress plant immune responses and, under certain conditions, activate plant defense mechanisms. Given the potential roles of HSP proteins in insect–plant interactions, their functions in most herbivorous insects remain largely unexplored, particularly in the cosmopolitan pest
Plutella xylostella, which is of significant economic importance due to its widespread infestation of cruciferous crops worldwide and its ability to rapidly adapt to various host plants.
In this study, we identified HSC70-3 in the gut regurgitant of P. xylostella. Additionally, it is secreted into plant wound sites during larval feeding. Short-term host transfer experiments revealed dynamic tissue-specific regulation of hsc70-3, indicating its differential regulation at transcriptional and post-translational levels in response to host plant shifts and suggesting its role in plant-induced stress responses and host adaptation. Furthermore, the knockout of hsc70-3 resulted in impaired larval growth, prolonged development, and reduced pupal weight on host plants, highlighting its role in host adaptation. These findings provide initial evidence that HSC70-3 functions as an effector in P. xylostella interactions with host plants, playing a crucial role in suppressing plant defenses.
2. Materials and Methods
2.1. Insect Strains and Rearing Conditions
The
P. xylostella strain used in this study was obtained from the Institute of Zoology, Chinese Academy of Sciences (Beijing, China), and has been maintained under insecticide-free and selection-free conditions since 2016 [
34]. The larvae were reared on an artificial diet (AD) under controlled conditions (25 ± 2 °C, 65 ± 5% relative humidity, and a 16:8 h light-dark photoperiod). Larvae were reared in 150 mm × 150 mm × 80 mm cardboard containers, and the diet was replenished every three days to ensure optimal development. The artificial diet was prepared by dissolving agar (12 g) and yeast powder (40 g) in 500 mL of distilled water with continuous stirring. After cooling to approximately 50 °C, the remaining ingredients were added, including wheat germ powder (75 g), vitamin powder (2 g), sorbic acid (2 g), ethyl
p-hydroxybenzoate (2 g), ascorbic acid (2 g), radish seed powder (6 g), sucrose (20 g), linoleic acid (0.2 mL), and rapeseed oil (2 mL), followed by thorough mixing [
34]. The homogenized mixture was poured into sterilized trays and allowed to solidify before use. Once pupation occurred, pupae were collected and transferred to 100 mm × 100 mm × 100 mm containers for adult emergence and mating. To promote oviposition, adult moths were provided with a 10% honey solution as a nutritional supplement.
2.2. Plant Materials and Growth Conditions
The radish variety ‘Nanpan Prefecture’ used in this experiment was provided by Kaiyisun Biotechnology Co., Ltd. (Shanghai, China) and was stored under dry conditions before sowing. Plants were grown in 450 mm × 300 mm × 70 mm plastic trays filled with a 1:1 (v/v) mixture of peat soil and vermiculite without additional fertilization to maintain controlled growth conditions. Trays were watered with sterile distilled water every two days to maintain optimal soil moisture. All plants were cultivated in a climate-controlled growth chamber (ZYL202310001, Fuzhou, Fujian Zhongyouling Technology Co., Ltd., China), under a 16:8 h light/dark cycle, 2000 lux light intensity, 23 ± 1 °C, and 65 ± 5% relative humidity. Radish seedlings at the fully expanded cotyledon stage, defined by the presence of two fully expanded cotyledons without visible true leaves (~7 days post-germination), were selected for insect feeding experiments.
2.3. Short-Term Host Transfer Experiment
The short-term host transfer experiment involved transferring first-instar P. xylostella larvae from an artificial diet to radish seedlings. The rearing conditions for the larvae on radish seedlings were identical to those on the artificial diet. Larvae were reared on the host plant for two weeks, during which fourth-instar individuals were collected for tissue dissection. The same procedure was followed for hsc70-3 mutant larvae, with the larvae transferred to the host plant for the same duration under identical rearing conditions.
2.4. Sample Collection, RNA Extraction, cDNA Synthesis, and Gene Cloning
Samples from various developmental stages of P. xylostella were collected, including eggs (E) (n = 600), first-instar larvae (L1) (n = 7 for each replicate), second-instar larvae (L2) (n = 7), third-instar larvae (L3) (n = 7), fourth-instar larvae (L4) (n = 7), pupae (P) (n = 7), and adults (A) (n = 7) (sex not differentiated). Fourth-instar larvae were rinsed in phosphate-buffered saline (PBS) and dissected under sterile conditions to minimize contamination. Each larva was placed in a Petri dish containing 500 µL sterile PBS and dissected under a stereomicroscope (SZ660, Chongqing Aote Optical Instrument Co., Ltd., Chongqing, China) using sterilized fine forceps (Dumont, Montignez, Switzerland). The dissected tissues, including the fat body (FB), gut contents (GCT), head (HD), Malpighian tubules (MT), gut (GUT), silk gland (SK), and remaining tissues (RT), were carefully isolated and washed twice with PBS to remove residual debris. For each experimental group, twenty remaining tissue samples were collected, while 50 larvae were used for each of the other tissue types. The collected tissues were transferred into separate RNase-free microcentrifuge tubes and immediately flash frozen in liquid nitrogen. Samples were stored in an ultra-low temperature freezer (MDF-U54V, Panasonic, Osaka, Japan) until further use, with three biological replicates per tissue type, each consisting of pooled tissues from multiple individuals.
Total RNA was isolated from P. xylostella using the Total RNA Extraction Kit (Promega, Madison, WI, USA) following the manufacturer’s protocol. RNA purity and concentration were measured using a NanoDrop™ 2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA) with A260/A280 ratios between 1.8 and 2.2 deemed acceptable. RNA integrity was assessed via 1% agarose gel electrophoresis, with only high-quality RNA samples selected for subsequent experiments.
First-strand cDNA was synthesized using the FastKing RT Kit (including gDNase treatment) (TIANGEN Biotech, Beijing, China). Each 20 μL reaction mixture consisted of 4.0 μL HiScript III enzyme mix, 14 μL RNase-free ddH2O, and 2 μL total RNA (500 ng–1 μg). To ensure efficient cDNA synthesis, the reaction mix included random hexamer primers and oligo(dT)18 primers. The reaction was carried out at 42 °C for 15 min, followed by heat inactivation at 95 °C for 3 min using a thermal cycler (S1000 Thermal Cycler, Bio-Rad Laboratories, Hercules, CA, USA).
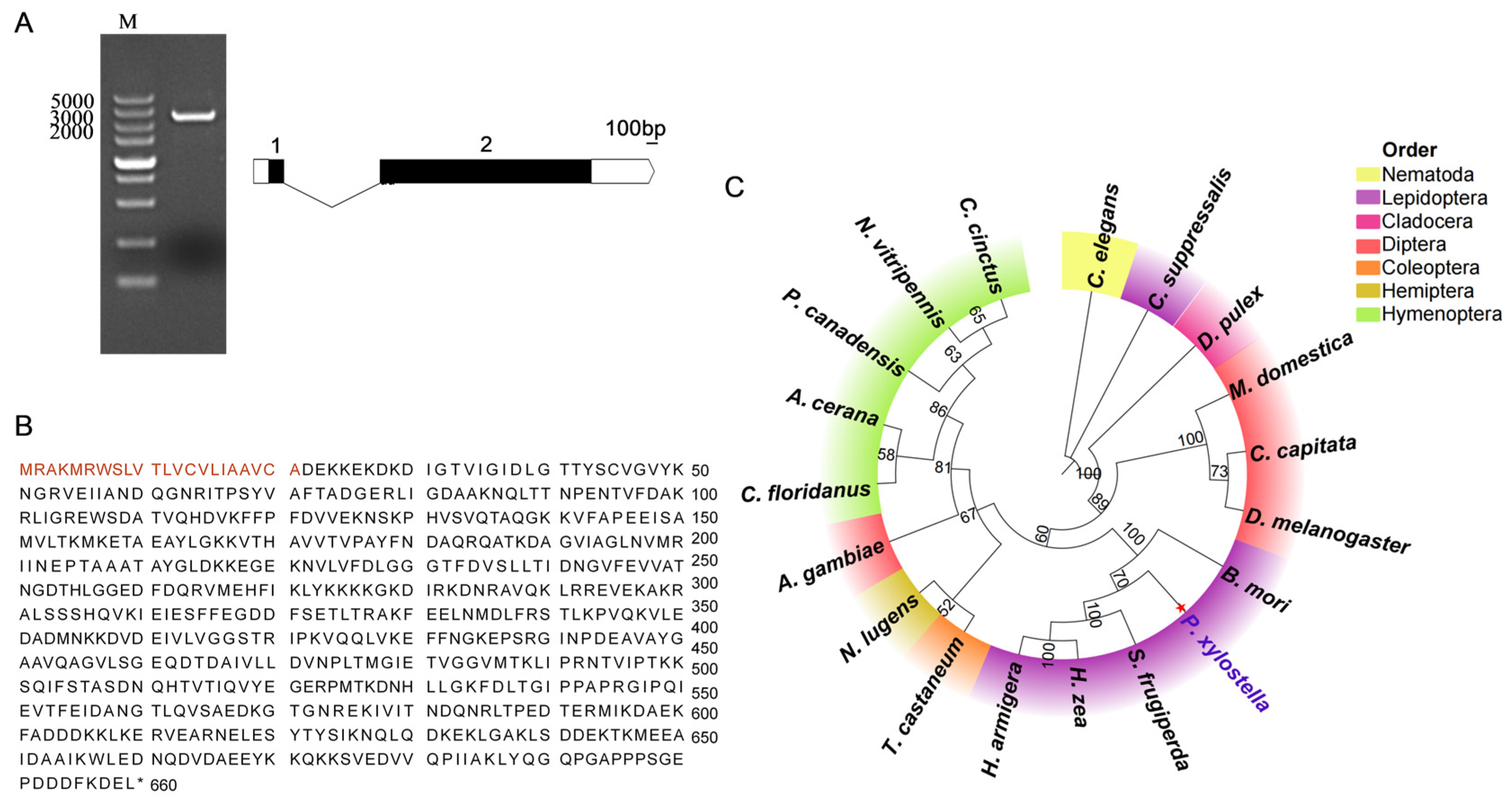
The
hsc70-3 gene sequence of
P. xylostella was identified from gut regurgitant transcriptomic data and retrieved from the DBM Genome Database (
http://iae.fafu.edu.cn/DBM/, accessed on 1 March 2025) and validated via NCBI BLAST online tool (
https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 1 March 2025) [
35]. Gene-specific primers for
hsc70-3 amplification were designed using Primer Premier software (version 6.0; PREMIER Biosoft, Palo Alto, CA, USA) (
Table A1). PCR was performed with 2× Rapid Taq Master Mix (Vazyme Biotech, Nanjing, China) in a 20 μL reaction mixture, consisting of 10 μL of 2× Master Mix, 0.5 μL of forward primer (10 μM), 0.5 μL of reverse primer (10 μM), 1 μL of cDNA template, and 8 μL of RNase-free ddH
2O. The PCR cycling conditions were as follows: initial denaturation at 95 °C for 3 min, followed by 35 cycles of 95 °C for 15 s, 60 °C for 15 s, and 72 °C for 30 s, with a final extension at 72 °C for 10 min.
The hsc70-3 PCR product was purified using the Gel Extraction Kit (Omega Bio-Tek, Norcross, GA, USA) and subsequently ligated into the pJET1.2/blunt vector using the CloneJET PCR Cloning Kit (Thermo Fisher Scientific, Waltham, MA, USA). The ligation reaction was conducted at 22 °C for 2 h in a total volume of 10 μL, consisting of 5 μL 2× Reaction Buffer, 2 μL purified PCR product, 1 μL T4 DNA ligase, 1 μL pJET1.2/blunt Cloning Vector, and 2 μL RNase-free ddH2O. The ligation mixture was transformed into chemically competent Escherichia coli DH5α cells via the heat-shock method (42 °C for 45 s, followed by immediate cooling on ice for 2 min). Transformed cells were spread onto LB agar supplemented with 100 μg/mL ampicillin and incubated at 37 °C overnight. Following incubation, colony PCR was performed using vector-specific primers to screen for positive clones. Six independent positive clones were selected and sent to Sangon Biotech (Shanghai) Co., Ltd. for Sanger sequencing to verify the presence and accuracy of the inserted sequences.
2.5. Sequence Analysis and Phylogenetic Tree Construction
Sanger sequencing was used for sequence validation, and the results were visualized with SnapGene software (version 6.0.2; Dotmatics, San Diego, CA, USA). The exon–intron structure of HSC70-3 was analyzed using the WormBase Exon–Intron Finder (
http://www.wormweb.org/exonintron, accessed on 1 March 2025). The physico-chemical properties of HSC70-3 were predicted using ExPASy ProtParam (
https://web.expasy.org/protparam/, accessed on 1 March 2025) based on the CDS. Amino acid composition, instability index, and aliphatic index were calculated to assess protein stability in vitro. The SMART tool (
https://smart.embl.de/, accessed on 1 March 2025) identified conserved HSP70 domains. Phylogenetic analysis was performed using MEGA10 software with the maximum likelihood method and 1000 bootstrap replications to ensure statistical robustness. The constructed phylogenetic tree was visualized using the Chiplot online platform (
https://www.chiplot.online/, accessed on 1 March 2025).
2.6. qRT-PCR
Primers specific to
hsc70-3 were designed based on its cDNA sequence using Primer Premier 6 software (PREMIER Biosoft, Palo Alto, CA, USA) and validated for specificity via BLAST analysis against the NCBI database (
Table A1). The
P. xylostella rpl32 gene (Genome Database ID:
Px008022) was used as the internal reference gene for normalization. Quantitative real-time PCR (qRT-PCR) was performed using the QuantStudio™ 6 Flex Real-Time PCR System (Applied Biosystems, Thermo Fisher Scientific, Waltham, MA, USA) in a 20 μL reaction mixture containing 10 μL GoTaq
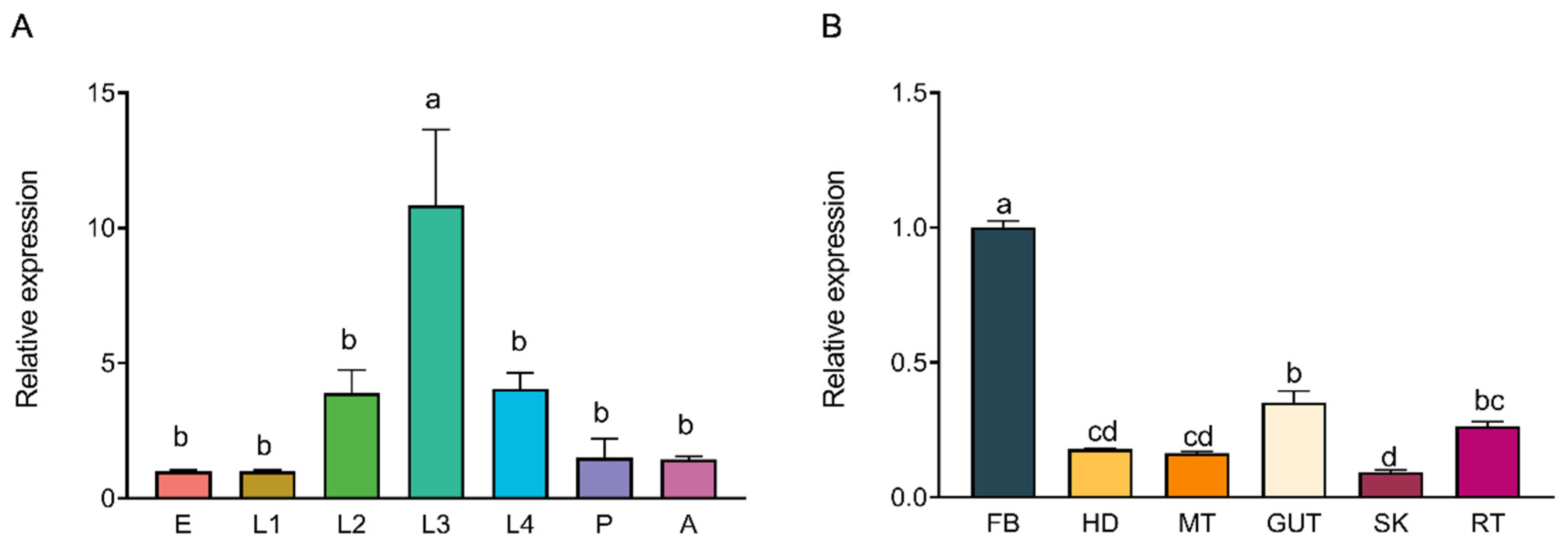
® qPCR Master Mix (Promega Corporation, Madison, WI, USA), 0.2 μL of each primer (10 μM), 2 μL cDNA template, and 7.6 μL nuclease-free water. The thermal cycling conditions consisted of an initial denaturation at 95 °C for 3 min, followed by 40 cycles of 95 °C for 15 s, 60 °C for 15 s, and 72 °C for 30 s. Melt curve analysis (65–95 °C, with 0.5 °C increments every 5 s) was conducted to confirm amplification specificity. Each sample was analyzed in three biological replicates, with each biological replicate consisting of three technical replicates. The results of the technical replicates were averaged for each biological replicate, and these averaged values were used for statistical analysis. The relative expression levels of
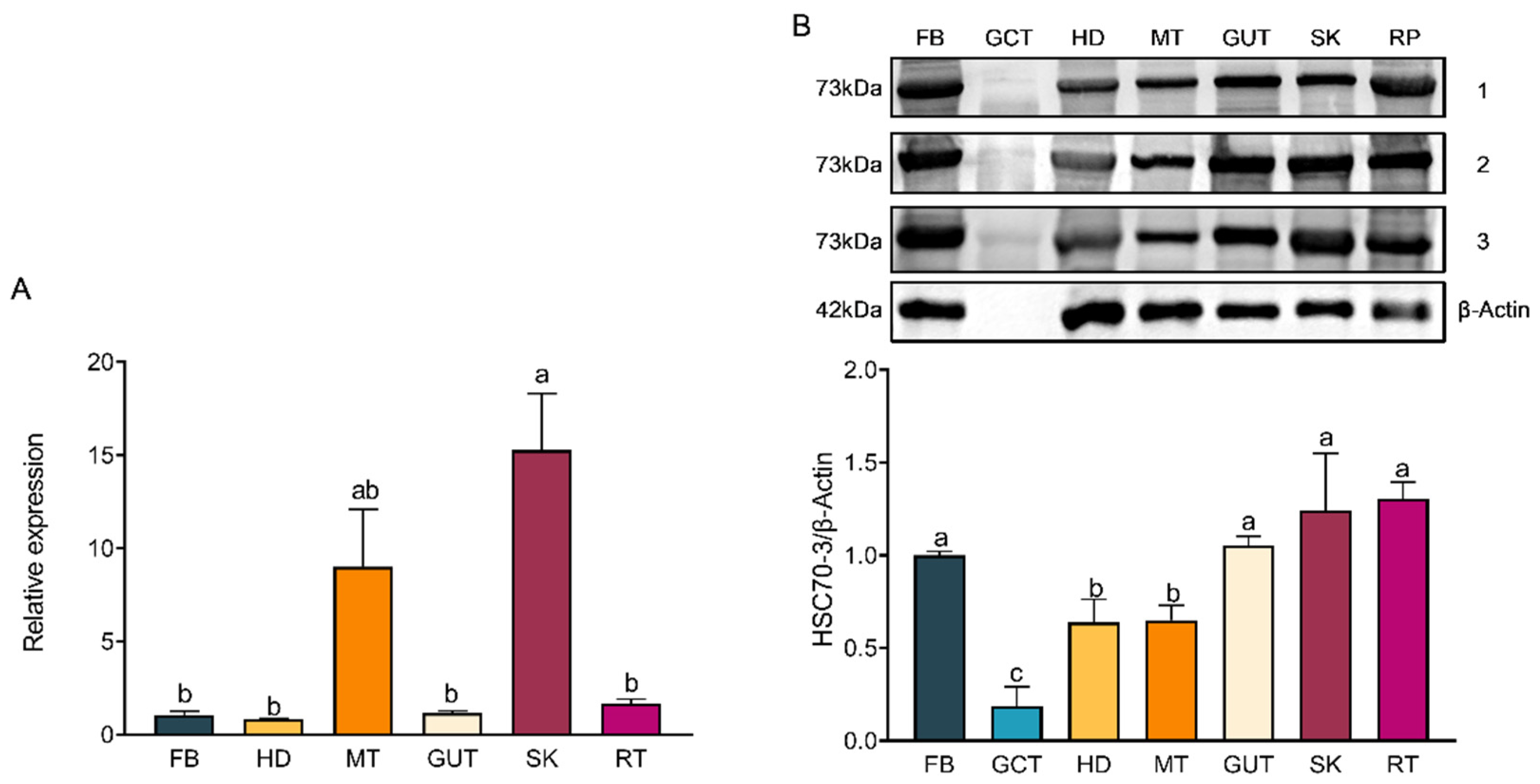
hsc70-3 across developmental stages (E, L1, L2, L3, L4, P, and A) and tissues (FB, GCT, HD, MT, GUT, SK, RT) were quantified using the 2
−ΔΔCt method.
2.7. Protein Extraction and Western Blot
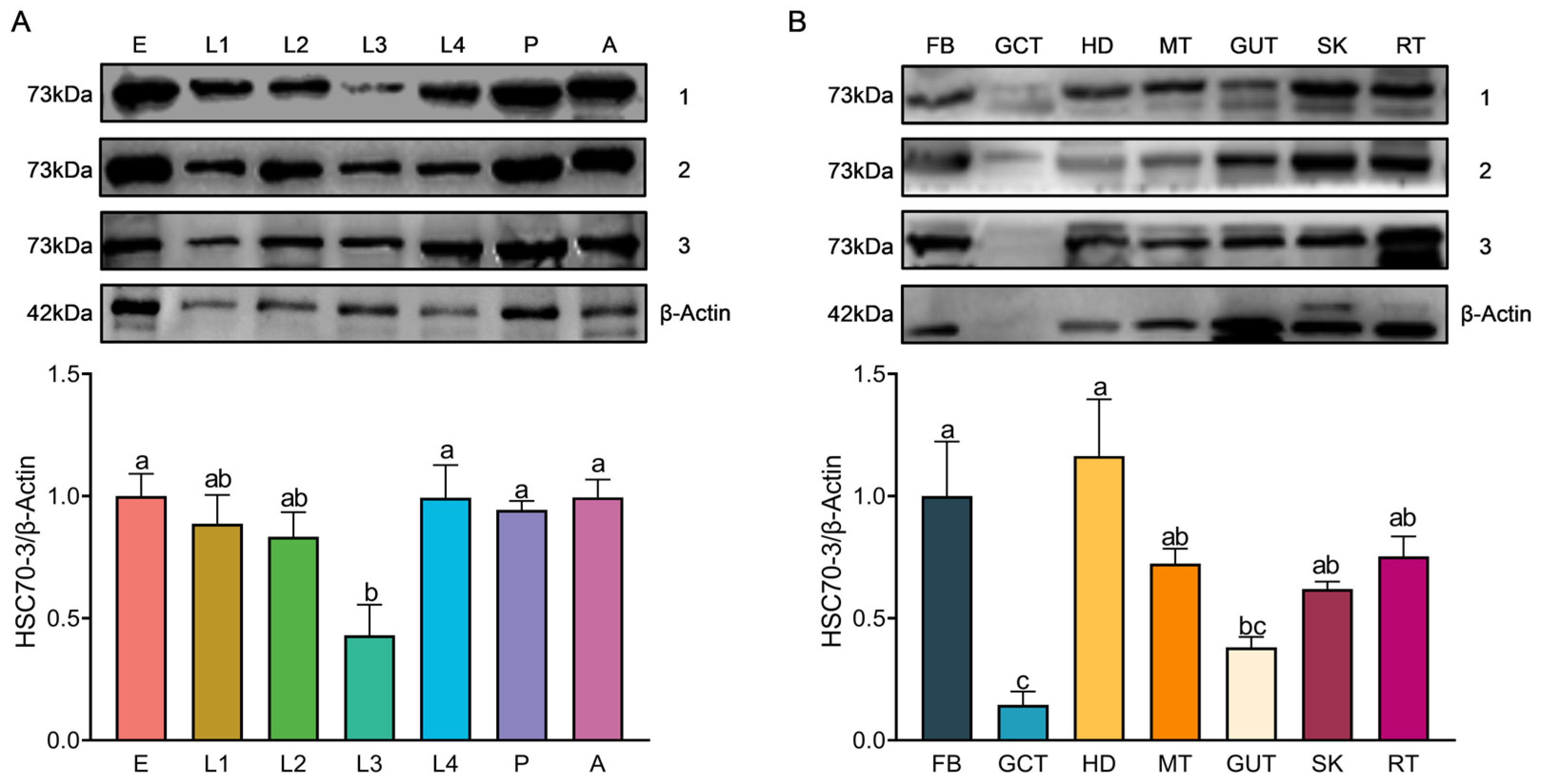
Total protein was extracted by homogenizing frozen samples on ice in 400 μL of radio immunoprecipitation assay (RIPA) lysis buffer (Beyotime Biotechnology, Shanghai, China) supplemented with a protease inhibitor (phenylmethylsulfonyl fluoride, PMSF; Solarbio, Beijing, China) and protease stabilizer (BestBio, Shanghai, China). The homogenate was gently vortexed and incubated at 4 °C for 30 min with intermittent shaking. The lysate was centrifuged at 14,000× g for 10 min at 4 °C to remove debris, and the supernatant was transferred into pre-chilled tubes. Protein concentration was determined using the Omni-Easy™ BCA Protein Quantification Kit (Epizyme Biomedical Technology Co., Ltd., Shanghai, China) following the manufacturer’s protocol. A 12.5% SDS-PAGE gel was prepared using the Omni-Easy™ One-Step PAGE Gel Preparation Kit (Epizyme Biomedical Technology Co., Ltd., Shanghai, China). Protein samples (20 μg per lane) were mixed with 5× loading buffer (Epizyme Biomedical Technology Co., Ltd., Shanghai, China), denatured at 100 °C for 10 min, briefly spun down, and then loaded onto the gel for electrophoresis. Electrophoresis was conducted at a constant voltage of 90 V for 20–30 min for protein stacking, followed by 130 V for approximately 80 min until complete separation. A pre-stained protein marker (Epizyme Biomedical Technology Co., Ltd., Shanghai, China) was included as a molecular weight reference. Proteins were transferred onto a polyvinylidene fluoride (PVDF) membrane (Millipore, Merck KGaA, Darmstadt, Germany) using a wet transfer system powered by the PowerPac™ Basic Power Supply (Bio-Rad, Bio-Rad Laboratories, Hercules, CA, USA) at 300 mA for 70 min at 4 °C. The membrane was blocked with TBST (Tris-buffered saline containing 0.5% Tween-20) supplemented with 3% skim milk at room temperature for 1 h and incubated overnight at 4 °C with the respective primary antibody (1:1000). Following three 10-min washes in TBST, the membrane was incubated with HRP-conjugated goat anti-rabbit IgG (1:5000, Affinity Biosciences, Changzhou, China) at room temperature for 2 h. Protein signals were detected using the Clarity™ Western ECL Substrate (Bio-Rad, Bio-Rad Laboratories, Hercules, CA, USA), and images were acquired using the Fusion FX5 imaging system (Vilber Lourmat, Collégien, France). Band intensities were quantified using ImageJ software (version 1.53a, National Institutes of Health, Bethesda, MD, USA), with relative protein levels normalized to β-actin expression.
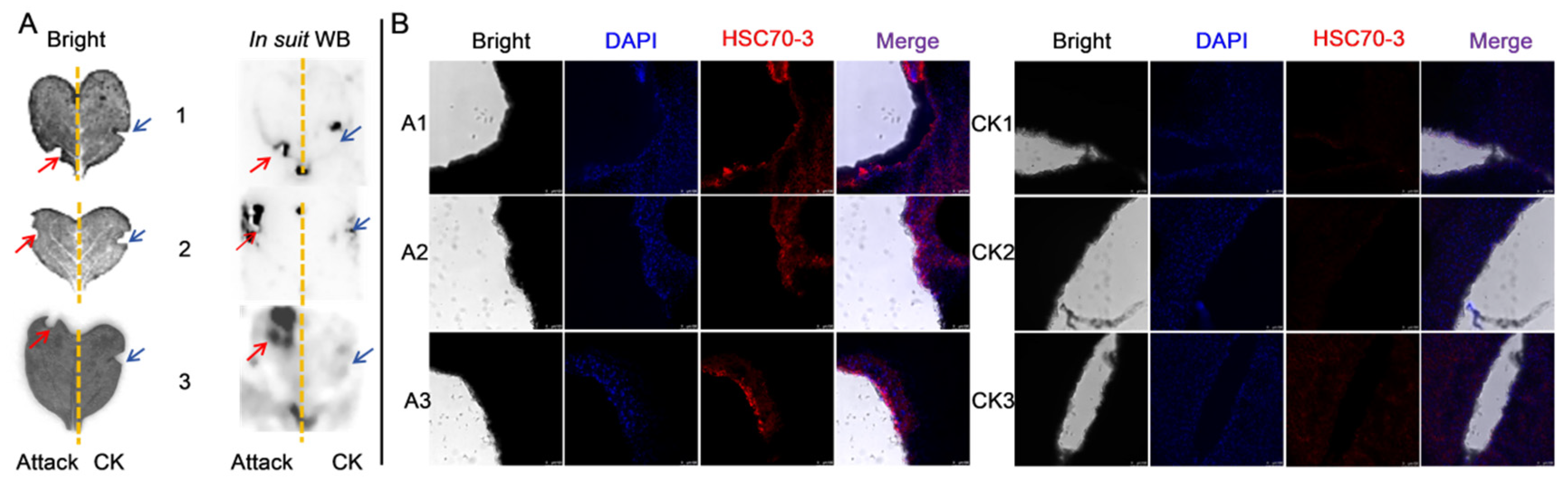
Western blot (WB) analysis was conducted to assess the expression of HSC70-3 in various developmental stages (E, L1, L2, L3, L4, P, and A) and tissues (FB, GCT, HD, MT, GUT, SK, RT), as well as to investigate its secretion at feeding sites on radish leaves. Fourth-instar P. xylostella larvae reared on an artificial diet were starved for 4 h to standardize feeding behavior before being allowed to feed on radish leaves for 6 h. To restrict the feeding area, half of the leaf surface was covered with aluminum foil during feeding. Following feeding, leaves were excised, and non-fed areas were trimmed to match the fed regions, which served as the mechanical damage control (CK). The treated leaves were then directly pressed onto a PVDF membrane for protein transfer, and HSC70-3 detection was performed using standard WB procedures.
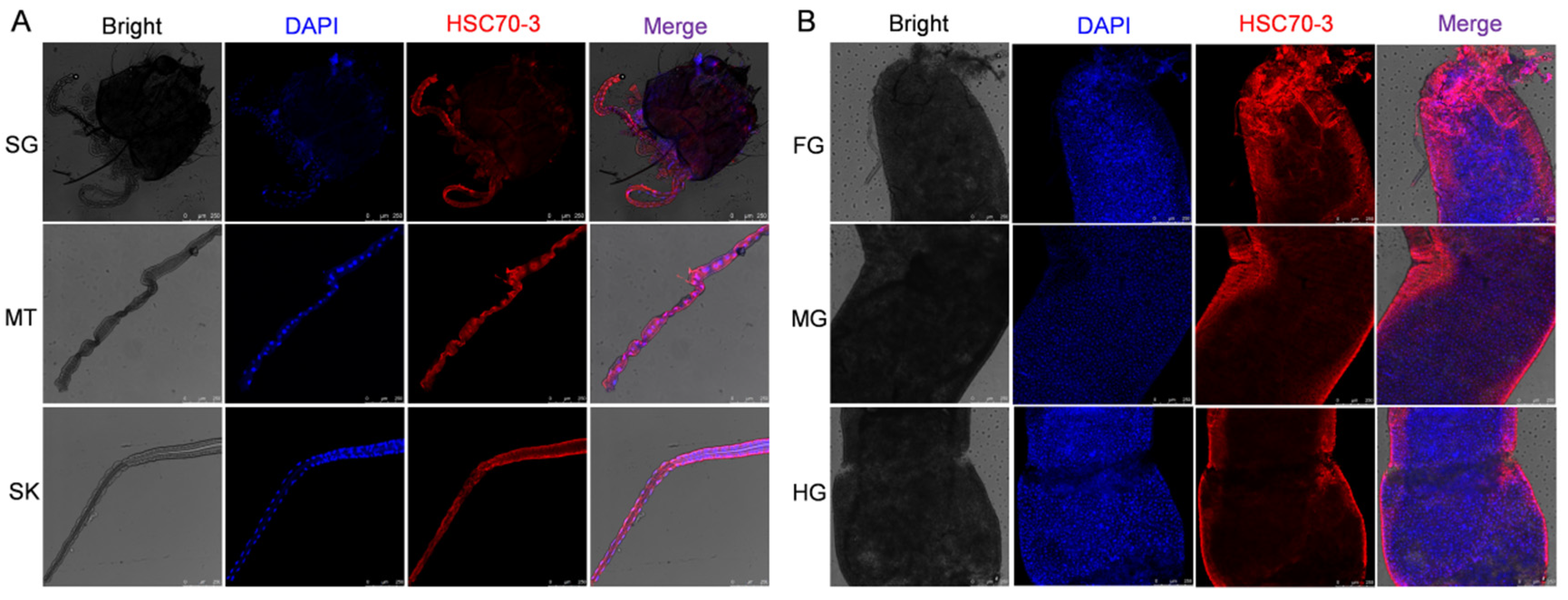
2.8. Immunofluorescent Localization of HSC70-3
To investigate the localization of HSC70-3 in feeding-related tissues, fourth-instar
P. xylostella larvae reared on an artificial diet were dissected to collect the salivary glands, silk glands, Malpighian tubules, foregut, midgut, and hindgut. The dissected tissues were immediately fixed in 1 mL of 4% paraformaldehyde (Solarbio, Beijing, China) at 4 °C for 1 h. After fixation, tissues were washed three times with 0.01 M TBST (10 min each) and permeabilized in 0.2% Triton X-100 (Solarbio, Beijing, China) for 1 h at room temperature. Samples were then washed again with 0.01 M TBST (three times, 10 min each). Tissues were incubated with an anti-HSC70-3 polyclonal antibody (1:100, GenScript, Nanjing, China) at room temperature for 2 h or overnight at 4 °C with gentle shaking in a humidified chamber. After three washes with 0.01 M TBST (10 min each), Alexa Fluor 594-conjugated goat anti-rabbit IgG (1:500, Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) was applied and incubated for 2 h at room temperature in the dark. After three additional washes with TBST, the tissues were mounted on slides using DAPI Fluoromount-G (SouthernBiotech, Birmingham, AL, USA) and incubated in the dark for 10 min. Coverslips were placed over the samples and sealed with neutral balsam (Solarbio, Beijing, China). Fluorescence imaging was performed using a Leica TCS SP8 confocal laser scanning microscope (Leica Microsystems, Wetzlar, Germany). To further investigate HSC70-3 secretion at plant feeding sites, as described in
Section 2.7, fourth-instar larvae were starved and then allowed to feed on radish leaves. The excised leaves were subsequently analyzed by immunofluorescence to detect HSC70-3 at the feeding sites.
2.9. Preparation of sgRNA and Embryo Microinjection
A 20 bp sgRNA targeting exon 1 of
hsc70-3 was designed using Benchling (
https://www.benchling.com/, accessed on 1 March 2025). Potential off-target sites were predicted using Cas-OFFinder (
http://www.rgenome.net/cas-offinder/, accessed on 1 March 2025) by screening the
P. xylostella genome for mismatches. The sgRNA oligonucleotide was synthesized with a T7 promoter and an N
20NGG sequence. To generate an in vitro transcription template, PCR amplification was performed using sgRNA-specific primers. Oligonucleotide sequences and primers for mutation detection were designed based on the genomic sequence (
Table A1). The PCR product was purified by agarose gel electrophoresis, extracted using the Gel Extraction Kit (Omega Bio-Tek, Norcross, GA, USA), and used as a template for in vitro transcription. The sgRNA transcription reaction (10 µL) was assembled using the MEGAscript
® T7 High Yield Transcription Kit (Thermo Fisher Scientific, Ambion, Waltham, MA, USA) and contained 1 µL of 10× transcription buffer, 1 µL of enzyme mix, 1 µL ATP, 1 µL CTP, 1 µL UTP, 1 µL GTP, 2 µL purified PCR product, and 2 µL RNase-free ddH
2O. The reaction mixture was incubated overnight at 37 °C. Following transcription, the reaction volume (10 µL) was diluted to 300 µL with nuclease-free water. An equal volume of phenol: chloroform: isoamyl alcohol (25:24:1) was added, gently mixed by inversion, and incubated at room temperature for 2 min. The mixture was then centrifuged at 14,000×
g for 10 min at 4 °C, and the supernatant was transferred to a fresh tube. RNA was precipitated by adding 25 µL of 3 M sodium acetate and 750 µL of absolute ethanol, followed by incubation at −20 °C for 4 h. The RNA pellet was collected by centrifugation at 14,000×
g for 15 min at 4 °C and washed twice with 1 mL of absolute ethanol. After air-drying, the pellet was resuspended in 20 µL of RNase-free water. RNA integrity was assessed via agarose gel electrophoresis, while concentration and purity were measured using a NanoDrop™ 2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA). The RNA sample was subsequently stored at −80 °C for future use.
Following sgRNA and Cas9 protein preparation, the injection mixture was assembled by combining 150 ng/μL sgRNA and 300 ng/μL Cas9 in a nuclease-free PCR tube, followed by incubation at 37 °C for 30 min. A 3 μL aliquot was loaded into a sterile glass capillary needle (GD-1, Narishige, Tokyo, Japan) using a micropipette (Eppendorf Research® plus, Eppendorf, Hamburg, Germany), and the needle tip was beveled under a stereomicroscope (SZX16, Olympus, Tokyo, Japan) to ensure precise penetration during microinjection. Freshly laid P. xylostella eggs (collected within 15 min of oviposition) were carefully transferred onto a clean microscope slide. Microinjections were performed using a stereomicroscope (SZX16, Olympus, Tokyo, Japan) in conjunction with a microinjection system (IM-300, Narishige, Tokyo, Japan). For each injection session, 300 embryos were injected. To minimize desiccation, each injection session was completed within 30 min per slide. Injected embryos were maintained at 25 °C for 24 h before being transferred to an artificial diet for further rearing. In cases where no mutations were detected after the first round of injections, a second round was performed, and the process was repeated until homozygous mutants were obtained.
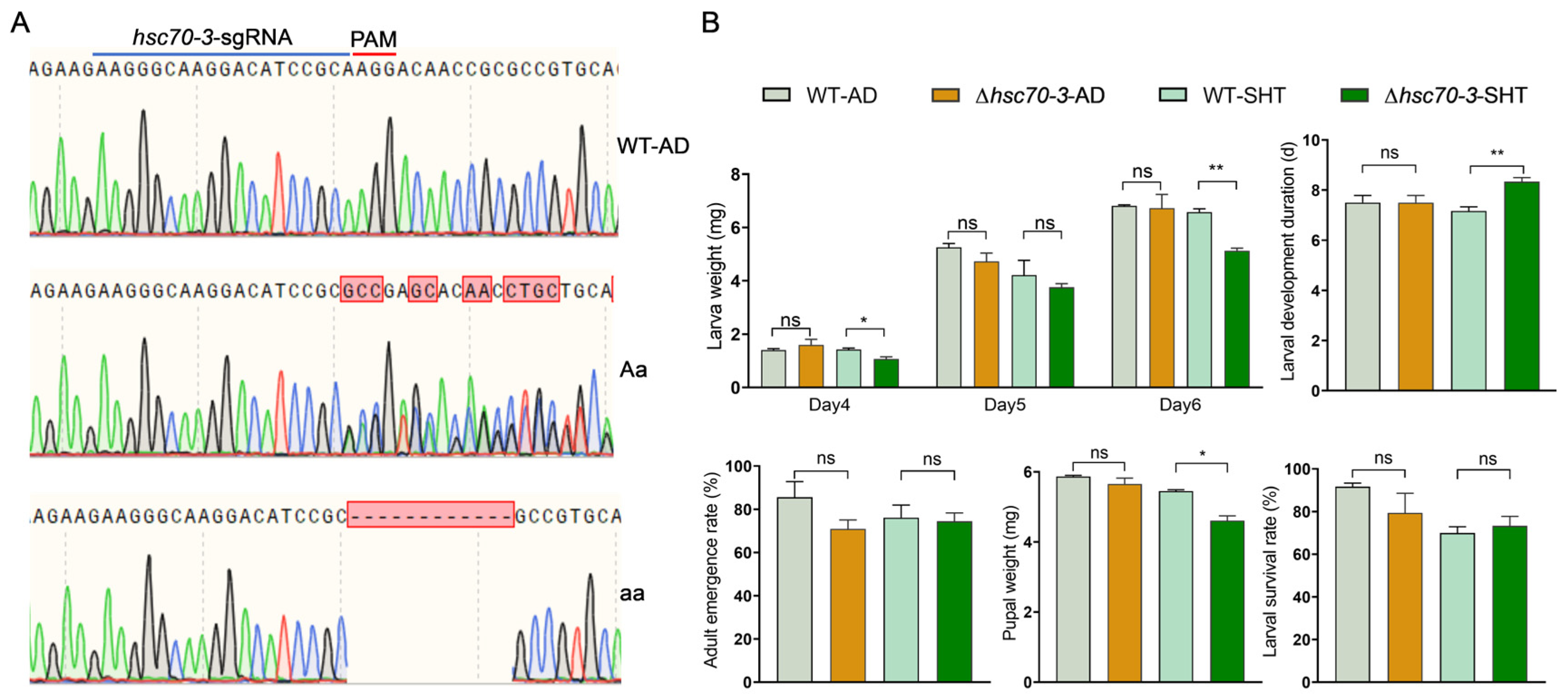
2.10. Establishment of Mutant Strains
The hsc70-3 mutant strain was established in our laboratory via CRISPR/Cas9 technology. Injected embryos developed into G0 larvae, which were reared to pupation and collected in sterile centrifuge tubes for subsequent analysis. Genomic DNA was extracted from G0 adults using the FastPure® Cell/Tissue DNA Isolation Mini Kit (Vazyme Biotech, Nanjing, China), according to the manufacturer’s instructions. Genotyping was performed via PCR amplification with locus-specific primers, followed by Sanger sequencing at Sangon Biotech (Shanghai, China). Overlapping peaks in sequencing chromatograms indicated successful genome editing, and individuals carrying mutations were retained as the G1 generation. G1 individuals were crossed with wild-type adults to generate the G2 generation, and PCR-based genotyping was performed on G2 adults after mating to identify mutations. Self-crossing was carried out over multiple generations until homozygous mutants were established. Homozygous mutants were maintained for 4–5 generations, with mutation screening conducted in each generation to ensure genetic stability.
2.11. Insect Bioassays
To investigate the role of hsc70-3 in the growth, development, and host adaptation of P. xylostella, newly hatched larvae (n = 120) from both the artificial-diet-reared wild-type (AD-WT) and hsc70-3 mutant strains were collected and randomly assigned to six replicate groups per strain (20 larvae per group). Three groups per strain were reared on an artificial diet, while the remaining three groups were fed fresh radish seedlings. The artificial diet and radish seedlings were replenished every 48 h to ensure a continuous food supply. Mutant newly hatched larvae were transferred to the host plant for one generation (approximately two weeks), alongside the AD-WT, under identical rearing conditions. During the short-term host transfer experiment, biological parameters, including survival rate; larval weight on days 4, 5, and 6 after hatching; developmental duration from larva to pupa; pupal weight; and eclosion rate, were measured in parallel.
2.12. Statistical Analysis
All statistical analyses were performed using SPSS 21.0 (IBM Corp., Armonk, NY, USA), and data visualization was performed in GraphPad Prism 9.0 (GraphPad Software, San Diego, CA, USA). One-way ANOVA followed by Tukey’s post hoc test was used for multiple group comparisons, including qRT-PCR expression levels and protein expression levels across different developmental stages and tissues. Independent sample t-tests were applied to assess pairwise differences, such as larval weight, larval development duration, adult emergency rate, pupal weight, and larval survival rate between the wild-type and mutant strains. Statistical significance was set at p < 0.05. Results are presented as the mean ± standard error (SE).
4. Discussion
Over evolutionary timescales, herbivorous insects have evolved diverse feeding strategies and effector secretion systems to circumvent host plant defenses, thereby enhancing their adaptability [
36,
37,
38]. The co-evolutionary “arms race” between plants and herbivores has driven the specialization of insect effectors, enabling them to suppress plant immune responses and enhance feeding success [
3,
37,
39]. While insect saliva has been extensively studied in plant–insect interactions, particularly in piercing–sucking insects such as aphids and mirids, research on effectors present in the gut regurgitant of Lepidoptera remains poorly characterized [
38,
40,
41]. This study demonstrates that HSC70-3, traditionally recognized as a molecular chaperone involved in cellular stress responses, may function as an effector in
P. xylostella, revealing a novel mechanism of host adaptation.
Short-term host transfer of
P. xylostella from an artificial diet to radish seedlings resulted in significant changes in HSC70-3 expression across various larval tissues. Despite the relatively low mRNA levels in gut tissues, the corresponding protein abundance remained high, suggesting potential posttranslational regulation or tissue-specific variations in protein stability. The observed discrepancy between mRNA and protein levels may result from multiple regulatory mechanisms, as transcript levels alone often fail to precisely reflect protein abundance [
42,
43,
44]. Interestingly, although HSC70-3 protein levels were relatively high in gut tissues, its abundance in the gut contents was markedly reduced. This spatial discrepancy indicates that HSC70-3 is likely retained within gut epithelial cells rather than being passively released into the gut lumen. This retention is likely linked to its role as a molecular chaperone, where it facilitates protein folding, stabilization, and intracellular transport [
24]. Consequently, only a small proportion of HSC70-3 is secreted into plant wounds through gut regurgitant during feeding. Therefore, the secretion of HSC70-3 into plant tissues is likely a regulated process, consistent with its well-known functions in protein stabilization and stress responses [
24]. Gut regurgitant, as a critical interface between insect and host plant, could serve as a potential vehicle for the delivery of HSC70-3, which supports the notion that HSC70-3 may contribute to insect–plant interactions. Furthermore, the significantly higher HSC70-3 levels in FB tissues relative to its mRNA levels suggest that the fat body may function as a reservoir or transport hub for HSC70-3. As a key tissue involved in protein metabolism, the fat body plays a pivotal role in insect environmental adaptation [
45,
46]. The elevated expression of HSC70-3 in this tissue may contribute to an enhanced immune capacity, facilitating adaptation to physiological stress imposed by the host plant. In conclusion, following short-term host plant transfer, tissue-specific expression patterns of HSC70-3 exhibited noticeable changes, albeit modest in magnitude. This may be attributed to the focus on short-term responses, where changes may not have fully manifested in the early stages following transfer. Alternatively, the host plant transfer may have induced an initial stress response that was not fully captured in the present study. Future studies should explore the long-term effects of host plant feeding on HSC70-3 expression, especially by incorporating a control group continuously feeding on host plants. This would enable a more comprehensive comparison of HSC70-3 expression between short-term host plant transfer and continuous feeding on host plants. Moreover, the observed changes in HSC70-3 expression could result from stress induced by host plant transfer or plant-derived challenges, potentially contributing to the increase in HSC70-3 levels. Further investigations are needed to elucidate the mechanisms underlying these stress responses and their impact on HSC70-3 expression.
Immunofluorescence and in situ WB analyses confirmed that HSC70-3 is secreted and accumulates at plant wound sites during
P. xylostella larval feeding. This finding suggests that HSC70-3 may play a dual role, functioning not only in insect stress responses but also in the modulation of plant defense mechanisms. As a multifunctional molecular chaperone, HSC70-3 facilitates protein folding and stress regulation within insect cells and may interact with plant membrane receptors or signaling molecules, thereby influencing host immune responses [
47,
48]. Previous studies have presented indirect evidence supporting the extracellular function of HSC70-3 in plant–insect interactions [
32]. In
N. lugens, the HSP70 family member NlHSC70-3 significantly suppresses Flg22-induced ROS accumulation and downregulates the expression of key pathogenesis-related genes (
NbPR1,
NbPR2,
NbPR3, and
NbPR4) in
Nicotiana benthamiana, thereby interfering with rice immune responses and promoting insect feeding [
32]. The identification of HSC70-3 as a potential effector in
P. xylostella provides compelling evidence that gut regurgitant plays a crucial role in insect adaptation to host plants. This finding broadens the current understanding of effector-mediated interactions in Lepidoptera and highlights a novel avenue for investigating the interplay between gut-derived secretions and plant signaling pathways.
CRISPR/Cas9-mediated knockout of
hsc70-3 provided functional insights into the essential role of HSC70-3 in
P. xylostella larval adaptation. Mutant larvae exhibited significantly reduced body weight, delayed development, and decreased pupal weight when reared on radish seedlings, suggesting that
hsc70-3 facilitates host plant adaptation by modulating plant defense responses and optimizing nutrient assimilation. However, it is important to note that plant defense was not directly measured in this study. These findings are based on the observed developmental impairments in mutant larvae, which may be linked to an impaired ability to modulate plant defense. The previous studies on
H. zea, in which the effector-mediated suppression of plant immunity promotes larval growth and survival, support this potential role of HSC70-3 in overcoming plant resistance mechanisms [
49]. Notably,
hsc70-3 knockout larvae exhibited no significant developmental defects when reared on artificial diets, supporting the notion that
hsc70-3 primarily modulates plant-induced defense responses rather than directly affecting insect metabolism or development. This finding highlights the critical role of effectors in overcoming plant resistance mechanisms, although a direct measurement of plant defense was not conducted in this study. Previous studies have shown that HSC70 family proteins are involved in nutrient metabolism and stress responses in various insect species [
48,
50]. Accordingly, we do not exclude the possibility that HSC70-3 may influence larval development by modulating the absorption or utilization of plant-derived nutrients. Further investigation is warranted to determine whether HSC70-3 contributes to nutrient acquisition in addition to its role in suppressing plant defense responses. Although only three biological replicates were used for survival and adult emergence rate assays, this limitation was due to the restricted number of eggs produced by CRISPR-edited lines and the need to maintain their genetic stability across generations. To ensure reliable interpretation, we additionally measured larval weight, developmental duration, and pupal weight, which are widely recognized as robust indicators of insect fitness. These complementary data consistently supported the biological role of
hsc70-3 in host adaptation.
Although this study presents strong evidence supporting the role of HSC70-3 as an effector in
P. xylostella, the precise molecular mechanisms underlying its role remain to be fully elucidated. Growing evidence indicates that herbivorous insects utilize effectors to interfere with plant defense pathways via multiple mechanisms [
1,
51,
52]. While some insect effectors have been shown to interact with specific plant receptors or signaling molecules to suppress plant defense pathways, whether HSC70-3 adopts a similar strategy in
P. xylostella remains unknown. Future research should investigate the specific interactions between HSC70-3 and plant immune receptors or signaling pathways to elucidate its role in host adaptation. The identification of HSC70-3 as an immune-suppressing effector in
P. xylostella suggests novel strategies for pest control. Specifically, plant defense responses could be measured using qRT-PCR to analyze the expression of defense-related genes or by assessing ROS accumulation and pathogenesis-related proteins to evaluate the impact of HSC70-3 on plant immunity. If HSC70-3 significantly attenuates plant defenses, its targeted inhibition could enhance crop resistance towards herbivorous insects. Furthermore, identifying the receptors or downstream targets of HSC70-3 in plants could facilitate the development of genetically modified pest-resistant crops, thereby improving plant resilience against
P. xylostella.