Unusual Genetic Diversity Within Thereuopoda clunifera (Wood, 1862) (Chilopoda: Scutigeromorpha) Revealed by Phylogeny and Divergence Times Using Mitochondrial Genomes
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Taxon Collection and DNA Extraction
2.2. Mitogenome Generation and Sequence Analyses
2.3. Species Delimitation
2.4. Dataset Selection and Phylogenetic Analyses
2.5. Divergence Time Estimation
3. Results
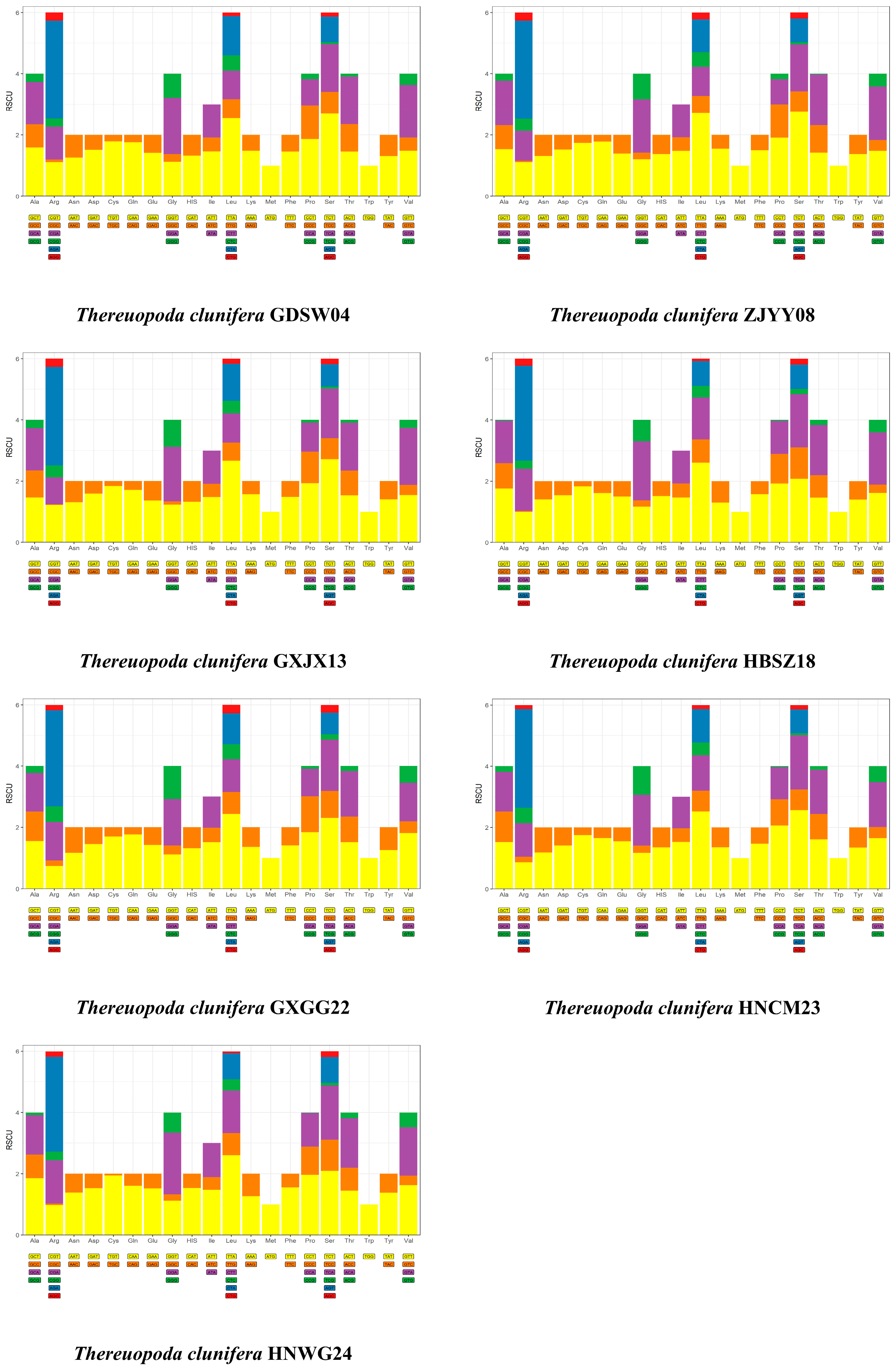
3.1. Composition and Organization of Seven Mitogenomes of T. clunifera
3.2. Species Delimitation
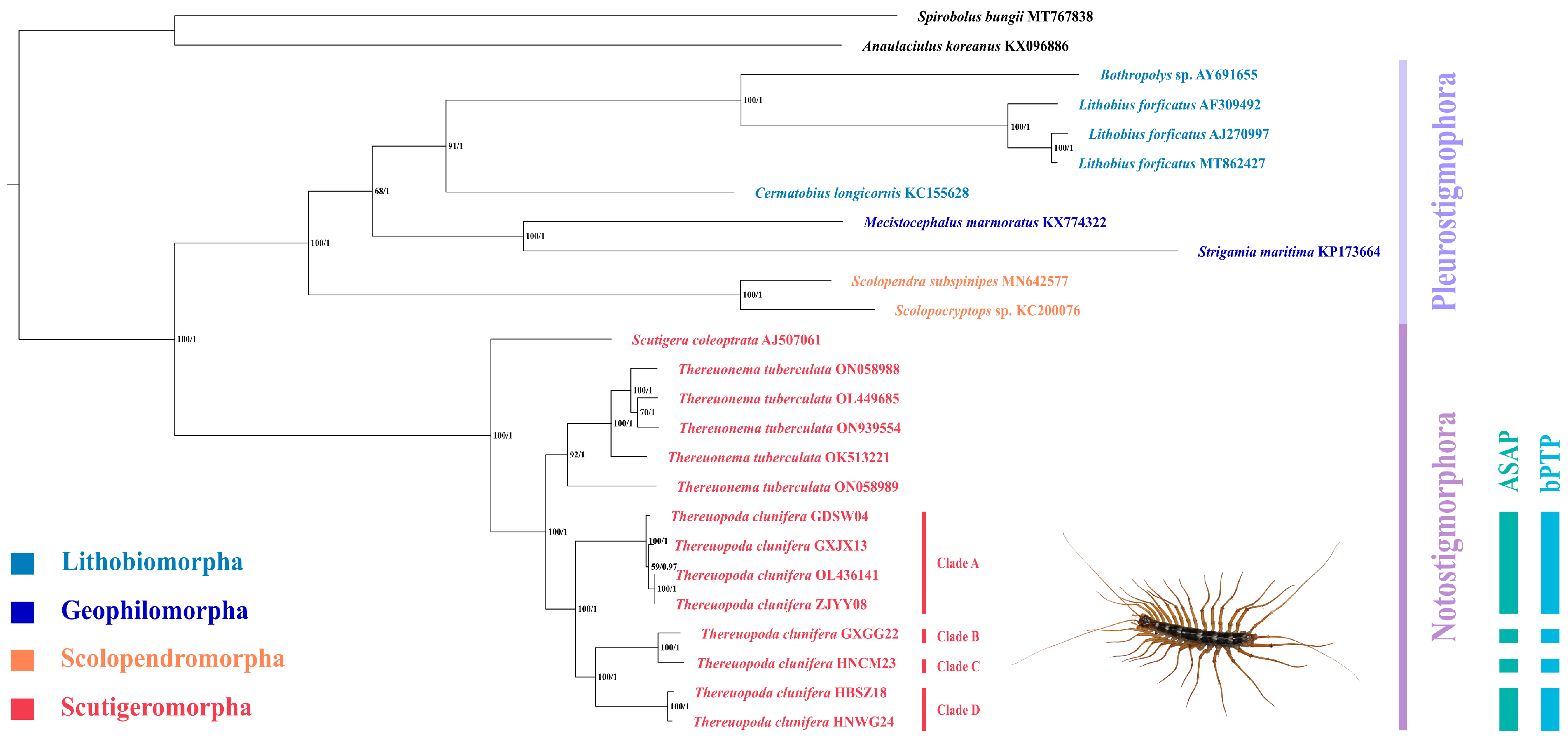
3.3. Phylogenetic Analyses
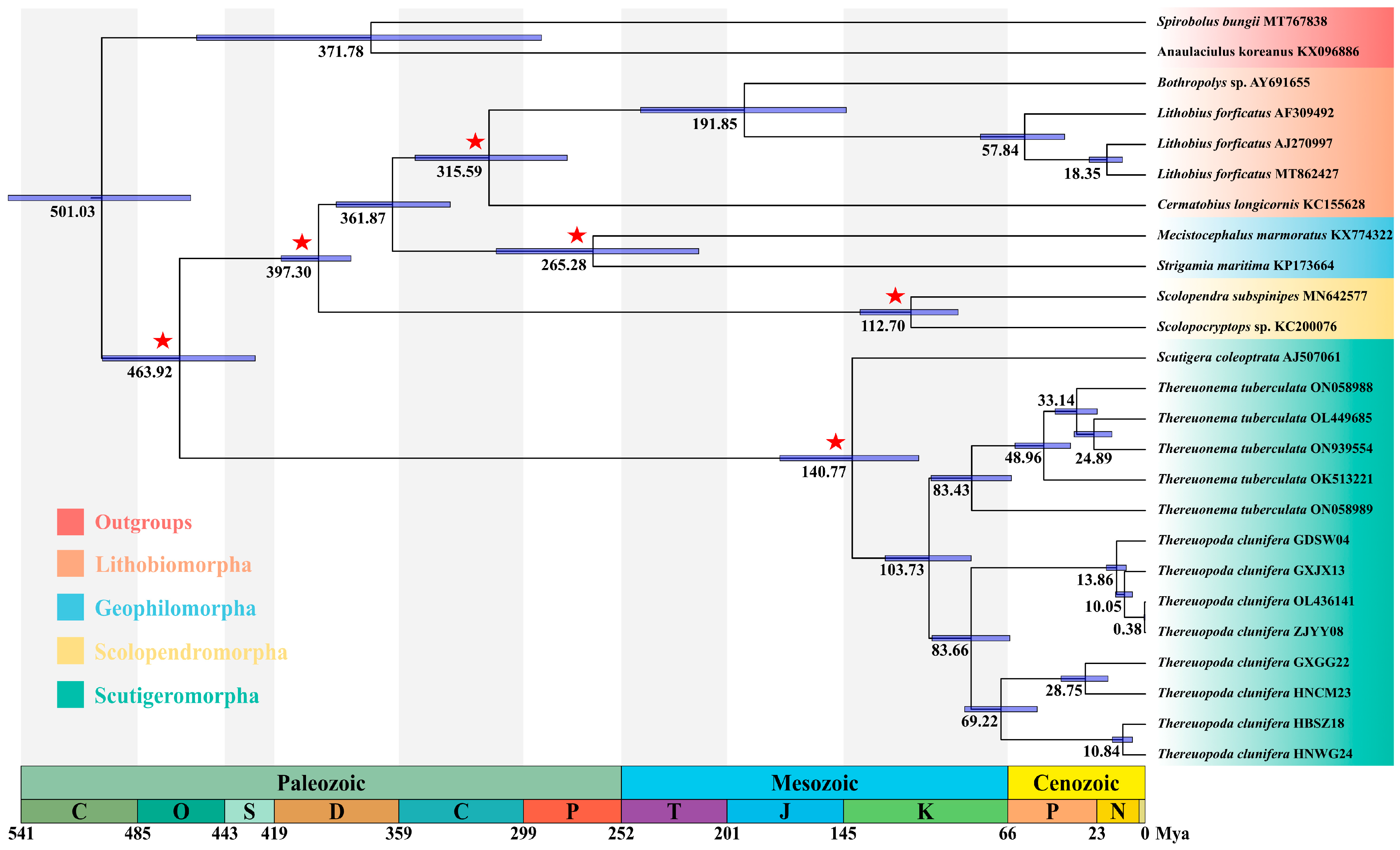
3.4. Divergence Time Estimation
4. Discussion
4.1. Structure of Seven Mitogenomes of T. clunifera
4.2. Phylogenetic Relationship of Chilopoda
4.3. Divergence Time of Chilopoda
4.4. Identification of Cryptic Species
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Edgecombe, G.D.; Giribet, G. The molecularization of centipede systematics. In Perspectives on Evolutionary and Developmental Biology: Essays for Alessandro Minelli; Fusco, G., Ed.; Padova University Press: Padova, Italy, 2019; pp. 153–165. [Google Scholar]
- Dohle, W. Phylogenetic pathways in the Chilopoda. Bijdr. Tot Dierkd. 1985, 55, 55–66. [Google Scholar]
- Shear, W.A.; Edgecombe, G.D. The geological record and phylogeny of the Myriapoda. Arthropod Struct. Dev. 2010, 39, 174–190. [Google Scholar] [CrossRef] [PubMed]
- Pocock, R.I. A new and annectant type of chilopod. Quart. J. Microscop. Sci. 1902, 45, 417–448. [Google Scholar] [CrossRef]
- Verhoeff, K.W. Chilopoda. In Klassen und Ordnungen des Tierreichs, 5, Abteilung 2, Buch 1; Bronn, H.G., Ed.; C.F. Winter’sche Verlagshandlung: Leipzig, Germany, 1902–1925; pp. 1–725. [Google Scholar]
- Shear, W.A.; Bonamo, P.M. Devonobiomorpha, a new order of centipeds (Chilopoda) from the Middle Devonian of Gilboa, New York State, USA, and the phylogeny of centiped orders. Am. Mus. Novit. 1988, 2927, 1–30. [Google Scholar]
- Giribet, G.; Carranza, S.; Riutort, M.; Baguñà, J.; Ribera, C. Internal phylogeny of the Chilopoda (Myriapoda, Arthropoda) using complete 18S rDNA and partial 28S rDNA sequences. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1999, 354, 215–222. [Google Scholar] [CrossRef]
- Edgecombe, G.; Giribet, G. Adding mitochondrial sequence data (16S rRNA and cytochrome c oxidase subunit I) to the phylogeny of centipedes (Myriapoda: Chilopoda): An analysis of morphology and four molecular loci. J. Zool. Syst. Evol. Res. 2004, 42, 89–134. [Google Scholar] [CrossRef]
- Regier, J.C.; Wilson, H.M.; Shultz, J.W. Phylogenetic analysis of Myriapoda using three nuclear protein-coding genes. Mol. Phylogenet. Evol. 2005, 34, 147–158. [Google Scholar] [CrossRef]
- Regier, J.C.; Shultz, J.W.; Zwick, A.; Hussey, A.; Ball, B.; Wetzer, R.; Martin, J.W.; Cunningham, C.W. Arthropod relationships revealed by phylogenomic analysis of nuclear protein-coding sequences. Nature 2010, 463, 1079–1083. [Google Scholar] [CrossRef]
- Murienne, J.; Edgecombe, G.D.; Giribet, G. Including secondary structure, fossils and molecular dating in the centipede tree of life. Mol. Phylogenet. Evol. 2010, 57, 301–313. [Google Scholar] [CrossRef]
- Benavides, L.R.; Edgecombe, G.D.; Giribet, G. Re-evaluating and dating myriapod diversification with phylotranscriptomics under a regime of dense taxon sampling. Mol. Phylogenet. Evol. 2023, 178, 107621. [Google Scholar] [CrossRef]
- Fernández, R.; Edgecombe, G.D.; Giribet, G. Exploring phylogenetic relationships within Myriapoda and the effects of matrix composition and occupancy on phylogenomic reconstruction. Syst. Biol. 2016, 65, 871–889. [Google Scholar] [CrossRef] [PubMed]
- Fernández, R.; Laumer, C.E.; Vahtera, V.; Libro, S.; Kaluziak, S.; Sharma, P.P.; Pérez-Porro, A.R.; Edgecombe, G.D.; Giribet, G. Evaluating topological conflict in centipede phylogeny using transcriptomic data sets. Mol. Biol. Evol. 2014, 31, 1500–1513. [Google Scholar] [CrossRef] [PubMed]
- Fernández, R.; Edgecombe, G.D.; Giribet, G. Phylogenomics illuminates the backbone of the Myriapoda tree of life and reconciles morphological and molecular phylogenies. Sci. Rep. 2018, 8, 83. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.J.; Bai, Y.; Zhao, H.; Mu, R.; Dong, Y. Reinvestigating the phylogeny of Myriapoda with more extensive taxon sampling and novel genetic perspective. PeerJ 2021, 9, e12691. [Google Scholar] [CrossRef]
- Hu, C.; Wang, S.; Huang, B.; Liu, H.; Xu, L.; Hu, Z.; Liu, Y. The complete mitochondrial genome sequence of Scolopendra mutilans L. Koch, 1878 (Scolopendromorpha, Scolopendridae), with a comparative analysis of other centipede genomes. ZooKeys 2020, 925, 73–88. [Google Scholar] [CrossRef]
- Yang, Y.M.; Zhang, L.H.; Lin, Y.J.; Zheng, Y.M.; Jin, W.T.; Storey, K.B.; Yu, D.N.; Zhang, J.Y. The genetic diversity in Thereuonema tuberculata (Wood, 1862) (Scutigeromorpha: Scutigeridae) and the phylogenetic relationship of Scutigeromorpha using the mitochondrial genome. Insects 2022, 13, 620. [Google Scholar] [CrossRef]
- Wang, J.J.; Bai, Y.; Dong, Y. A rearrangement of the mitochondrial genes of centipedes (Arthropoda, Myriapoda) with a phylogenetic analysis. Genes 2022, 13, 1787. [Google Scholar] [CrossRef]
- Zhao, X.; Lu, Y.; Fan, S.; Xu, W.; Wang, J.; Wang, G.; Liu, H. The complete mitochondrial genome of Thereuopoda clunifera (Chilopoda: Scutigeridae) and phylogenetic implications within Chilopoda. Zootaxa 2022, 5174, 165–180. [Google Scholar] [CrossRef]
- Ding, J.; Lan, H.; Xu, W.; Chen, Y.; Wu, H.; Jiang, H.; Wang, J.; Wu, Y.; Liu, H. Two complete mitochondrial genomes in Scolopendra and a comparative analysis of tRNA rearrangements in centipedes. Mol. Biol. Rep. 2022, 49, 6173–6180. [Google Scholar] [CrossRef]
- Edgecombe, G.D. Chilopoda—Taxonomic overview. Order Scutigeromorpha. In Treatise on Zoology—Anatomy, Taxonomy, Biology. The Myriapoda; Minelli, A., Ed.; Brill: Leiden, The Netherlands, 2011; Volume 1, pp. 363–370. [Google Scholar]
- Pérez-Gelabert, D.E.; Edgecombe, G.D. Scutigeromorph centipedes (Chilopoda: Scutigeromorpha) of the Dominican Republic, Hispaniola. Novit. Caribaea 2013, 6, 36–44. [Google Scholar] [CrossRef]
- Edgecombe, G.D.; Giribet, G. A century later—A total evidence re-evaluation of the phylogeny of scutigeromorph centipedes (Myriapoda: Chilopoda). Invertebr. Syst. 2006, 20, 503–525. [Google Scholar] [CrossRef]
- Edgecombe, G.D.; Giribet, G. Phylogenetics of scutigeromorph centipedes (Myriapoda: Chilopoda) with implications for species delimitation and historical biogeography of the Australian and New Caledonian faunas. Cladistics 2009, 25, 406–427. [Google Scholar] [CrossRef] [PubMed]
- Giribet, G.; Edgecombe, G.D. Stable phylogenetic patterns in scutigeromorph centipedes (Myriapoda: Chilopoda: Scutigeromorpha): Dating the diversification of an ancient lineage of terrestrial arthropods. Invertebr. Syst. 2013, 27, 485–501. [Google Scholar] [CrossRef]
- Manivannan, M.; Gurung, N.; Edgecombe, G.D.; Joshi, J. A passage through India: The biotic ferry model supports the build-up of Indo-Australian biodiversity of an ancient soil arthropod clade. J. Biogeogr. 2024, 51, 2395–2411. [Google Scholar] [CrossRef]
- Porta, A.O.; Giribet, G. A new genus of scutigerid centipede from southern South America with the description of two new species and an updated molecular phylogeny of the myriapod order Scutigeromorpha (Myriapoda: Chilopoda). Invertebr. Syst. 2024, 38, IS24006. [Google Scholar] [CrossRef]
- Edgecombe, G.D. Centipede systematics: Progress and problems. Zootaxa 2007, 1668, 327–341. [Google Scholar] [CrossRef]
- Edgecombe, G.D.; Barrow, L.J. A new genus of scutigerid centipedes (Chilopoda) from Western Australia, with new characters for morphological phylogenetics of Scutigeromorpha. Zootaxa 2007, 1409, 23–50. [Google Scholar] [CrossRef]
- Würmli, M. Taxonomic problems in the genus Thereuopoda (Chilopoda Scutigeromorpha: Scutigeridae): The role of postmaturational moultings. In Myriapod Biology; Camatini, M., Ed.; Academic Press: London, UK, 1979; pp. 39–48. [Google Scholar]
- Struck, T.H.; Feder, J.L.; Bendiksby, M.; Birkeland, S.; Cerca, J.; Gusarov, V.I.; Kistenich, S.; Larsson, K.H.; Liow, L.H.; Nowak, M.D.; et al. Finding evolutionary processes hidden in cryptic species. Trends Ecol. Evol. 2018, 33, 153–163. [Google Scholar] [CrossRef]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef]
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef]
- Boore, J.L. The use of genome-level characters for phylogenetic reconstruction. Trends Ecol. Evol. 2006, 21, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Moritz, C.; Dowling, T.E.; Brown, W.M. Evolution of animal mitochondrial DNA: Relevance for population biology and systematics. Annu. Rev. Ecol. Evol. Syst. 1987, 18, 269–292. [Google Scholar] [CrossRef]
- Tong, Y.; Wu, L.; Ayivi, S.P.G.; Storey, K.B.; Ma, Y.; Yu, D.N.; Zhang, J.Y. Cryptic species exist in Vietnamella sinensis Hsu, 1936 (Insecta: Ephemeroptera) from studies of complete mitochondrial genomes. Insects 2022, 13, 412. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.Q.; Shen, C.Y.; Cheng, H.Y.; Chen, Y.X.; Wu, H.Y.; Storey, K.B.; Yu, D.N.; Zhang, J.Y. Mitogenome-based phylogeny with divergence time estimates revealed the presence of cryptic species within Heptageniidae (Insecta, Ephemeroptera). Insects 2024, 15, 745. [Google Scholar] [CrossRef]
- Tong, Y.; Shen, C.Y.; Zhao, Y.Y.; Lin, Y.J.; Wu, L.; Storey, K.B.; Yu, D.N.; Zhang, J.Y. The genetic diversity and the divergence time in extant primitive mayfly, Siphluriscus chinensis Ulmer, 1920 using the mitochondrial genome. Genes 2022, 13, 1780. [Google Scholar] [CrossRef]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; dePamphilis, C.W.; Yi, T.S.; Li, D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Laslett, D.; Canbäck, B. ARWEN: A program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 2008, 24, 172–175. [Google Scholar] [CrossRef]
- Negrisolo, E.; Minelli, A.; Valle, G. Extensive gene order rearrangement in the mitochondrial genome of the centipede Scutigera coleoptrata. J. Mol. Evol. 2004, 58, 413–423. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.Y.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef] [PubMed]
- Kerpedjiev, P.; Hammer, S.; Hofacker, I.L. Forna (force-directed RNA): Simple and effective online RNA secondary structure diagrams. Bioinformatics 2015, 31, 3377–3379. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Puillandre, N.; Brouillet, S.; Achaz, G. ASAP: Assemble species by automatic partitioning. Mol. Ecol. Resour. 2021, 21, 609–620. [Google Scholar] [CrossRef]
- Zhang, J.; Kapli, P.; Pavlidis, P.; Stamatakis, A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 2013, 29, 2869–2876. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Lladó Fernández, S.; Větrovský, T.; Baldrian, P. The concept of operational taxonomic units revisited: Genomes of bacteria that are regarded as closely related are often highly dissimilar. Folia Microbiol. 2019, 64, 19–23. [Google Scholar] [CrossRef]
- Lavrov, D.V.; Brown, W.M.; Boore, J.L. A novel type of RNA editing occurs in the mitochondrial tRNAs of the centipede Lithobius forficatus. Proc. Natl. Acad. Sci. USA 2000, 97, 13738–13742. [Google Scholar] [CrossRef]
- Hwang, U.W.; Friedrich, M.; Tautz, D.; Park, C.J.; Kim, W. Mitochondrial protein phylogeny joins myriapods with chelicerates. Nature 2001, 413, 154–157. [Google Scholar] [CrossRef]
- Gai, Y.; Junye, M.; Li, C.-X.; Yang, Q. The complete mitochondrial genome of Scolopocryptops sp. (Chilopoda: Scolopendromorpha: Scolopocryptopidae). Mitochondrial DNA 2013, 25, 192–193. [Google Scholar] [CrossRef] [PubMed]
- Gai, Y.; Sun, X.; Junye, M.; Li, C.-X.; Yang, Q. The complete mitochondrial genome of Cermatobius longicornis (Chilopoda: Lithobiomorpha: Henicopidae). Mitochondrial DNA 2013, 24, 331–332. [Google Scholar] [CrossRef] [PubMed]
- Robertson, H.E.; Lapraz, F.; Rhodes, A.C.; Telford, M.J. The complete mitochondrial genome of the geophilomorph centipede Strigamia maritima. PLoS ONE 2015, 10, e0121369. [Google Scholar] [CrossRef] [PubMed]
- Woo, H.; Nguyen, D.A.; Jang, K.; Choi, E.H.; Ryu, S.; Hwang, U.W. The complete mitochondrial genome of the Korean endemic millipede Anaulaciulus koreanus (Verhoeff, 1937), with notes on the gene arrangement of millipede orders. Zootaxa 2017, 4329, 574–583. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef]
- Lanfear, R.; Calcott, B.; Ho, S.Y.W.; Guindon, S. PartitionFinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 2012, 29, 1695–1701. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Rambaut, A. Figtree Version 1.4.4. 2019. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 27 November 2024).
- Edgecombe, G.D. The fossil history. In Treatise on Zoology—Anatomy, Taxonomy, Biology. The Myriapoda; Minelli, A., Ed.; Brill: Leiden, The Netherlands, 2011; Volume 1, pp. 355–361. [Google Scholar]
- Le Cadre, J.; Melzer, R.R.; Müller, P.; Haug, C.; Haug, J.T. Three new lithobiomorphan centipede specimens from mid-Cretaceous Myanmar amber, a clue on the geological record of Lithobiomorpha. Mesozoic 2024, 1, 493–505. [Google Scholar] [CrossRef]
- Bonato, L.; Drago, L.; Murienne, J. Phylogeny of Geophilomorpha (Chilopoda) inferred from new morphological and molecular evidence. Cladistics 2014, 30, 485–507. [Google Scholar] [CrossRef]
- Bonato, L.; Edgecombe, G.D.; Minelli, A. Geophilomorph centipedes from the Cretaceous amber of Burma. Palaeontology 2014, 57, 97–110. [Google Scholar] [CrossRef]
- Wolfe, J.M.; Daley, A.C.; Legg, D.A.; Edgecombe, G.D. Fossil calibrations for the arthropod tree of life. Earth Sci. Rev. 2016, 160, 43–110. [Google Scholar] [CrossRef]
- Wilson, H.M. First Mesozoic scutigeromorph centipede, from the Lower Cretaceous of Brazil. Palaeontology 2001, 44, 489–495. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 214. [Google Scholar] [CrossRef]
- Lavrov, D.V.; Boore, J.L.; Brown, W.M. The complete mitochondrial DNA sequence of the horseshoe crab Limulus polyphemus. Mol. Biol. Evol. 2000, 17, 813–824. [Google Scholar] [CrossRef]
- Wolstenholme, D.R. Animal mitochondrial DNA: Structure and evolution. Int. Rev. Cytol. 1992, 141, 173–216. [Google Scholar]
- Machida, R.J.; Miya, M.U.; Nishida, M.; Nishida, S. Complete mitochondrial DNA sequence of Tigriopus japonicus (Crustacea: Copepoda). Mar. Biotechnol. 2002, 4, 406–417. [Google Scholar] [CrossRef]
- Hassanin, A.; Léger, N.; Deutsch, J. Evidence for multiple reversals of asymmetric mutational constraints during the evolution of the mitochondrial genome of metazoa, and consequences for phylogenetic inferences. Syst. Biol. 2005, 54, 277–298. [Google Scholar] [CrossRef]
- Ojala, D.; Montoya, J.; Attardi, G. tRNA punctuation model of RNA processing in human mitochondria. Nature 1981, 290, 470–474. [Google Scholar] [CrossRef]
- Hanada, T.; Suzuki, T.; Yokogawa, T.; Takemoto-Hori, C.; Sprinzl, M.; Watanabe, K. Translation ability of mitochondrial tRNAsSer with unusual secondary structures in an in vitro translation system of bovine mitochondria. Genes Cells 2001, 6, 1019–1030. [Google Scholar] [CrossRef] [PubMed]
- McClain, W.H. Surprising contribution to aminoacylation and translation of non-Watson–Crick pairs in tRNA. Proc. Natl. Acad. Sci. USA 2006, 103, 4570–4575. [Google Scholar] [CrossRef] [PubMed]
- Edgecombe, G.D. Chilopoda—Phylogeny. In Treatise on Zoology—Anatomy, Taxonomy, Biology. The Myriapoda; Minelli, A., Ed.; Brill: Leiden, The Netherlands, 2011; Volume 1, pp. 339–354. [Google Scholar]
- Edgecombe, G.D. Phylogenetic relationships of Myriapoda. In Treatise on Zoology—Anatomy, Taxonomy, Biology. The Myriapoda; Minelli, A., Ed.; Brill: Leiden, The Netherlands, 2011; Volume 1, pp. 1–20. [Google Scholar]
- Jamieson, B. The spermatozoa of the Chilopoda (Uniramia): An ultrastructural review with data on dimorphism in Ethmostigmus rubripes and phylogenetic discussion. J. Submicrosc. Cytol. 1986, 18, 543–558. [Google Scholar]
- Shear, W.A.; Jeram, A.J.; Selden, P.A. Centiped legs (Arthropoda, Chilopoda, Scutigeromorpha) from the Silurian and Devonian of Britain and the Devonian of North America. Am. Mus. Novit. 1998, 3231, 1–16. [Google Scholar]
- Wang, Y.; Huang, C.; Sun, B.; Quan, C.; Wu, J.; Lin, Z. Paleo-CO2 variation trends and the Cretaceous greenhouse climate. Earth Sci. Rev. 2014, 129, 136–147. [Google Scholar] [CrossRef]
- Shao, L.; Li, S. Early Cretaceous greenhouse pumped higher taxa diversification in spiders. Mol. Phylogenet. Evol. 2018, 127, 146–155. [Google Scholar] [CrossRef]
- Fernández, R.; Hormiga, G.; Giribet, G. Phylogenomic analysis of spiders reveals nonmonophyly of orb weavers. Curr. Biol. 2014, 24, 1772–1777. [Google Scholar] [CrossRef]
- Vieites, D.R.; Min, M.-S.; Wake, D.B. Rapid diversification and dispersal during periods of global warming by plethodontid salamanders. Proc. Natl. Acad. Sci. USA 2007, 104, 19903–19907. [Google Scholar] [CrossRef]
- Wahlberg, N.; Wheat, C.W.; Peña, C. Timing and patterns in the taxonomic diversification of Lepidoptera (butterflies and moths). PLoS ONE 2013, 8, e80875. [Google Scholar] [CrossRef]
- Evangelista, D.A.; Wipfler, B.; Béthoux, O.; Donath, A.; Fujita, M.; Kohli, M.K.; Legendre, F.; Liu, S.; Machida, R.; Misof, B.; et al. An integrative phylogenomic approach illuminates the evolutionary history of cockroaches and termites (Blattodea). Proc. R. Soc. B Biol. Sci. 2019, 286, 20182076. [Google Scholar] [CrossRef]
- Liu, J.; May-Collado, L.J.; Pekár, S.; Agnarsson, I. A revised and dated phylogeny of cobweb spiders (Araneae, Araneoidea, Theridiidae): A predatory Cretaceous lineage diversifying in the era of the ants (Hymenoptera, Formicidae). Mol. Phylogenet. Evol. 2016, 94, 658–675. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.G.E. The Biology of Centipedes; Cambridge University Press: Cambridge, UK, 1981; pp. 183–185. [Google Scholar]
- Robertson, D.S.; McKenna, M.C.; Toon, O.B.; Hope, S.; Lillegraven, J.A. Survival in the first hours of the Cenozoic. Geol. Soc. Am. Bull. 2004, 116, 760–768. [Google Scholar] [CrossRef]
- Tapponnier, P.; Peltzer, G.; Le Dain, A.Y.; Armijo, R.; Cobbold, P. Propagating extrusion tectonics in Asia: New insights from simple experiments with plasticine. Geology 1982, 10, 611–616. [Google Scholar] [CrossRef]
- Zhu, H. New insight into the origin and evolution of the flora and fauna of Hainan Island, China. Sci. China Earth Sci. 2025, 68, 52–61. [Google Scholar] [CrossRef]
- Replumaz, A.; Tapponnier, P. Reconstruction of the deformed collision zone between India and Asia by backward motion of lithospheric blocks. J. Geophys. Res. Solid Earth 2003, 108, 2285. [Google Scholar] [CrossRef]
- Wesener, T.; Voigtländer, K.; Decker, P.; Oeyen, J.P.; Spelda, J. Barcoding of central european Cryptops centipedes reveals large interspecific distances with ghost lineages and new species records from Germany and Austria (Chilopoda, Scolopendromorpha). ZooKeys 2016, 564, 21–46. [Google Scholar] [CrossRef]
- Bonato, L.; Chagas-Junior, A.; Edgecombe, G.D.; Lewis, J.G.E.; Minelli, A.; Pereira, L.A.; Shelley, R.M.; Stoev, P.; Zapparoli, M. ChiloBase 2.0—A Word Catalogue of Centipedes (Chilopoda). Available online: https://chilobase.biologia.unipd.it/ (accessed on 27 November 2024).
- Bolton, S.J.; Macleod, N.; Edgecombe, G.D. Geometric approaches to the taxonomic analysis of centipede gonopods (Chilopoda: Scutigeromorpha). Zool. J. Linn. Soc. 2009, 156, 239–259. [Google Scholar] [CrossRef]
- Gutierrez, B.L.; Macleod, N.; Edgecombe, G.D. Detecting taxonomic signal in an under-utilised character system: Geometric morphometrics of the forcipular coxae of Scutigeromorpha (Chilopoda). ZooKeys 2011, 156, 49–66. [Google Scholar]

| Number | Species | Sampling Localities | Accession No. |
|---|---|---|---|
| ZJYY08 | T. clunifera | Ningbo, Zhejiang, China | PQ595907 |
| GDSW04 | T. clunifera | Shanwei, Guangdong, China | PQ595908 |
| GXGG22 | T. clunifera | Guigang, Guangxi, China | PQ595909 |
| GXJX13 | T. clunifera | Laibin, Guangxi, China | PQ595910 |
| HBSZ18 | T. clunifera | Suizhou, Hubei, China | PQ595911 |
| HNCM23 | T. clunifera | Chengmai, Hainan, China | PQ595912 |
| HNWG24 | T. clunifera | Pingdingshan, Henan, China | PQ595913 |
| Dated Crown Group | Calibration Fossil | Bound for MCMCTree | Phylogenetic Justification | Age Justification |
|---|---|---|---|---|
| Crown Chilopoda | Crussolum sp. | B (4.16, 5.28) | [13] | [68] |
| Crown Scutigeromorpha | Fulmenocursor tenax | B (1.12, 4.16) | [69] | [68] |
| Crown Pleurostigmophora | Devonobius delta | B (3.82, 4.16) | [13] | [68] |
| Crown Lithobiomorpha | Henicopidae sp. | B (0.98, 3.82) | [65] | [65] |
| Crown Geophilomorpha | Kachinophilus pereirai | B (0.98, 3.09) | [66,67] | [68] |
| Crown Scolopendromorpha | Cratoraricrus oberlii | B (1.12, 3.09) | [64] | [68] |
| Number | GDSW04 | ZJYY08 | GXJX13 | OL436141 | GXGG22 | HNCM23 | HBSZ18 |
|---|---|---|---|---|---|---|---|
| GDSW04 | |||||||
| ZJYY08 | 0.036 | ||||||
| GXJX13 | 0.032 | 0.033 | |||||
| OL436141 | 0.036 | 0.001 | 0.033 | ||||
| GXGG22 | 0.183 | 0.184 | 0.179 | 0.184 | |||
| HNCM23 | 0.187 | 0.187 | 0.185 | 0.187 | 0.089 | ||
| HBSZ18 | 0.179 | 0.178 | 0.177 | 0.178 | 0.180 | 0.183 | |
| HNWG24 | 0.179 | 0.177 | 0.176 | 0.177 | 0.177 | 0.182 | 0.033 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, J.-H.; Wu, H.-Y.; Gao, Y.-X.; Shen, C.-Y.; Yang, Z.-W.; Storey, K.B.; Yu, D.-N.; Zhang, J.-Y. Unusual Genetic Diversity Within Thereuopoda clunifera (Wood, 1862) (Chilopoda: Scutigeromorpha) Revealed by Phylogeny and Divergence Times Using Mitochondrial Genomes. Insects 2025, 16, 486. https://doi.org/10.3390/insects16050486
Ji J-H, Wu H-Y, Gao Y-X, Shen C-Y, Yang Z-W, Storey KB, Yu D-N, Zhang J-Y. Unusual Genetic Diversity Within Thereuopoda clunifera (Wood, 1862) (Chilopoda: Scutigeromorpha) Revealed by Phylogeny and Divergence Times Using Mitochondrial Genomes. Insects. 2025; 16(5):486. https://doi.org/10.3390/insects16050486
Chicago/Turabian StyleJi, Jie-Hong, Hui-Yuan Wu, Yi-Xin Gao, Chen-Yang Shen, Zi-Wen Yang, Kenneth B. Storey, Dan-Na Yu, and Jia-Yong Zhang. 2025. "Unusual Genetic Diversity Within Thereuopoda clunifera (Wood, 1862) (Chilopoda: Scutigeromorpha) Revealed by Phylogeny and Divergence Times Using Mitochondrial Genomes" Insects 16, no. 5: 486. https://doi.org/10.3390/insects16050486
APA StyleJi, J.-H., Wu, H.-Y., Gao, Y.-X., Shen, C.-Y., Yang, Z.-W., Storey, K. B., Yu, D.-N., & Zhang, J.-Y. (2025). Unusual Genetic Diversity Within Thereuopoda clunifera (Wood, 1862) (Chilopoda: Scutigeromorpha) Revealed by Phylogeny and Divergence Times Using Mitochondrial Genomes. Insects, 16(5), 486. https://doi.org/10.3390/insects16050486







