Host Specificity of Snodgrassella in Eastern and Western Honeybees and Its Effects on Naturally Occurring Deformed Wing Virus Titers
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain Cultivation and DNA Extraction
2.2. Library Construction and Illumina HiSeq Sequencing
2.3. Genome Assembly
2.4. Genome Annotation and Phylogenetic Construction
2.5. Colonization Experiments
2.6. DNA Extraction from the Gut and Bacterial Quantification
2.7. RNA Extraction, Immune Gene Expression, and Virus Titer Measurement
2.8. Statistical Analysis
3. Results
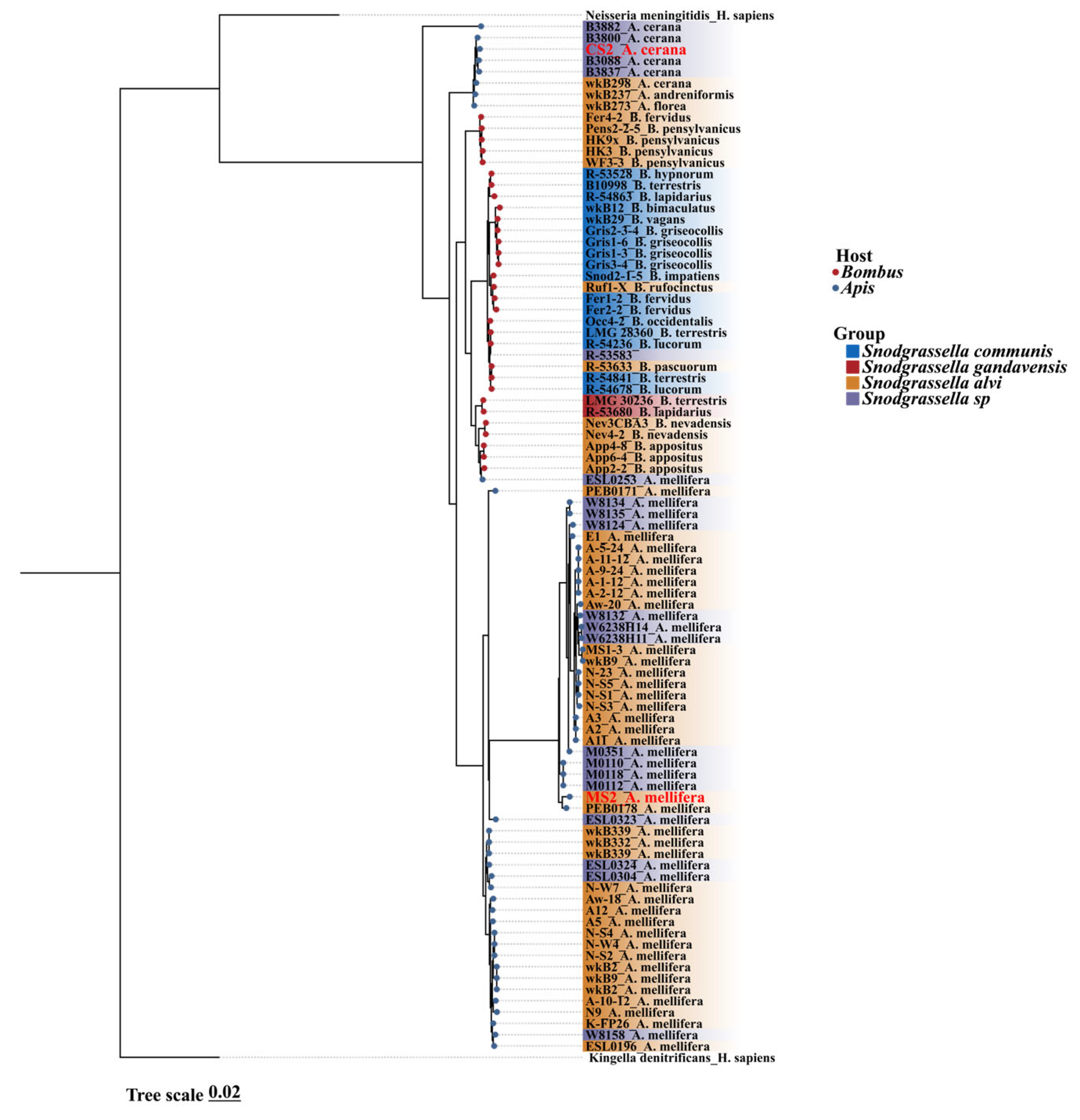
3.1. Snodgrassella Phylogenetic Trees Based on Core Gene Sequences Were Clustered by Host Type
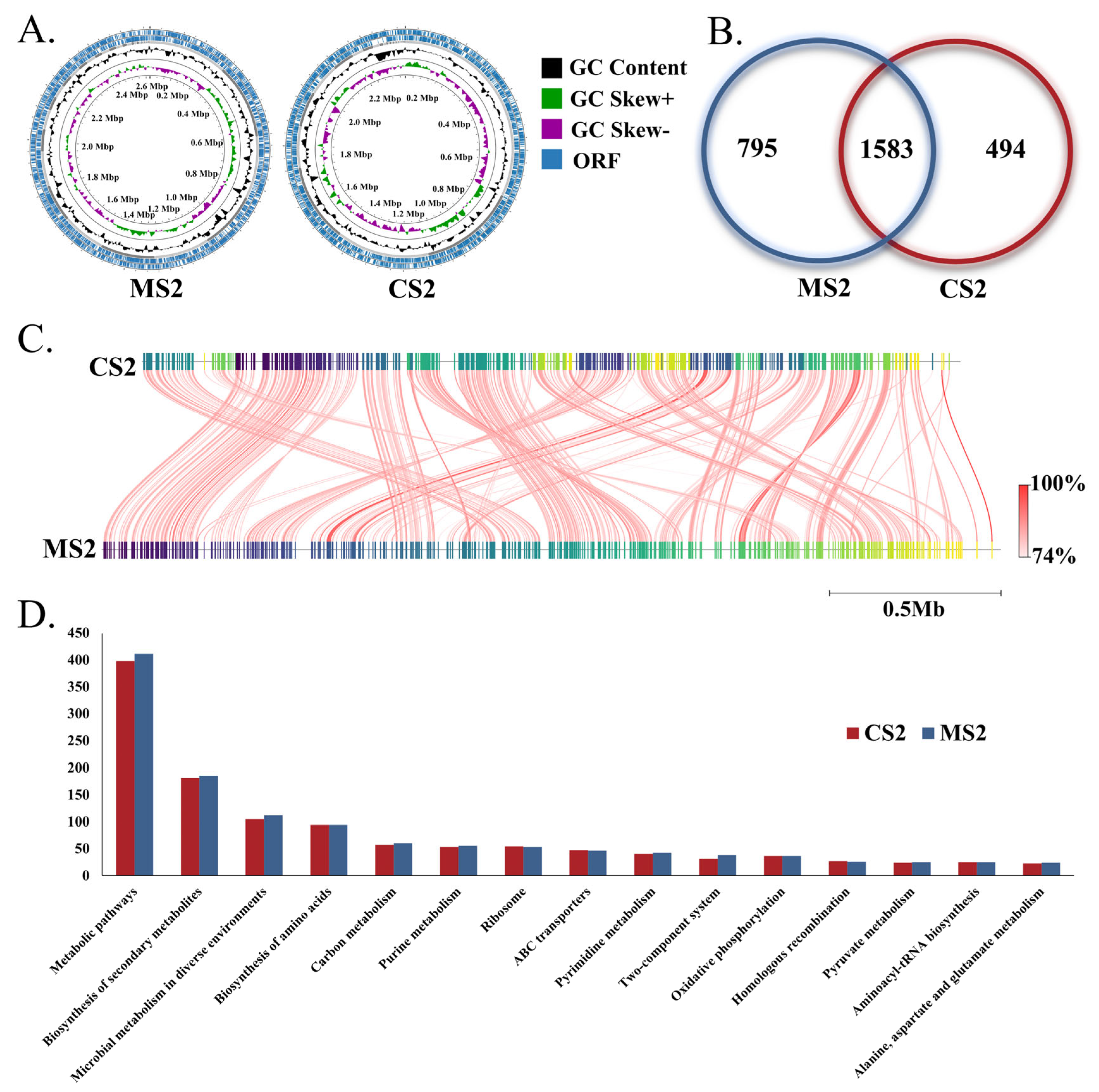
3.2. Genome Characteristics of Snodgrassella
3.3. Gut Colonization of Snodgrassella Strains
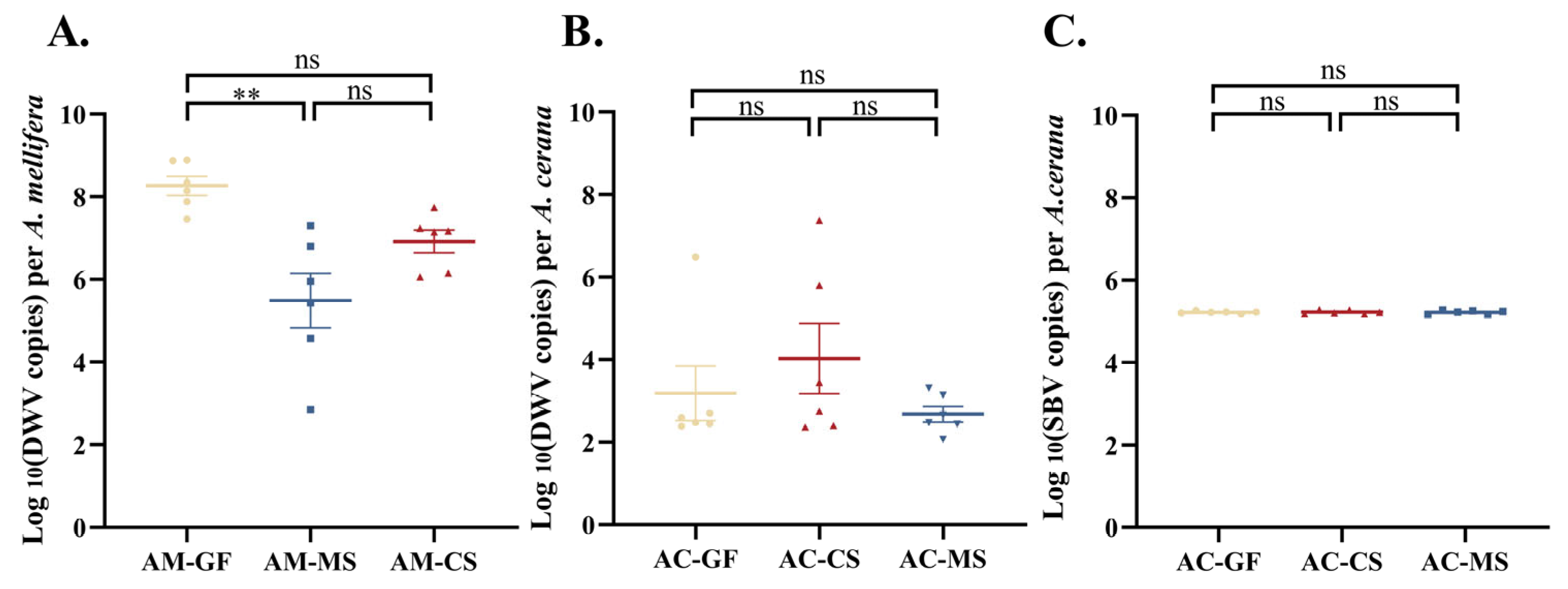
3.4. Colonization of Native Snodgrassella Strain Reduces Naturally Occurring Virus Titers in A. mellifera
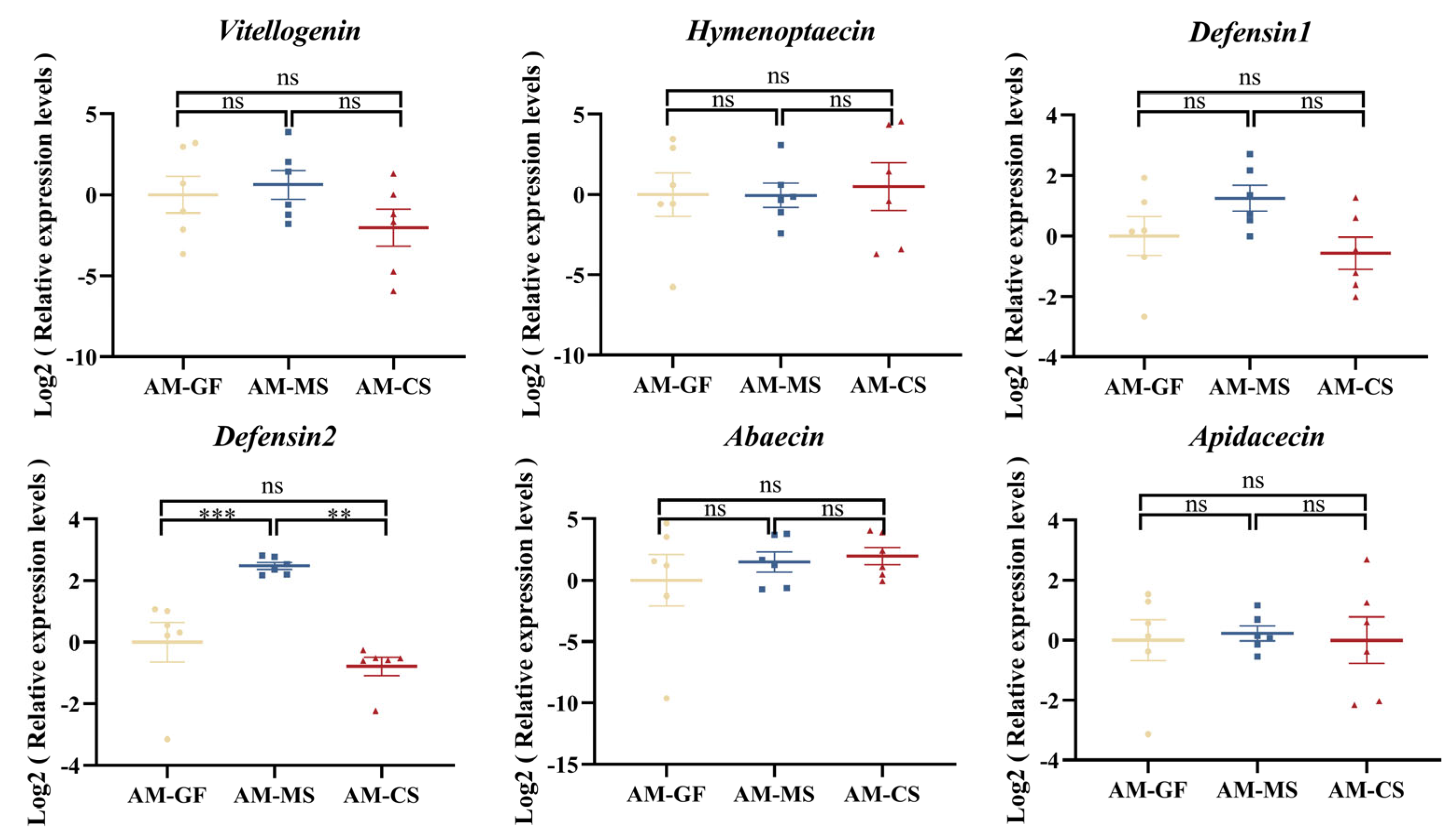
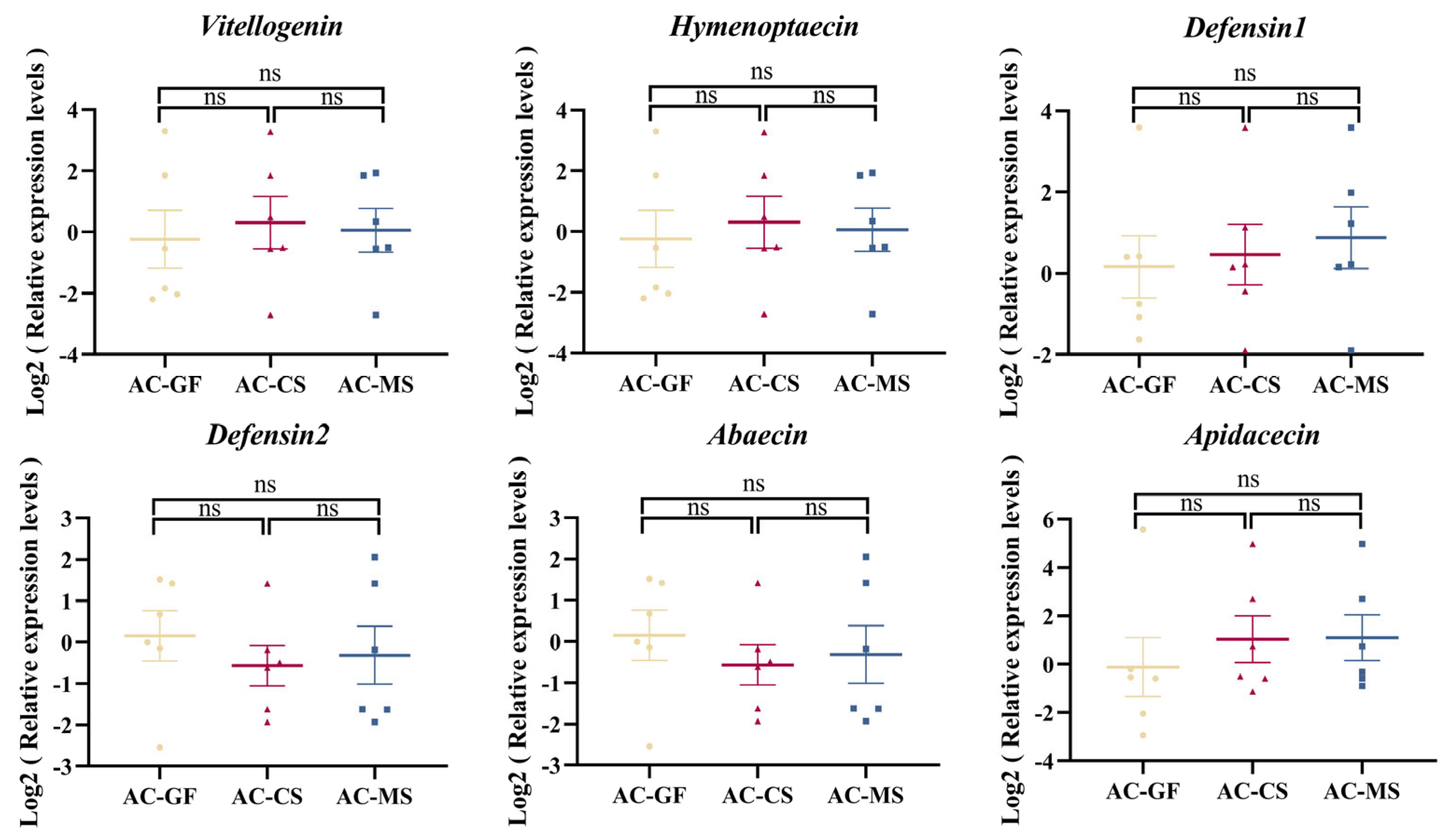
3.5. Colonization of Native Snodgrassella Strain Promotes the Expression of the Defensin 2 Gene in A. mellifera
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bäckhed, F.; Ley, R.E.; Sonnenburg, J.L.; Peterson, D.A.; Gordon, J.I. Host-Bacterial Mutualism in the Human Intestine. Science 2005, 307, 1915–1920. [Google Scholar] [CrossRef]
- Brestoff, J.R.; Artis, D. Commensal Bacteria at the Interface of Host Metabolism and the Immune System. Nat. Immunol. 2013, 14, 676–684. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An Obesity-Associated Gut Microbiome with Increased Capacity for Energy Harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef] [PubMed]
- Sprockett, D.D.; Price, J.D.; Juritsch, A.F.; Schmaltz, R.J.; Real, M.V.F.; Goldman, S.L.; Sheehan, M.; Ramer-Tait, A.E.; Moeller, A.H. Home-Site Advantage for Host Species–Specific Gut Microbiota. Sci. Adv. 2023, 9, eadf5499. [Google Scholar] [CrossRef]
- Moeller, A.H.; Gomes-Neto, J.C.; Mantz, S.; Kittana, H.; Segura Munoz, R.R.; Schmaltz, R.J.; Ramer-Tait, A.E.; Nachman, M.W. Experimental Evidence for Adaptation to Species-Specific Gut Microbiota in House Mice. mSphere 2019, 4, e00387-19. [Google Scholar] [CrossRef] [PubMed]
- Zmora, N.; Zilberman-Schapira, G.; Suez, J.; Mor, U.; Dori-Bachash, M.; Bashiardes, S.; Kotler, E.; Zur, M.; Regev-Lehavi, D.; Brik, R.B.-Z.; et al. Personalized Gut Mucosal Colonization Resistance to Empiric Probiotics Is Associated with Unique Host and Microbiome Features. Cell 2018, 174, 1388–1405.e21. [Google Scholar] [CrossRef]
- Brooks, A.W.; Kohl, K.D.; Brucker, R.M.; van Opstal, E.J.; Bordenstein, S.R. Correction: Phylosymbiosis: Relationships and Functional Effects of Microbial Communities across Host Evolutionary History. PLoS Biol. 2017, 15, e1002587. [Google Scholar] [CrossRef]
- Zheng, H.; Steele, M.I.; Leonard, S.P.; Motta, E.V.S.; Moran, N.A. Honey Bees as Models for Gut Microbiota Research. Lab. Anim. 2018, 47, 317–325. [Google Scholar] [CrossRef]
- Martinson, V.G.; Danforth, B.N.; Minckley, R.L.; Rueppell, O.; Tingek, S.; Moran, N.A. A Simple and Distinctive Microbiota Associated with Honey Bees and Bumble Bees. Mol. Ecol. 2011, 20, 619–628. [Google Scholar] [CrossRef]
- Corby-Harris, V.; Maes, P.; Anderson, K.E. The Bacterial Communities Associated with Honey Bee (Apis mellifera) Foragers. PLoS ONE 2014, 9, e95056. [Google Scholar] [CrossRef]
- Jeyaprakash, A.; Hoy, M.A.; Allsopp, M.H. Bacterial Diversity in Worker Adults of Apis mellifera capensis and Apis mellifera scutellata (Insecta: Hymenoptera) Assessed Using 16S rRNA Sequences. J. Invertebr. Pathol. 2003, 84, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Kwong, W.K.; Engel, P.; Koch, H.; Moran, N.A. Genomics and Host Specialization of Honey Bee and Bumble Bee Gut Symbionts. Proc. Natl. Acad. Sci. USA 2014, 111, 11509–11514. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Leonard, S.P.; Powell, J.E.; Moran, N.A. Species Divergence in Gut-Restricted Bacteria of Social Bees. Proc. Natl. Acad. Sci. USA 2022, 119, e2115013119. [Google Scholar] [CrossRef]
- Zhang, Z.; Guo, Y.; Yang, F.; Li, J. Pan-Genome Analysis Reveals Functional Divergences in Gut-Restricted Gilliamella and Snodgrassella. Bioengineering 2022, 9, 544. [Google Scholar] [CrossRef]
- Zhang, Z.; Mu, X.; Cao, Q.; Shi, Y.; Hu, X.; Zheng, H. Honeybee Gut Lactobacillus Modulates Host Learning and Memory Behaviors via Regulating Tryptophan Metabolism. Nat. Commun. 2022, 13, 2037. [Google Scholar] [CrossRef] [PubMed]
- Liberti, J.; Kay, T.; Quinn, A.; Kesner, L.; Frank, E.T.; Cabirol, A.; Richardson, T.O.; Engel, P.; Keller, L. The Gut Microbiota Affects the Social Network of Honeybees. Nat. Ecol. Evol. 2022, 6, 1471–1479. [Google Scholar] [CrossRef]
- Jones, J.C.; Fruciano, C.; Marchant, J.; Hildebrand, F.; Forslund, S.; Bork, P.; Engel, P.; Hughes, W.O.H. The Gut Microbiome Is Associated with Behavioural Task in Honey Bees. Insect. Soc. 2018, 65, 419–429. [Google Scholar] [CrossRef]
- Sauers, L.A.; Sadd, B.M. An Interaction between Host and Microbe Genotypes Determines Colonization Success of a Key Bumble Bee Gut Microbiota Member. Evolution 2019, 73, 2333–2342. [Google Scholar] [CrossRef]
- Koch, H.; Abrol, D.P.; Li, J.; Schmid-Hempel, P. Diversity and Evolutionary Patterns of Bacterial Gut Associates of Corbiculate Bees. Mol. Ecol. 2013, 22, 2028–2044. [Google Scholar] [CrossRef]
- Kwong, W.K.; Moran, N.A. Evolution of Host Specialization in Gut Microbes: The Bee Gut as a Model. Gut Microbes 2015, 6, 214–220. [Google Scholar] [CrossRef]
- Powell, E.; Ratnayeke, N.; Moran, N.A. Strain Diversity and Host Specificity in a Specialized Gut Symbiont of Honeybees and Bumblebees. Mol. Ecol. 2016, 25, 4461–4471. [Google Scholar] [CrossRef] [PubMed]
- Oh, P.L.; Benson, A.K.; Peterson, D.A.; Patil, P.B.; Moriyama, E.N.; Roos, S.; Walter, J. Diversification of the Gut Symbiont Lactobacillus reuteri as a Result of Host-Driven Evolution. ISME J. 2010, 4, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Amiri, N.; Keady, M.M.; Lim, H.C. Honey Bees and Bumble Bees Occupying the Same Landscape Have Distinct Gut Microbiomes and Amplicon Sequence Variant-Level Responses to Infections. PeerJ 2023, 11, e15501. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Tang, J.; Tang, M.; Luo, S.; Zhou, X. Reactive Oxygen Species Are Regulated by Immune Deficiency and Toll Pathways in Determining the Host Specificity of Honeybee Gut Bacteria. Proc. Natl. Acad. Sci. USA 2023, 120, e2219634120. [Google Scholar] [CrossRef]
- Steele, M.I.; Kwong, W.K.; Whiteley, M.; Moran, N.A. Diversification of Type VI Secretion System Toxins Reveals Ancient Antagonism among Bee Gut Microbes. mBio 2017, 8, e01630-17. [Google Scholar] [CrossRef]
- Steele, M.I.; Moran, N.A. Evolution of Interbacterial Antagonism in Bee Gut Microbiota Reflects Host and Symbiont Diversification. mSystems 2021, 6, e00063-21. [Google Scholar] [CrossRef]
- Motta, E.V.S.; Lariviere, P.J.; Jones, K.R.; Song, Y.; Moran, N.A. Type VI Secretion Systems Promote Intraspecific Competition and Host Interactions in a Bee Gut Symbiont. Proc. Natl. Acad. Sci. USA 2024, 121, e2414882121. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zheng, Y.; Wang, S.; Chen, Y.; Tao, J.; Chen, Y.; Chen, G.; Zhao, H.; Wang, K.; Dong, K.; et al. Genetic Divergence and Functional Convergence of Gut Bacteria between the Eastern Honey Bee Apis cerana and the Western Honey Bee Apis mellifera. J. Adv. Res. 2022, 37, 19–31. [Google Scholar] [CrossRef]
- Ellegaard, K.M.; Suenami, S.; Miyazaki, R.; Engel, P. Vast Differences in Strain-Level Diversity in the Gut Microbiota of Two Closely Related Honey Bee Species. Curr. Biol. 2020, 30, 2520–2531.e7. [Google Scholar] [CrossRef]
- Zhou, N.; Zheng, Q.; Liu, Y.; Huang, Z.; Feng, Y.; Chen, Y.; Hu, F.; Zheng, H. Strain Diversity and Host Specificity of the Gut Symbiont Gilliamella in Apis mellifera, Apis cerana and Bombus terrestris. Microbiol. Res. 2025, 293, 128048. [Google Scholar] [CrossRef]
- Grozinger, C.M.; Flenniken, M.L. Bee Viruses: Ecology, Pathogenicity, and Impacts. Annu. Rev. Entomol. 2019, 64, 205–226. [Google Scholar] [CrossRef] [PubMed]
- Highfield, A.C.; El Nagar, A.; Mackinder, L.C.M.; Noël, L.M.-L.J.; Hall, M.J.; Martin, S.J.; Schroeder, D.C. Deformed Wing Virus Implicated in Overwintering Honeybee Colony Losses. Appl. Environ. Microbiol. 2009, 75, 7212–7220. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.; Cao, L.; Feng, Y.; Chen, Y.; Chen, G.; Zheng, H. Sacbrood Virus: A Growing Threat to Honeybees and Wild Pollinators. Viruses 2022, 14, 1871. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Wu, Y.; Deng, J.; Wen, Z.; Wang, S.; Chen, Y.; Hu, F.; Zheng, H. Seasonal Variation of Viral Infections between the Eastern Honey Bee (Apis cerana) and the Western Honey Bee (Apis mellifera). Microbiol. Open 2021, 10, e1162. [Google Scholar] [CrossRef]
- Evans, J.D.; Aronstein, K.; Chen, Y.P.; Hetru, C.; Imler, J.-L.; Jiang, H.; Kanost, M.; Thompson, G.J.; Zou, Z.; Hultmark, D. Immune Pathways and Defence Mechanisms in Honey Bees Apis mellifera. Insect Mol. Biol. 2006, 15, 645–656. [Google Scholar] [CrossRef]
- Chen, Y.P.; Pettis, J.S.; Corona, M.; Chen, W.P.; Li, C.J.; Spivak, M.; Visscher, P.K.; DeGrandi-Hoffman, G.; Boncristiani, H.; Zhao, Y.; et al. Israeli Acute Paralysis Virus: Epidemiology, Pathogenesis and Implications for Honey Bee Health. PLoS Pathog. 2014, 10, e1004261. [Google Scholar] [CrossRef]
- Mookhploy, W.; Krongdang, S.; Chantawannakul, P. Effects of Deformed Wing Virus Infection on Expressions of Immune- and Apoptosis-Related Genes in Western Honeybees (Apis mellifera). Insects 2021, 12, 82. [Google Scholar] [CrossRef]
- Wu, Y.; Zheng, Y.; Chen, Y.; Chen, G.; Zheng, H.; Hu, F. Apis cerana Gut Microbiota Contribute to Host Health Though Stimulating Host Immune System and Strengthening Host Resistance to Nosema ceranae. R. Soc. Open Sci. 2020, 7, 192100. [Google Scholar] [CrossRef]
- Leonard, S.P.; Powell, J.E.; Perutka, J.; Geng, P.; Heckmann, L.C.; Horak, R.D.; Davies, B.W.; Ellington, A.D.; Barrick, J.E.; Moran, N.A. Engineered Symbionts Activate Honey Bee Immunity and Limit Pathogens. Science 2020, 367, 573–576. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Simpson, J.T.; Wong, K.; Jackman, S.D.; Schein, J.E.; Jones, S.J.M.; Birol, I. ABySS: A Parallel Assembler for Short Read Sequence Data. Genome Res. 2009, 19, 1117–1123. [Google Scholar] [CrossRef]
- Xu, M.; Guo, L.; Gu, S.; Wang, O.; Zhang, R.; Peters, B.A.; Fan, G.; Liu, X.; Xu, X.; Deng, L.; et al. TGS-GapCloser: A Fast and Accurate Gap Closer for Large Genomes with Low Coverage of Error-Prone Long Reads. GigaScience 2020, 9, giaa094. [Google Scholar] [CrossRef] [PubMed]
- Schwengers, O.; Jelonek, L.; Dieckmann, M.A.; Beyvers, S.; Blom, J.; Goesmann, A. Bakta: Rapid and Standardized Annotation of Bacterial Genomes via Alignment-Free Sequence Identification. Microb. Genom. 2021, 7, 685. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A Program for Improved Detection of Transfer RNA Genes in Genomic Sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Stærfeldt, H.-H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and Rapid Annotation of Ribosomal RNA Genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef]
- Brown, C.L.; Mullet, J.; Hindi, F.; Stoll, J.E.; Gupta, S.; Choi, M.; Keenum, I.; Vikesland, P.; Pruden, A.; Zhang, L. mobileOG-Db: A Manually Curated Database of Protein Families Mediating the Life Cycle of Bacterial Mobile Genetic Elements. Appl. Environ. Microbiol. 2022, 88, e00991-22. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic Orthology Inference for Comparative Genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Sato, Y.; Furumichi, M.; Tanabe, M. KEGG for Integration and Interpretation of Large-Scale Molecular Data Sets. Nucleic Acids Res. 2012, 40, D109–D114. [Google Scholar] [CrossRef]
- Tian, R.; Imanian, B. VBCG: 20 Validated Bacterial Core Genes for Phylogenomic Analysis with High Fidelity and Resolution. Microbiome 2023, 11, 247. [Google Scholar] [CrossRef]
- Blanchard, P.; Guillot, S.; Antùnez, K.; Köglberger, H.; Kryger, P.; de Miranda, J.R.; Franco, S.; Chauzat, M.-P.; Thiéry, R.; Ribière, M. Development and Validation of a Real-Time Two-Step RT-qPCR TaqMan® Assay for Quantitation of Sacbrood Virus (SBV) and Its Application to a Field Survey of Symptomatic Honey Bee Colonies. J. Virol. Methods 2014, 197, 7–13. [Google Scholar] [CrossRef]
- Xie, J.; Chen, Y.; Cai, G.; Cai, R.; Hu, Z.; Wang, H. Tree Visualization By One Table (tvBOT): A Web Application for Visualizing, Modifying and Annotating Phylogenetic Trees. Nucleic Acids Res. 2023, 51, W587–W592. [Google Scholar] [CrossRef] [PubMed]
- Nannan, L.; Huamiao, L.; Yan, J.; Xingan, L.; Yang, L.; Tianjiao, W.; Jinming, H.; Qingsheng, N.; Xiumei, X. Geometric Morphology and Population Genomics Provide Insights into the Adaptive Evolution of Apis cerana in Changbai Mountain. BMC Genom. 2022, 23, 64. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Wei, R.; Chen, Q.; Shang, T.; Zhou, N.; Wang, Z.; Chen, Y.; Chen, G.; Zhang, G.; Dong, K.; et al. Host Specificity and Cophylogeny in the “Animal-Gut Bacteria-Phage” Tripartite System. npj Biofilms Microbiomes 2024, 10, 72. [Google Scholar] [CrossRef]
- Kwong, W.K.; Medina, L.A.; Koch, H.; Sing, K.-W.; Soh, E.J.Y.; Ascher, J.S.; Jaffé, R.; Moran, N.A. Dynamic Microbiome Evolution in Social Bees. Sci. Adv. 2017, 3, e1600513. [Google Scholar] [CrossRef] [PubMed]
- Raymann, K.; Moran, N.A. The Role of the Gut Microbiome in Health and Disease of Adult Honey Bee Workers. Curr. Opin. Insect Sci. 2018, 26, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Dosch, C.; Manigk, A.; Streicher, T.; Tehel, A.; Paxton, R.J.; Tragust, S. The Gut Microbiota Can Provide Viral Tolerance in the Honey Bee. Microorganisms 2021, 9, 871. [Google Scholar] [CrossRef]
- Lang, H.; Wang, H.; Wang, H.; Zhong, Z.; Xie, X.; Zhang, W.; Guo, J.; Meng, L.; Hu, X.; Zhang, X.; et al. Engineered Symbiotic Bacteria Interfering Nosema Redox System Inhibit Microsporidia Parasitism in Honeybees. Nat. Commun. 2023, 14, 2778. [Google Scholar] [CrossRef]
- Miller, D.L.; Smith, E.A.; Newton, I.L.G. A Bacterial Symbiont Protects Honey Bees from Fungal Disease. Mbio 2021, 12, e00503-21. [Google Scholar] [CrossRef]
- Deng, Y.; Yang, S.; Zhao, H.; Diao, Q.; Hou, C. IAPV-Induced Paralytic Symptoms Associated with Tachypnea via Impaired Tracheal System Function. Int. J. Mol. Sci. 2021, 22, 10078. [Google Scholar] [CrossRef]
- Liu, Y.; Jia, S.; Wu, Y.; Zhou, N.; Xie, Y.; Wei, R.; Huang, Z.; Chen, Y.; Hu, F.; Zheng, H. Tetracycline-Induced Gut Community Dysbiosis and Israeli Acute Paralysis Virus Infection Synergistically Negatively Affect Honeybees. Ecotoxicol. Environ. Saf. 2024, 282, 116706. [Google Scholar] [CrossRef]
- Deng, Y.; Yang, S.; Zhang, L.; Chen, C.; Cheng, X.; Hou, C. Chronic Bee Paralysis Virus Exploits Host Antimicrobial Peptides and Alters Gut Microbiota Composition to Facilitate Viral Infection. ISME J. 2024, 18, wrae051. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Lariviere, P.J.; Powell, J.E.; Moran, N.A. Engineered Gut Symbiont Inhibits Microsporidian Parasite and Improves Honey Bee Survival. Proc. Natl. Acad. Sci. USA 2023, 120, e2220922120. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Chen, G.; Lin, Z.; Wu, Y.; Hu, F.; Zheng, H. Occurrence of Multiple Honeybee Viruses in the Ectoparasitic Mites Varroa Spp. in Apis cerana Colonies. J. Invertebr. Pathol. 2019, 166, 107225. [Google Scholar] [CrossRef] [PubMed]
- McMahon, D.P.; Natsopoulou, M.E.; Doublet, V.; Fürst, M.; Weging, S.; Brown, M.J.F.; Gogol-Döring, A.; Paxton, R.J. Elevated Virulence of an Emerging Viral Genotype as a Driver of Honeybee Loss. Proc. R. Soc. B Biol. Sci. 2016, 283, 20160811. [Google Scholar] [CrossRef]
- Hasegawa, N.; Techer, M.A.; Adjlane, N.; al-Hissnawi, M.S.; Antúnez, K.; Beaurepaire, A.; Christmon, K.; Delatte, H.; Dukku, U.H.; Eliash, N.; et al. Evolutionarily Diverse Origins of Deformed Wing Viruses in Western Honey Bees. Proc. Natl. Acad. Sci. USA 2023, 120, e2301258120. [Google Scholar] [CrossRef]
- Li, J.H.; Evans, J.D.; Li, W.F.; Zhao, Y.Z.; DeGrandi-Hoffman, G.; Huang, S.K.; Li, Z.G.; Hamilton, M.; Chen, Y.P. Lactobacillus Spp. Attenuate Antibiotic-Induced Immune and Microbiota Dysregulation in Honey Bees. PLoS ONE 2017, 12, e0187505. [Google Scholar] [CrossRef]
- Iorizzo, M.; Lombardi, S.J.; Ganassi, S.; Testa, B.; Ianiro, M.; Letizia, F.; Succi, M.; Tremonte, P.; Vergalito, F.; Cozzolino, A.; et al. Antagonistic Activity against Ascosphaera apis and Functional Properties of Lactobacillus Kunkeei Strains. Antibiotics 2020, 9, 262. [Google Scholar] [CrossRef]
- Iorizzo, M.; Testa, B.; Ganassi, S.; Lombardi, S.J.; Ianiro, M.; Letizia, F.; Succi, M.; Tremonte, P.; Vergalito, F.; Cozzolino, A.; et al. Probiotic Properties and Potentiality of Lactiplantibacillus plantarum Strains for the Biological Control of Chalkbrood Disease. J. Fungi 2021, 7, 379. [Google Scholar] [CrossRef]
- Sattayawat, P.; Inwongwan, S.; Noirungsee, N.; Li, J.; Guo, J.; Disayathanoowat, T. Engineering Gut Symbionts: A Way to Promote Bee Growth? Insects 2024, 15, 369. [Google Scholar] [CrossRef]
- Wu, Y.; Zheng, Y.; Chen, Y.; Wang, S.; Chen, Y.; Hu, F.; Zheng, H. Honey Bee (Apis mellifera) Gut Microbiota Promotes Host Endogenous Detoxification Capability via Regulation of P450 Gene Expression in the Digestive Tract. Microb. Biotechnol. 2020, 13, 1201–1212. [Google Scholar] [CrossRef]
- Motta, E.V.S.; Moran, N.A. The Honeybee Microbiota and Its Impact on Health and Disease. Nat. Rev. Microbiol. 2024, 22, 122–137. [Google Scholar] [CrossRef] [PubMed]
- Kwong, W.K.; Mancenido, A.L.; Moran, N.A. Immune System Stimulation by the Native Gut Microbiota of Honey Bees. R. Soc. Open Sci. 2017, 4, 170003. [Google Scholar] [CrossRef] [PubMed]
- Emery, O.; Schmidt, K.; Engel, P. Immune System Stimulation by the Gut Symbiont Frischella perrara in the Honey Bee (Apis mellifera). Mol. Ecol. 2017, 26, 2576–2590. [Google Scholar] [CrossRef] [PubMed]
- Horak, R.D.; Leonard, S.P.; Moran, N.A. Symbionts Shape Host Innate Immunity in Honeybees. Proc. R. Soc. B 2020, 287, 20201184. [Google Scholar] [CrossRef] [PubMed]
- Lang, H.; Duan, H.; Wang, J.; Zhang, W.; Guo, J.; Zhang, X.; Hu, X.; Zheng, H. Specific Strains of Honeybee Gut Lactobacillus Stimulate Host Immune System to Protect against Pathogenic Hafnia alvei. Microbiol. Spectrum. 2022, 10, e01896-21. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, N.; Yang, S.; Wei, R.; Hu, F.; Liu, D.; Zheng, H. Host Specificity of Snodgrassella in Eastern and Western Honeybees and Its Effects on Naturally Occurring Deformed Wing Virus Titers. Insects 2025, 16, 478. https://doi.org/10.3390/insects16050478
Zhou N, Yang S, Wei R, Hu F, Liu D, Zheng H. Host Specificity of Snodgrassella in Eastern and Western Honeybees and Its Effects on Naturally Occurring Deformed Wing Virus Titers. Insects. 2025; 16(5):478. https://doi.org/10.3390/insects16050478
Chicago/Turabian StyleZhou, Nihong, Shangning Yang, Ruike Wei, Fuliang Hu, Dandan Liu, and Huoqing Zheng. 2025. "Host Specificity of Snodgrassella in Eastern and Western Honeybees and Its Effects on Naturally Occurring Deformed Wing Virus Titers" Insects 16, no. 5: 478. https://doi.org/10.3390/insects16050478
APA StyleZhou, N., Yang, S., Wei, R., Hu, F., Liu, D., & Zheng, H. (2025). Host Specificity of Snodgrassella in Eastern and Western Honeybees and Its Effects on Naturally Occurring Deformed Wing Virus Titers. Insects, 16(5), 478. https://doi.org/10.3390/insects16050478







