The First Cavernicolous Species of Arrhopalites (Collembola, Symphypleona, Arrhopalitidae) from China and Its Phylogenetic Position †
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples Collection
2.2. Taxonomic Identification and Description
2.3. DNA Isolation and Sequencing
2.4. Mitogenomes Assemblies and Annotations
2.5. Matrix Generation and Phylogenetic Analyses
3. Results
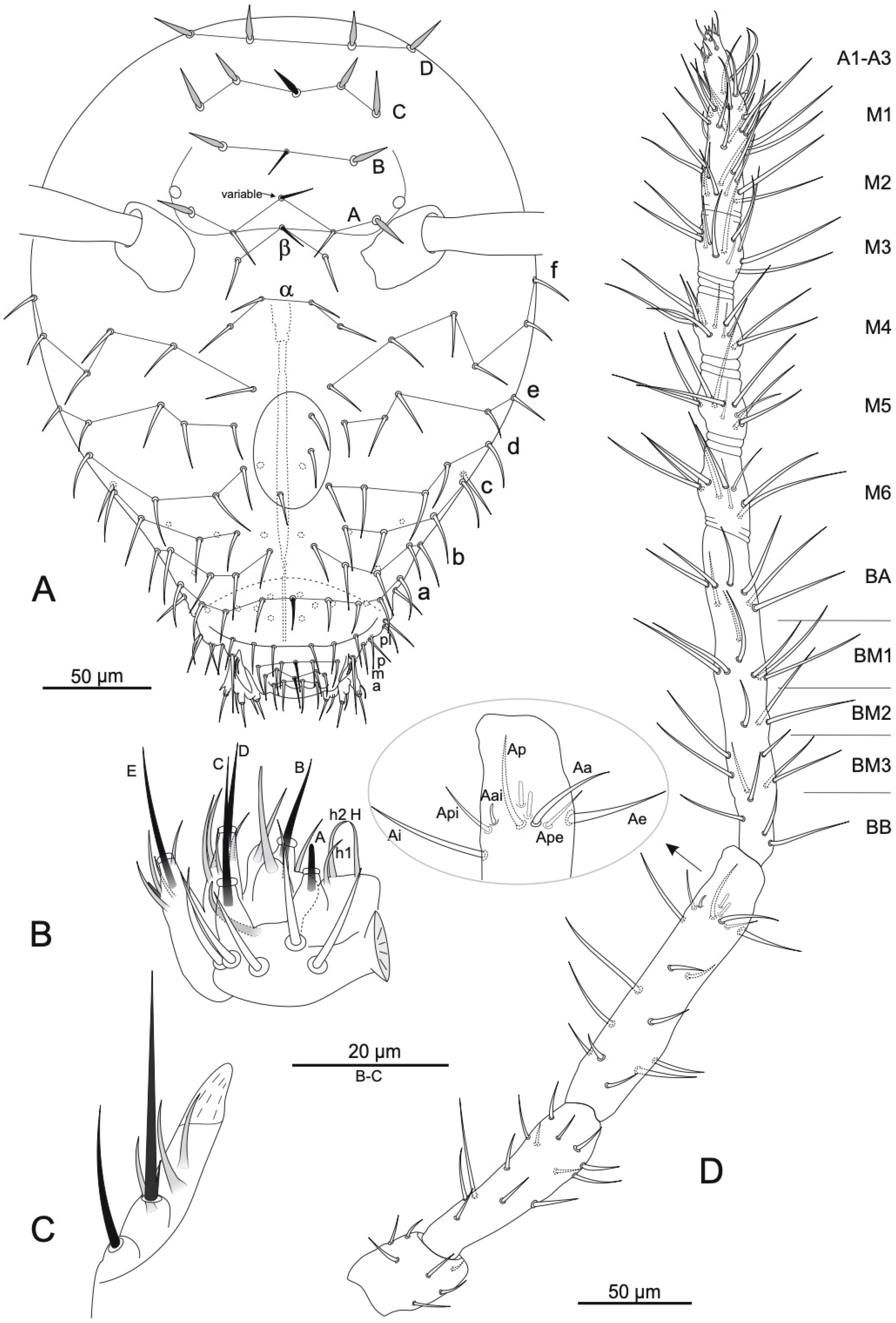
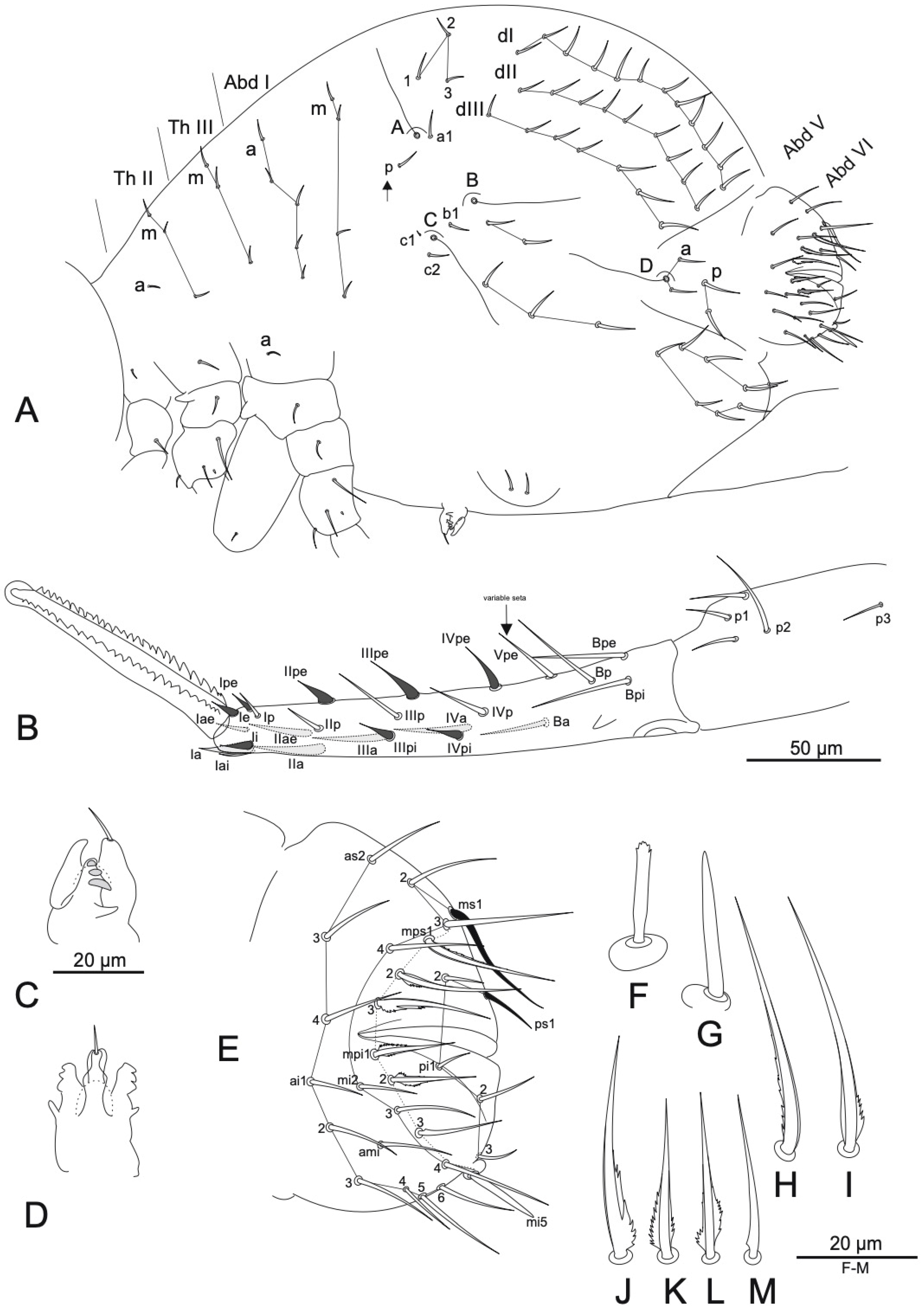
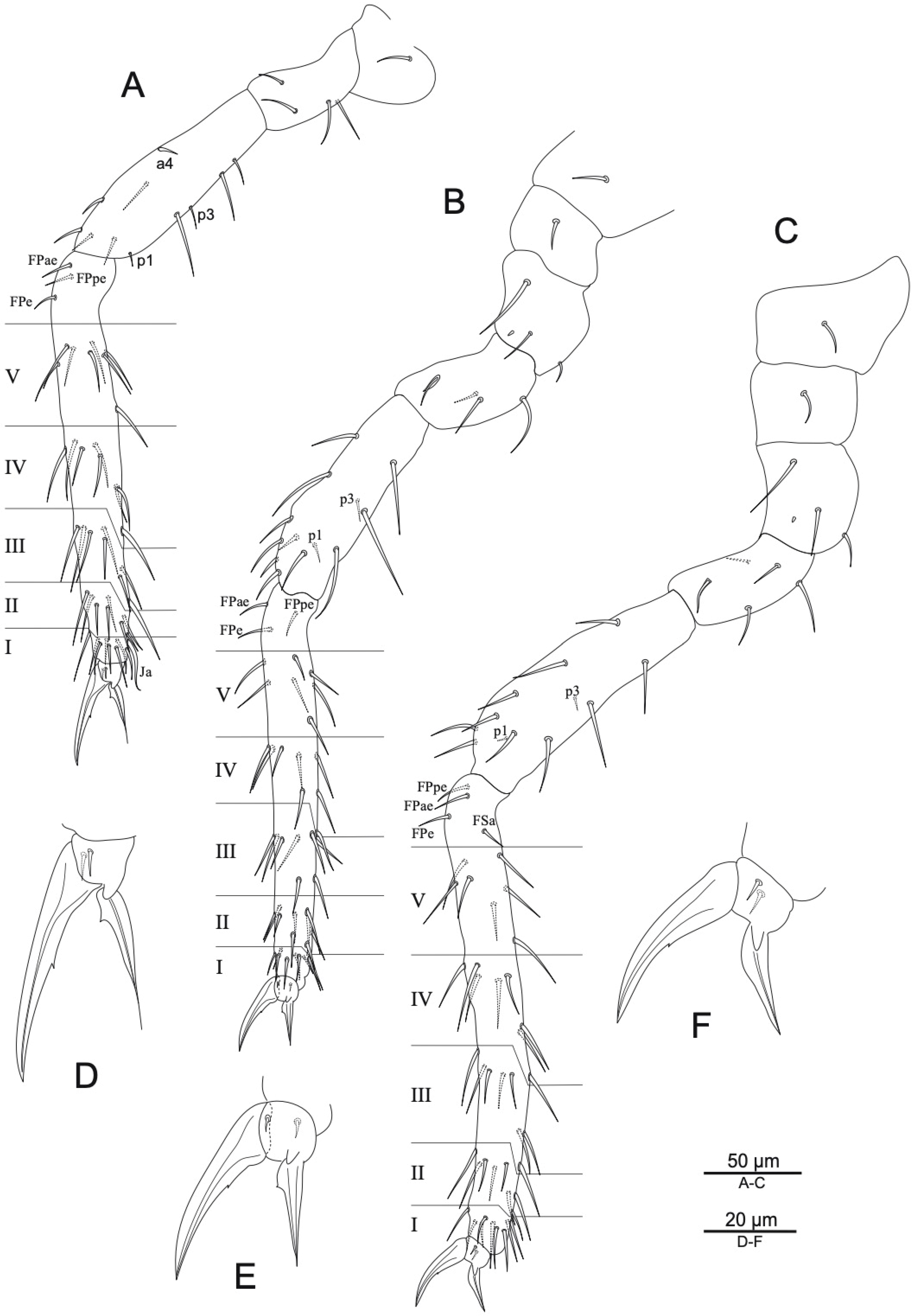
3.1. Taxonomy
3.2. Mitochondrial Genomes
3.3. Phylogenetic Placement of the New Mitogenomes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ye, M.; Huang, C.; Yu, D.; Sun, X.; Godeiro, N.N.; Jiang, J.; Li, Z.; Luan, Y.; Wu, D.; Zhang, F. The first checklist of the Collembola of China in the 21st century. Zool. Syst. 2025, 50, 1–94. [Google Scholar] [CrossRef]
- Börner, C. Das System der Collembolen nebst Beschreibung neuer Collembolen des Hamburger Naturhistorischen Museums. Mitteilungen Naturhistorischen Mus. Hambg. 1906, 23, 147–188. [Google Scholar]
- Godeiro, N.N.; Zhang, F.; Bellini, B.C. A new species of Arrhopalites Börner (Collembola, Symphypleona, Arrhopalitidae) from China, with a key to the Asian species of the caecus group. ZooKeys 2022, 1102, 163–176. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Christiansen, K. A new species of Arrhopalites from China (Collembola: Sminthuridae). Fla. Entomolo. 1997, 80, 266–269. [Google Scholar] [CrossRef]
- D’Haese, C.A. Were the first springtails semi-aquatic? A phylogenetic approach by means of 28S rDNA and optimization alignment. Proc. R. Soc. London 2002, 269, 1143–1151. [Google Scholar] [CrossRef]
- Bellini, B.C.; Zhang, F.; de Souza, P.G.C.; Santos-Costa, R.C.; Medeiros, G.S.; Godeiro, N.N. The evolution of Collembola higher taxa (Arthropoda, Hexapoda) based on mitogenome data. Diversity 2023, 15, 7. [Google Scholar] [CrossRef]
- Yu, D.; Du, S.; Wei, X.; Zhu, J.; Ding, Y.; Hu, F. Whole-genome-based phylogenetic analyses provide new insights into the evolution of springtails (Hexapoda: Collembola). Mol. Phyl. Evol. 2024, 200, 108169. [Google Scholar] [CrossRef]
- Vargovitsh, R.S. Cavernicolous Arrhopalites abchasicus sp. nov. (Collembola: Symphypleona: Arrhopalitidae) from the West Caucasus with a key to the World species of the genus. Zootaxa 2013, 3666, 16–30. [Google Scholar] [CrossRef]
- Christiansen, K.; Bellinger, P. The Collembola of North America, North of the Rio Grande: A Taxonomic Analysis; Grinnell College: Grinnell, IA, USA, 1980; p. 1520. [Google Scholar]
- Wu, M.; Chen, J.X. A new species of the subgenus Papirioides from China (Collembola: Dicyrtomidae). Entomolo. Sin. 1996, 3, 138–144. [Google Scholar] [CrossRef]
- Li, Z.H.; Chen, J.X.; Li, J. A new species of the genus Papirioides (Collembola: Dicyrtomidae) from China. Zootaxa 2007, 1499, 61–68. [Google Scholar] [CrossRef]
- Bellinger, P.F.; Christiansen, K.A.; Janssens, F. (1996–2025) Checklist of the Collembola of the World. Available online: http://www.collembola.org (accessed on 18 January 2025).
- Betsch, J.M.; Waller, A. Chaetotaxic nomenclature of the head, thorax and abdomen in Symphypleona (Insecta, Collembola). Acta Zool. Fennica 1994, 195, 5–12. [Google Scholar]
- Fjellberg, A. The labial palp in Collembola. Zoolo. Anz. 1999, 237, 309–330. [Google Scholar]
- Vargovitsh, R.S. New cave Arrhopalitidae (Collembola: Symphypleona) from the Crimea (Ukraine). Zootaxa 2009, 2047, 1–47. [Google Scholar] [CrossRef]
- Vargovitsh, R.S. New troglomorphic Arrhopalitidae (Collembola: Symphypleona) from the Western Caucasus. Zootaxa 2012, 3174, 1–21. [Google Scholar] [CrossRef]
- Bretfeld, G. The chaetotaxy of the small abdomen of the Symphypleona (Insecta, Collembola) and its phylogenetic interpretation. Acta Zool. Fennica 1994, 195, 13–17. [Google Scholar]
- Betsch, J.M. An ontogenetically focused chaetotaxical scheme in Symphypleona (Collembola): The 6th abdominal segment. Pedobiologia 1997, 41, 13–18. [Google Scholar] [CrossRef]
- Nayrolles, P. Chétotaxie tibiotarsale des Collemboles Symphypléones. Trav. Lab. D’ecobiologie Arthropodes Edaphiques Toulouse 1988, 5, 1–19. [Google Scholar]
- Nayrolles, P. Chétotaxie furcale des Collemboles Symphypléones. Trav. Lab. D’ecobiologie Arthropodes Edaphiques Toulouse 1990, 6, 27–50. [Google Scholar]
- Nayrolles, P. La chétotaxie antennaire des Collemboles Symphypléones. Trav. Lab. D’ecobiologie Arthropodes Edaphiques Toulouse 1991, 6, 1–94. [Google Scholar]
- Meng, G.; Li, Y.; Yang, C.; Liu, S. MitoZ: A toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic. Acids Res. 2019, 47, e63. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, T.J.; Eddy, S.R. nhmmer: DNA homology search with profile HMMs. Bioinformatics 2013, 29, 2487–2489. [Google Scholar] [CrossRef]
- Gertz, E.M.; Yu, Y.K.; Agarwala, R.; Schäffer, A.A.; Altschul, S.F. Composition-based statistics and translated nucleotide searches: Improving the TBLASTN module of BLAST. BMC Biol. 2006, 4, 41. [Google Scholar] [CrossRef] [PubMed]
- Birney, E.; Clamp, M.; Durbin, R. GeneWise and Genomewise. Genome Res. 2004, 14, 988–995. [Google Scholar] [CrossRef] [PubMed]
- Nawrocki, E.P.; Eddy, S.R. Infernal 1.1:100-fold faster RNA homology searches. Bioinformatics 2013, 29, 2933–2935. [Google Scholar] [CrossRef]
- Juhling, F.; Putz, J.; Bernt, M.; Donath, A.; Middendorf, M.; Florentz, C.; Stadler, P.F. Improved systematic tRNA gene annotation allows new insights into the evolution of mitochondrial tRNA structures and into the mechanisms of mitochondrial genome rearrangements. Nucleic. Acids Res. 2012, 40, 2833–2845. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup.The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Serra, F.; Bork, P. ETE 3: Reconstruction, analysis and visualization of phylogenomic data. Mol. Biol. Evo. 2016, 33, 1635–1638. [Google Scholar] [CrossRef]
- Wu, J.; Chen, K. Characterization of the complete mitochondrial genome of Homidia socia (Collembola: Entomobryidae). Mitochondrial DNA Part B 2019, 4, 3941–3942. [Google Scholar] [CrossRef]
- Jiang, L.; Dong, J.; Jin, J.; Zhang, F. Mitochondrial genome of Brachystomella parvula (Collembola: Brachystomellidae). Mitochondrial DNA Part B 2019, 5, 104–105. [Google Scholar] [CrossRef]
- Nardi, F.; Cucini, C.; Leo, C.; Frati, F.; Fanciulli, P.P.; Carapelli, A. The complete mitochondrial genome of the springtail Allacma fusca, the internal phylogenetic relationships and gene order of Symphypleona. Mitochondrial DNA B Resour. 2020, 5, 3103 3105. [Google Scholar] [CrossRef]
- Collins, G.; Schneider, C.; Boštjančić, L.L.; Burkhardt, U.; Christian, A.; Decker, P.; Ebersberger, I.; Hohberg, K.; Lecompte, O.; Merges, D.; et al. The MetaInvert soil invertebrate genome resource provides insights into below-ground biodiversity and evolution. Commun. Biol 2023, 6, 1241. [Google Scholar] [CrossRef] [PubMed]
- Leo, C.; Nardi, F.; Frati, F.; Fanciulli, P.P.; Cucini, C.; Vitale, M.; Brunetti, C.; Carapelli, A. The mitochondrial genome of the springtail Bourletiella arvalis (Symphypleona, Collembola). Mitochondrial DNA Part B 2019, 4, 2978–2979. [Google Scholar] [CrossRef] [PubMed]
- Leo, C.; Carapelli, A.; Cicconardi, F.; Frati, F.; Nardi, F. Mitochondrial Genome Diversity in Collembola: Phylogeny, Dating and Gene Order. Diversity 2019, 11, 169. [Google Scholar] [CrossRef]
- Sun, X.; Yu, D.; Xie, Z.; Dong, J.; Ding, Y.; Yao, H.; Greenslade, P. Phylomitogenomic analyses on collembolan higher taxa with enhanced taxon sampling and discussion on method selection. PLoS ONE 2020, 15, e0230827. [Google Scholar] [CrossRef]
- Schneider, C.; Woehle, C.; Greve, C.; Haese, A.D.; Wolf, M.; Hiller, M.; Janke, A.; Balint, M.; Huettel, B. Two high-quality de novo genomes from single ethanol-preserved specimens of tiny metazoans (Collembola). GigaScience 2021, 10, giab035. [Google Scholar] [CrossRef] [PubMed]
- Carapelli, A.; Lio, P.; Nardi, F.; VanDerWath, E.; Frati, F. Phylogenetic analysis of mitochondrial protein coding genes confirms the reciprocal paraphyly of Hexapoda and Crustacea. BMC Evol. Biol. 2007, 7, S8. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Kück, P.; Longo, G.C. FASconCAT-G: Extensive functions for multiple sequence alignment preparations concerning phylogenetic studies. Front. Zool. 2014, 11, 81. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evo. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evo. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Lartillot, N.; Rodrigues, N.; Stubbs, D.; Richer, J. PhyloBayes MPI: Phylogenetic reconstruction with Infinite mixtures of profiles in a parallel environment. Syst. Biol. 2013, 62, 611–615. [Google Scholar] [CrossRef]
- Kottek, M.; Grieser, J.; Beck, C.; Rudolf, B.; Rubel, F. World Map of the Köppen–Geiger climate classification updated. Meteorol. Z. 2006, 15, 259–263. [Google Scholar] [CrossRef]
- Sánchez-García, A.; Engel, M.S. Long-term stasis in a diverse fauna of Early Cretaceous springtails (Collembola: Symphypleona). J. Syst. Palaeontol. 2016, 15, 513–537. [Google Scholar] [CrossRef]
- Bretfeld, G. Phylogenetic systematics of the higher taxa of Symphypleona Börner, 1901 (Insecta, Entognatha, Collembola). In Proceedings of 2nd International Seminary of Apterygota; University of Siena: Siena, Italy, 1986; Volume 1, pp. 302–311. [Google Scholar]
- Vargovitsh, R.S. Deep troglomorphy: New Arrhopalitidae (Collembola: Symphypleona) of different life forms from the Snezhnaya Cave System in the Caucasus. Diversity 2022, 14, 678. [Google Scholar] [CrossRef]
- Cucini, C.; Fanciulli, P.P.; Frati, F.; Convey, P.; Nardi, F.; Carapelli, A. Re-evaluating the internal phylogenetic relationships of collembola by means of mitogenome data. Genes 2021, 12, 44. [Google Scholar] [CrossRef]

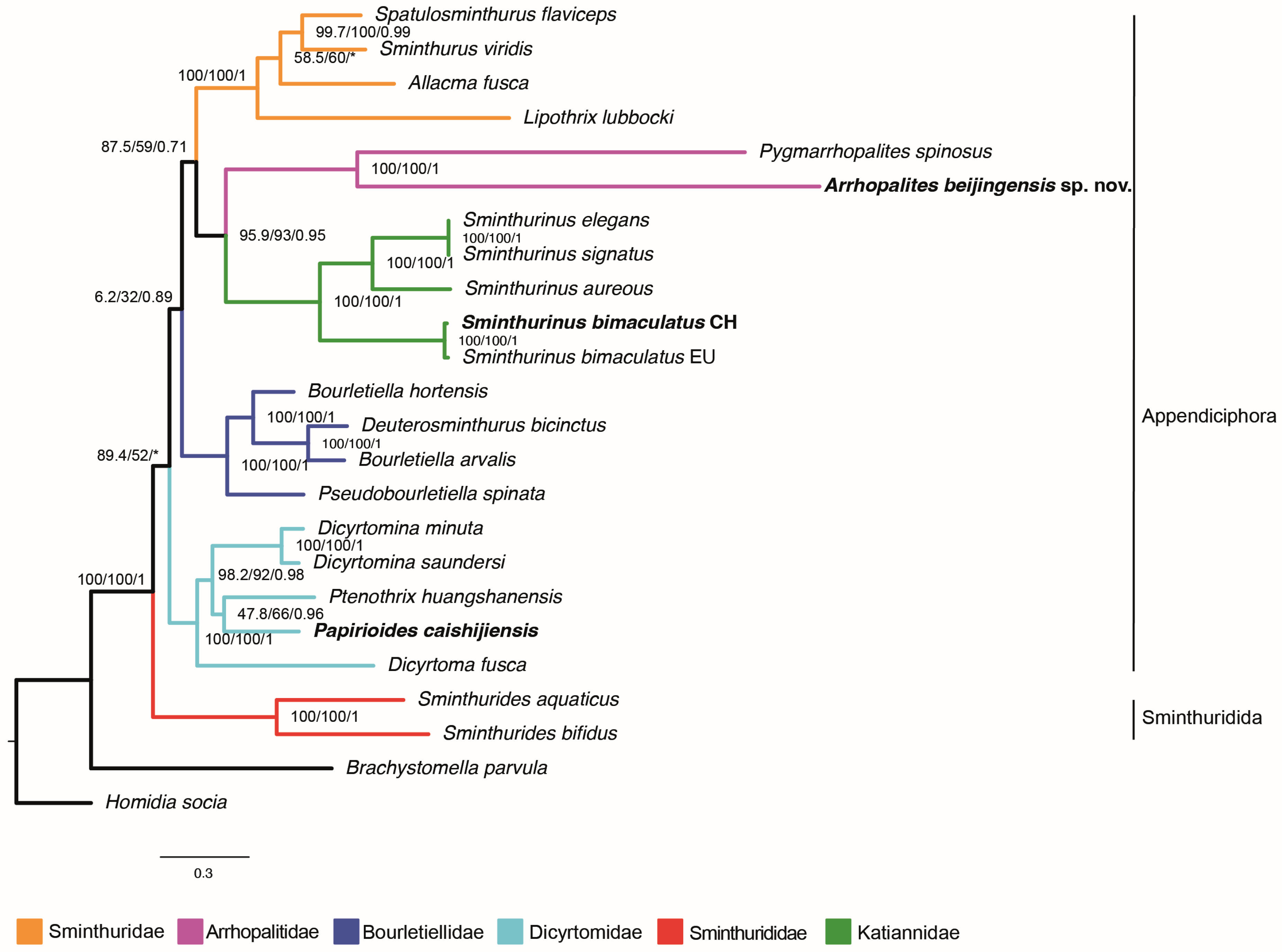
| Species | Order | Suborder | Superfamily | Family | Subfamily | NCBI Number | Country | Source | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Homidia socia | Entomobryomorpha | ND | Entomobryoidea | Entomobryidae | Entomobryinae | MN480464.1 | China | Wu and Chen [33] |
| 2 | Brachistomella parvula | Poduromorpha | ND | Neanuroidea | Brachistomellidae | ND | MN660050.1 | China | Jiang et al. [34] |
| 3 | Allacma fusca | Symphypleona | Appendiciphora | Sminthuroidea | Sminthuridae | Sminthurinae | MT547779.1 | Italy | Nardi et al. [35] |
| 4 | Arrhopalites beijingensis sp. nov. | Symphypleona | Appendiciphora | Katiannoidea | Arrhopalitidae | ND | PQ046244.1 | China | This study |
| 5 | Pygmarrhopalites spinosus | Symphypleona | Appendiciphora | Katiannoidea | Arrhopalitidae | ND | SRR17308015 | NA | Collins et al. [36] |
| 6 | Bourletiella arvalis | Symphypleona | Appendiciphora | Sminthuroidea | Bourletiellidae | Bourletiellinae | NC039558.1 | Italy | Leo et al. [37] |
| 7 | Bourletiella hortensis | Symphypleona | Appendiciphora | Sminthuroidea | Bourletiellidae | Bourletiellinae | SRR17308033 | NA | Collins et al. [36] |
| 8 | Deuterosminthurus bicinctus | Symphypleona | Appendiciphora | Sminthuroidea | Bourletiellidae | Parabourletiellinae | SRR21208389 | NA | Collins et al. [36] |
| 9 | Dicyrtoma fusca | Symphypleona | Appendiciphora | Dicyrtomoidea | Dicyrtomidae | Dicyrtominae | SRR22586363 | NA | Collins et al. [36] |
| 10 | Dicyrtomina minuta | Symphypleona | Appendiciphora | Dicyrtomoidea | Dicyrtomidae | Dicyrtominae | SRR22586362 | NA | Collins et al. [36] |
| 11 | Dicyrtomina saundersi | Symphypleona | Appendiciphora | Dicyrtomoidea | Dicyrtomidae | Dicyrtominae | NC044134.1 | Italy | Leo et al. [38] |
| 12 | Lipothrix lubbocki | Symphypleona | Appendiciphora | Sminthuroidea | Sminthuridae | Sphyrothecinae | MK431899.1 | France | Sun et al. [39] |
| 13 | Papirioides caishijiensis | Symphypleona | Appendiciphora | Dicyrtomoidea | Dicyrtomidae | Ptenothricinae | PQ035987.1 | China | This study |
| 14 | Pseudobourletiella spinata | Symphypleona | Appendiciphora | Sminthuroidea | Bourletiellidae | Bourletiellinae | SRR9066814 | China | NP |
| 15 | Ptenothrix huangshanensis | Symphypleona | Appendiciphora | Dicyrtomoidea | Dicyrtomidae | Ptenothricinae | MK423965.1 | China | Sun et al. [39] |
| 16 | Sminthurides aquaticus | Symphypleona | Sminthuridida | Sminthuridoidea | Sminthurididae | ND | OD987621.1 | France | Schneider et al. [40] |
| 17 | Sminthurides bifidus | Symphypleona | Sminthuridida | Sminthuridoidea | Sminthurididae | ND | MK423964.1 | China | Sun et al. [39] |
| 18 | Sminthurinus aureous | Symphypleona | Appendiciphora | Katiannoidea | Katiannidae | ND | SRR22586355 | NA | Collins et al. [36] |
| 19 | Sminthurinus bimaculatus | Symphypleona | Appendiciphora | Katiannoidea | Katiannidae | ND | SRR22812166 | Europe | NP |
| 20 | Sminthurinus bimaculatus | Symphypleona | Appendiciphora | Katiannoidea | Katiannidae | ND | PQ035988.1 | China | This study |
| 21 | Sminthurinus signatus | Symphypleona | Appendiciphora | Katiannoidea | Katiannidae | ND | SRR17308055 | NA | Collins et al. [36] |
| 22 | Sminthurinus elegans | Symphypleona | Appendiciphora | Katiannoidea | Katiannidae | ND | SRR22764688 | NA | NP |
| 23 | Sminthurus viridis | Symphypleona | Appendiciphora | Sminthuroidea | Sminthuridae | Sminthurinae | EU016192.1 | Germany | Carapelli et al. [41] |
| 24 | Spatulosminthurus flaviceps | Symphypleona | Appendiciphora | Sminthuroidea | Sminthuridae | Sminthurinae | SRR17308044 | NA | Collins et al. [36] |
| Body Part | Arrhopalites beijingensis sp. nov. (7 Females) | |||
|---|---|---|---|---|
| Holotype | Min | Max | Mean | |
| Total (without appendages) | 1000 | 700 | 1300 | 961 |
| Head | 300 | 250 | 440 | 330 |
| Body (without head) | 740 | 500 | 900 | 681 |
| Head dorsum longest spine | 17.5 | 15.0 | 25.0 | 19.7 |
| Eye diameter | 7.5 | 7.4 | 7.6 | 7.5 |
| Antenna | 625 | 473 | 865 | 627 |
| Ant I | 37.5 | 32.0 | 55.0 | 40.3 |
| Ant II | 87.5 | 70.0 | 127.5 | 91.6 |
| Ant III | 125.0 | 106.3 | 182.5 | 133.8 |
| Ant IV | 375.0 | 257.5 | 500.0 | 364.1 |
| Ant III organ rods | 7.5 | 4.7 | 10.0 | 6.4 |
| Tibiotarsus I | 200 | 150 | 265 | 198.7 |
| Tibiotarsus II | 205 | 145 | 255 | 204.1 |
| Tibiotarsus III | 235 | 175 | 350 | 250.1 |
| Unguis I | 52.5 | 40.0 | 67.5 | 50.6 |
| Unguis II | 50.0 | 40.0 | 62.5 | 47.9 |
| Unguis III | 42.5 | 37.5 | 55.0 | 43.4 |
| Unguiculus I | 30.0 | 25.0 | 37.5 | 30.7 |
| Unguiculus II | 25.0 | 20.0 | 32.5 | 24.6 |
| Unguiculus III | 25.0 | 20.0 | 32.5 | 24.5 |
| Th II chaeta m1 | 12.5 | 12.5 | 20.0 | 16.3 |
| Th II sensillum a | 12.5 | 10.0 | 12.5 | 11.9 |
| Th III sensillum a | 12.5 | 10.0 | 12.5 | 11.7 |
| Trichobothria AB distance | 90.0 | 82.5 | 90.0 | 86.7 |
| Trichobothria BC distance | 60.0 | 55.0 | 62.5 | 59.2 |
| Abd IV longest chaeta | 30.0 | 30.0 | 61.0 | 46.7 |
| Abd VI longest chaeta | 50.0 | 42.5 | 62.5 | 51.7 |
| Subanal appendages | 25.0 | 25.0 | 31.7 | 27.0 |
| Manubrium | 175.0 | 150.0 | 200.0 | 174.9 |
| Dens | 187.5 | 161.0 | 270.0 | 195.8 |
| Mucro | 107.5 | 95.0 | 150.0 | 114.9 |
| Dens spine Ia: length | 15.0 | 15.0 | 25.7 | 19.7 |
| Dens spine Ia: width | 5.0 | 2.5 | 5.0 | 4.6 |
| Dens spine Ie: length | 15.0 | 12.5 | 27.5 | 17.2 |
| Dens spine Ie: width | 5.0 | 2.5 | 5.0 | 4.0 |
| Ratio | Arrhopalites beijingensis sp. nov. (7 Females) | |||
|---|---|---|---|---|
| Holotype | Min | Max | Mean | |
| Antenna/head | 2.08 | 1.52 | 2.08 | 1.90 |
| Ant IV/head | 1.25 | 0.86 | 1.25 | 1.19 |
| Ant II/Ant I | 2.33 | 1.87 | 2.59 | 2.28 |
| Ant III/Ant I | 3.33 | 2.87 | 3.50 | 3.32 |
| Ant IV/Ant I | 10.00 | 6.87 | 10.00 | 9.01 |
| Head/tibiotarsus I | 1.50 | 1.50 | 1.85 | 1.66 |
| Tibiotarsus II/I | 1.03 | 0.96 | 1.11 | 1.01 |
| Tibiotarsus III/I | 1.18 | 1.17 | 1.43 | 1.26 |
| Tibiotarsus I/unguis I | 3.81 | 3.75 | 4.53 | 4.01 |
| Tibiotarsus II/unguis II | 4.10 | 3.63 | 4.75 | 4.27 |
| Tibiotarsus III/unguis III | 5.53 | 4.38 | 6.67 | 5.74 |
| Unguis I/unguiculus I | 1.75 | 1.46 | 1.80 | 1.64 |
| Unguis II/unguiculus II | 2.00 | 1.70 | 2.13 | 1.96 |
| Unguis III/unguiculus III | 1.70 | 1.63 | 2.11 | 1.78 |
| Dens/mucro | 1.74 | 1.61 | 1.80 | 1.70 |
| Trichobothria: AB/BC distance | 1.50 | 1.32 | 1.59 | 1.47 |
| Abd IV dI-1/Th II m1 chaeta | 2.40 | 1.71 | 4.40 | 2.79 |
| Abd IV/Abd VI longest chaeta | 0.60 | 0.60 | 1.29 | 0.88 |
| Abd IV dI-1/unguis III | 1.18 | 0.89 | 1.47 | 1.18 |
| Body total/tibiotarsus III | 4.26 | 3.51 | 4.26 | 3.87 |
| Body total/unguis I | 19.05 | 17.50 | 20.64 | 19.21 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Godeiro, N.N.; Bu, Y.; Medeiros, G.d.S.; Gao, Y.; Vargovitsh, R.S. The First Cavernicolous Species of Arrhopalites (Collembola, Symphypleona, Arrhopalitidae) from China and Its Phylogenetic Position. Insects 2025, 16, 314. https://doi.org/10.3390/insects16030314
Godeiro NN, Bu Y, Medeiros GdS, Gao Y, Vargovitsh RS. The First Cavernicolous Species of Arrhopalites (Collembola, Symphypleona, Arrhopalitidae) from China and Its Phylogenetic Position. Insects. 2025; 16(3):314. https://doi.org/10.3390/insects16030314
Chicago/Turabian StyleGodeiro, Nerivania Nunes, Yun Bu, Gleyce da Silva Medeiros, Yan Gao, and Robert S. Vargovitsh. 2025. "The First Cavernicolous Species of Arrhopalites (Collembola, Symphypleona, Arrhopalitidae) from China and Its Phylogenetic Position" Insects 16, no. 3: 314. https://doi.org/10.3390/insects16030314
APA StyleGodeiro, N. N., Bu, Y., Medeiros, G. d. S., Gao, Y., & Vargovitsh, R. S. (2025). The First Cavernicolous Species of Arrhopalites (Collembola, Symphypleona, Arrhopalitidae) from China and Its Phylogenetic Position. Insects, 16(3), 314. https://doi.org/10.3390/insects16030314








